Abstract
Vitamin B6 is an essential metabolite in all organisms, being required as a cofactor for a wide variety of biochemical reactions. De novo biosynthesis of the vitamin occurs in microorganisms and plants, but animals must obtain it from their diet. Two distinct and mutually exclusive de novo pathways have been identified to date, namely deoxyxylulose 5-phosphate dependent, which is restricted to a subset of eubacteria, and deoxyxylulose 5-phosphate independent, present in archaea, fungi, plants, protista, and most eubacteria. In these organisms, pyridoxal 5′-phosphate (PLP) formation is catalyzed by a single glutamine amidotransferase (PLP synthase) composed of a glutaminase domain, PDX2, and a synthase domain, PDX1. Despite plants being an important source of vitamin B6, very little is known about its biosynthesis. Here, we provide information for Arabidopsis thaliana. The functionality of PDX2 is demonstrated, using both in vitro and in vivo analyses. The expression pattern of PDX2 is assessed at both the RNA and protein level, providing insight into the spatial and temporal pattern of vitamin B6 biosynthesis. We then provide a detailed biochemical analysis of the plant PLP synthase complex. While the active sites of PDX1 and PDX2 are remote from each other, coordination of catalysis is much more pronounced with the plant proteins than its bacterial counterpart, Bacillus subtilis. Based on a model of the PDX1/PDX2 complex, mutation of a single residue uncouples enzyme coordination and in turn provides tangible evidence for the existence of the recently proposed ammonia tunnel through the core of PDX1.
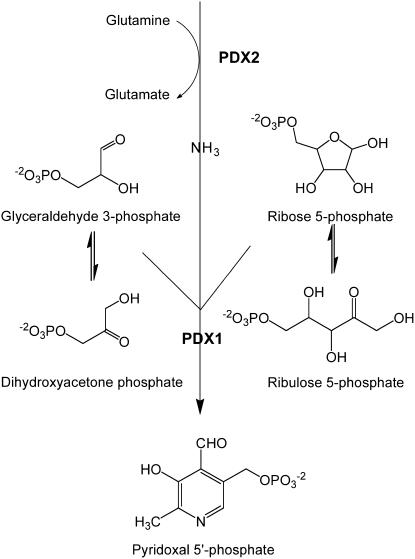
Vitamin B6 is the generic name for pyridoxal, pyridoxol, pyridoxamine, and their respective 5′-phosphorylated forms. This vitamin is essential for all organisms and its functional forms, pyridoxal 5′-P (PLP) and pyridoxamine 5′-P, play a crucial role in a broad range of biochemical reactions. Furthermore, vitamin B6 has recently been identified as a potent antioxidant (Ehrenshaft et al., 1999; Osmani et al., 1999; Bilski et al., 2000). Prokaryotes, fungi, protista, and plants are able to synthesize the vitamin de novo, but animals must obtain it from dietary sources. The biosynthetic pathway of vitamin B6 might therefore be an important target for drug design (Wrenger et al., 2005; Gengenbacher et al., 2006). Two distinctive pathways for its de novo synthesis have been identified to date. In the deoxyxylulose 5-P (DXP)-dependent pathway (Tambasco-Studart et al., 2005), present in only a small subset of eubacteria, pyridoxol 5′-P biosynthesis is mediated by the proteins PdxA and PdxJ from the precursors DXP and 4-phosphohydroxy-l-Thr (Spenser and Hill, 1995; Cane et al., 1999; Laber et al., 1999; Drewke and Leistner, 2001). The second pathway, referred to as DXP independent (Tambasco-Studart et al., 2005), was only recently discovered and appears to be present in all archaea, fungi, plants, and most eubacteria (Ehrenshaft et al., 1999; Mittenhuber, 2001; Tanaka et al., 2005). The cofactor form of the vitamin, PLP, is synthesized directly from either Rib 5-P (R5P) or ribulose 5-P (Ru5P), in combination with either glyceraldehyde 3-P (G3P) or dihydroxyacetone phosphate (DHAP), and Gln as the nitrogen (N) donor (Burns et al., 2005; Raschle et al., 2005; Scheme 1). Remarkably, the reaction is catalyzed by a single Gln amidotransferase (PLP synthase), composed of the glutaminase subunit, PDX2, and the synthase or acceptor subunit, PDX1.
Scheme 1.
Vitamin biosynthesis by the DXP-independent pathway.
Glutamine amidotransferases in general provide an impetus for research not only because they play a central role in metabolism through the incorporation of N into amino acids, amino sugars, nucleotides, coenzymes, and antibiotics, but also because they coordinate catalysis at distinctive, and, in all cases examined thus far, remote active sites (Myers et al., 2005; Strohmeier et al., 2006; Zein et al., 2006). Substrate (ammonia) channeling between the two sites is achieved via a protein tunnel between the glutaminase and synthase active site, respectively (Zalkin and Smith, 1998; Raushel et al., 1999). Furthermore, in many cases, the glutaminase is activated only upon binding of the substrates in the associated synthase domain, thus preventing futile hydrolysis of Gln (Zalkin and Smith, 1998; Bera et al., 2000). Thus far, neither of these phenomena has been observed for PLP synthase, making it somewhat of an enigma in the Gln amidotransferase family. However, the structure of bacterial PLP synthase has recently been solved (Strohmeier et al., 2006; Zein et al., 2006) and demonstrates activation of Pdx2 when in complex with Pdx1. Furthermore, a Met-rich hydrophobic tunnel has been proposed for the transfer of ammonia between the two active sites of bacterial PLP synthase (Strohmeier et al., 2006).
The polymorphic synthetic ability of PLP synthase has attracted considerable interest. PLP synthases from various organisms are being studied in the expectation that questions unanswered for one of the synthases can be revealed in another (Belitsky, 2004; Dong et al., 2004; Burns et al., 2005; Chen and Xiong, 2005; Raschle et al., 2005; Tambasco-Studart et al., 2005; Wrenger et al., 2005; Gengenbacher et al., 2006; Wagner et al., 2006). Currently, the functionality of both the Pdx1 and Pdx2 subunits as a complex in vitro and reconstitution of PLP biosynthesis have been demonstrated for the bacterium Bacillus subtilis and the apicomplexan Plasmodium falciparum (Burns et al., 2005; Raschle et al., 2005; Gengenbacher et al., 2006). In the model plant Arabidopsis (Arabidopsis thaliana), we have recently identified three homologs of PDX1, named PDX1.1, PDX1.2, and PDX1.3, and one homolog of PDX2 (Tambasco-Studart et al., 2005). Even though the disruption of PDX2 was shown to be lethal for the plant (Tambasco-Studart et al., 2005), the glutaminase activity of this protein and its direct correlation with vitamin B6 biosynthesis could not be demonstrated in that study. Moreover, the enzymatic activities of the PDX1 proteins could be verified in vitro only by using ammonium sulfate as the N source and in the absence of PDX2, which does not reflect the presumed in vivo scenario.
In this study, we provide a detailed description of the role of PDX2 in the biosynthesis of PLP in the model plant Arabidopsis. A comprehensive expression analysis at both the RNA and protein level and its comparison with that of PDX1 has allowed us to describe the spatial and temporal formation of vitamin B6 in plants. The functionality of PDX2 in vitamin B6 biosynthesis has been established in vivo by the ability of the vitamin to rescue pdx2 seeds and in vitro by the discovery of glutaminase activity of PDX2 in the presence of PDX1. A unique feature of plant PDX2 proteins is a C-terminal extension of approximately 30 amino acids that we show here to be important for catalysis. We also report a detailed biochemical analysis of the PLP synthase protein complex and compare it mechanistically to its prokaryotic counterparts. Lastly, we provide evidence that the coordination, or coupling, between the glutaminase and synthase domains is more pronounced with the plant as compared to the bacterial proteins and that this may be due to an alteration at the base of the proposed ammonia tunnel in PDX1.
RESULTS
In Vivo Evidence for the Involvement of PDX2 in Vitamin B6 Biosynthesis in Planta
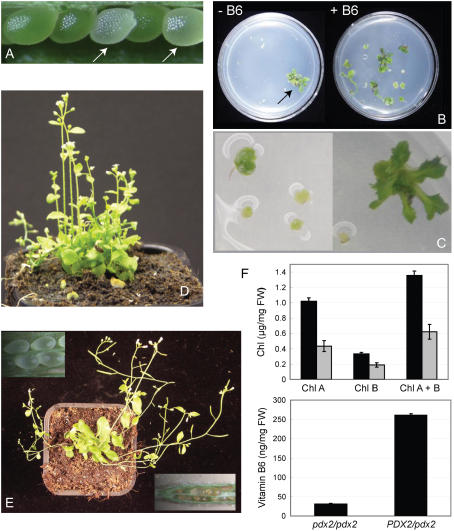
We recently reported the isolation and characterization of the mutants pdx2.1 (SALK_072168) and pdx2.2 (53-2381-1) from Arabidopsis (Tambasco-Studart et al., 2005). These mutants are arrested at the globular stage of embryo development, a notoriously difficult stage from which to rescue the seed (Patton et al., 1998), thus complicating analyses of gene functionality. After many previous unsuccessful attempts, we can now provide direct evidence for the involvement of PDX2 in vitamin B6 metabolism in planta. Culturing of albino seeds produced by PDX2/pdx2 plants (Fig. 1A) in the presence of vitamin B6, as described in “Materials and Methods,” led to growth after 14 d (Fig. 1B, right side), albeit in a heterogeneous fashion (Fig. 1C). pdx2 seeds did not respond on medium lacking the vitamin, whereas wild-type seeds developed and germinated (Fig. 1B, left). Successful rescue of pdx2 was only achieved if seeds were cultured at the time they had reached the globular stage of development and under low light conditions. Merely watering, spraying, or even vacuum infiltration of the vitamin did not result in an advancement of embryo development in the albino seeds beyond the globular stage. After 50 d, the teratomata that had resulted from the described successful rescue procedure were transferred to soil, and continuous watering with vitamin B6 allowed the plants to reach the reproductive stage of development (Fig. 1, D and E). Unless the pdx2 plants (the genotype of which was confirmed by PCR; data not shown) were continuously watered with the vitamin, they became chlorotic and died. A comparison of heterozygous and homozygous pdx2 plants, which had been deprived of vitamin B6 supplementation for 28 d, demonstrated that the latter had vitamin B6 and chlorophyll contents reduced 8.6- and 2.2-fold, respectively (Fig. 1F). Nonetheless, the entire seed set of all rescued/supplemented pdx2/pdx2 plants was albino (Fig. 1E, inset), demonstrating that vitamin supplementation is not sufficient to support the developing embryo and in turn highlighting the importance of the PDX2 protein in embryo development.
Figure 1.
Rescue of the homozygous pdx2 mutant by vitamin B6. A, An immature silique from a heterozygous PDX2/pdx2 plant, the arrows indicate albino pdx2 seeds. B, Immature albino seeds isolated from PDX2/pdx2 plants and cultured in the presence (right) and absence (left) of vitamin B6; pictures were taken 14 d after 16-h-day/8-h-night cycles with a light intensity of 35 μmol photons m−2 s−1; a single wild-type seed was included on the −B6 plate as a control (arrow). C, Samples similar to B in the presence of vitamin B6 but under a higher magnification to emphasize the heterogeneous growth between pdx2 teratomata. D and E, The development of rescued pdx2 plants 15 and 28 d, respectively, after transfer of teratomata to soil; vitamin B6 supplementation was maintained during this time. The insets in E show dissected siliques from a rescued pdx2 plant in which all seeds display an albino phenotype and are later aborted. F, The top panel shows the chlorophyll content of PDX2/pdx2 plants (black bars) and rescued pdx2 plants (gray bars) cultured under the same conditions; the bottom panel shows the vitamin B6 content of the same plants. The measurements were performed 28 d after discontinuing supplementation with vitamin B6.
Expression Analysis of PDX2
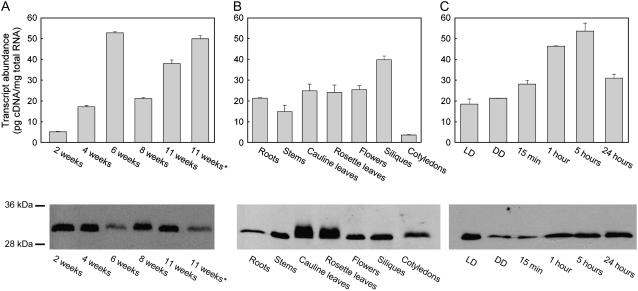
The dependence of the pdx2 mutant on vitamin B6 during development until seed set suggested that PDX2 is expressed throughout the life cycle. Both a quantitative reverse transcription (RT)-PCR and immunochemical analysis with a specific antibody of rosette leaves over an 11-week period revealed that this is indeed the case (Fig. 2A). PDX2 appears to be ubiquitously expressed throughout development, but there is some variation in the levels observed (Fig. 2A). Levels of transcript abundance were similar in almost all tissues examined, i.e. roots, stems, cauline leaves, rosette leaves, flowers, siliques, and cotyledons, with the lowest and highest levels being observed in cotyledons and siliques, respectively (Fig. 2B). Tissue levels of the PDX2 protein were positively correlated with the transcript levels, with the notable exception of the abundance of the protein in roots (Fig. 2B). The cause for the diffuse protein band observed for leaf extracts (Fig. 2B) is not known at present. In addition, similar to what has been observed with PDX1 (Titiz et al., 2006), it appears that expression of PDX2 is regulated by light, albeit in a slightly different manner. After transfer of etiolated seedlings to light, the level of PDX2 transcript gradually increases up to 5 h followed by a decline (Fig. 2C); this decrease is not observed with PDX1, which appears to continuously increase over the same period (Titiz et al., 2006). Light-induced PDX2 transcript levels correlated largely with protein levels, with the exception that the level of PDX2 in etiolated seedlings was much lower than that under long-day conditions (Fig. 2C). The latter observation may be indicative of a negative posttranscriptional regulation of PDX2 in the dark.
Figure 2.
Expression analysis of PDX2. The top segment shows transcript abundance of PDX2. A, Rosette leaves of 2-, 4-, 6-, 8-, and 11-week-old plants. The asterisk indicates senescent leaves of 11-week-old plants. B, Roots, stems, cauline leaves, rosette leaves, flowers, siliques, and cotyledons. C, Five-day-old etiolated seedlings 15 min and 1, 5, and 24 h after transfer to light, as well as long-day (LD; 16 h of light/8 h dark) and continuous dark (DD) controls. All values are presented as pg cDNA/mg total RNA. For absolute amounts of mRNA, standard curves were generated using known concentrations of a plasmid carrying the PDX2 gene. Plasmid DNA was diluted to 1 ng μL−1 followed by 10-fold serial dilutions to 1 fg μL−1. The results shown are the average of the experiment performed in triplicate. The bottom panel shows PDX2 protein abundance as assessed by an immunochemical analysis. Total protein was extracted from the same samples as described for the top panel and probed with an antibody raised and purified against the recombinant PDX2. Thirty micrograms of total protein was loaded per lane. Molecular mass standards are as indicated.
Glutaminase Activity of PDX2
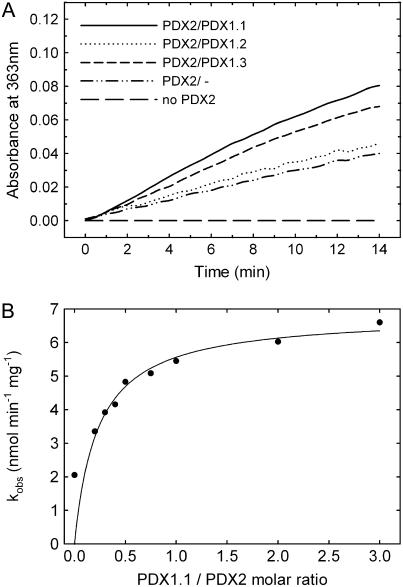
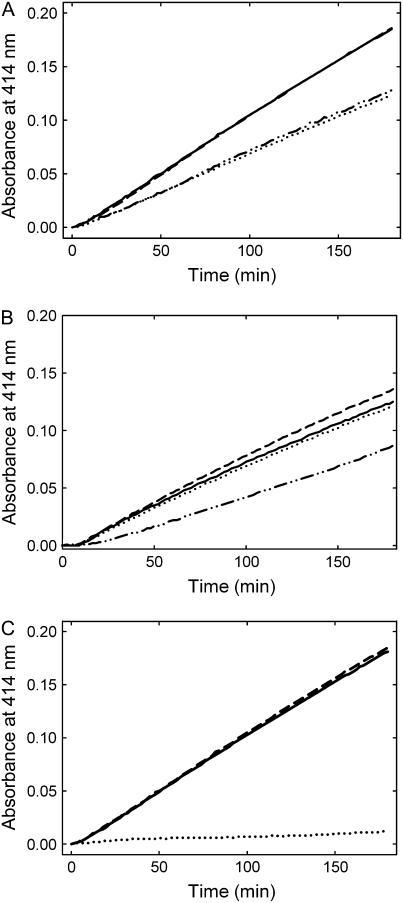
In certain lower organisms, PDX2 has been identified as the glutaminase domain of the Gln amidotransferase, PLP synthase (Belitsky, 2004; Dong et al., 2004; Gengenbacher et al., 2006). Here, we have established functionality of the plant protein by demonstrating glutaminase activity of the isolated recombinant protein (Fig. 3). Activity as a function of the Gln concentration followed typical Michaelis-Menten kinetics from which the catalytic constants could be estimated (turnover number [kcat] = 0.23 ± 0.01 min−1 and Km = 1.92 ± 0.32 mm; Supplemental Fig. S1). While the Km is within the range of those observed with the orthologous B. subtilis (Raschle et al., 2005) and P. falciparum (Gengenbacher et al., 2006) enzymes, the kcat is substantially lower (Table I). Furthermore, in contrast to what has been observed previously with the bacterial and apicomplexan proteins (Raschle et al., 2005; Gengenbacher et al., 2006), we could detect a basal glutaminase activity of the plant PDX2 in the absence of its partner protein PDX1 (0.04 min−1); in the presence of PDX1, the PDX2 activity increased approximately 4-fold (Fig. 3A). The level of activity was not appreciably different with either PDX1.1 or PDX1.3, whereas PDX1.2 did not significantly enhance the glutaminase activity of PDX2 (Fig. 3A). Optimal activity was observed when the functional PDX proteins were in a 1:1 molar ratio, indicating a stoichiometric protein complex (Fig. 3B).
Figure 3.
Glutaminase activity of PDX2. A, Initial rates of PDX2 glutaminase activity either in the absence of PDX1 (dashed and dotted line) or in the presence of PDX1.1 (solid line), PDX1.2 (dotted line), or PDX1.3 (dashed line). In every case, 10 μm of each protein and 20 mm Gln was used. B, Rate of PDX2 (10 μm) glutaminase activity as a function of the concentration of PDX1.1 (molar ratio 0–3.0).
Table I.
Steady-state kinetic constants of PDX2
Kinetic parameters were determined as a function of the Gln concentration by measuring the PDX2 glutaminase activity using a coupled assay (Raschle et al., 2005). In the case of Arabidopsis, 10 μm each of PDX1 and PDX2 were used.
| Species | kcat | Km | kcat/Km |
|---|---|---|---|
| min−1 | mm | mm min−1 | |
| B. subtilisa | 7.6 | 0.99 | 7.68 |
| P. falciparumb | 6.6 | 0.56 | 11.8 |
| Arabidopsis (full length) | 0.23 ± 0.01 | 1.92 ± 0.32 | 0.12 |
| Arabidopsis (ΔC30) | 0.13 ± 0.01 | 2.62 ± 0.35 | 0.05 |
Taken from Raschle et al. (2005).
Taken from Gengenbacher et al. (2006).
We noticed that plant PDX2s have a C-terminal extension of approximately 30 amino acids that is not present in the orthologous proteins (Supplemental Fig. S2A). The cytosolic localization of PDX2 (Tambasco-Studart et al., 2005) precludes this sequence from a targeting function within the compartmental context of the plant cell. In an attempt to define the function of this domain, we constructed a deletion mutant of PDX2 lacking the C-terminal 30 amino acids (PDX2ΔC30) and determined the kinetic parameters for Gln hydrolysis. A comparison between the full-length and truncated version of the protein, respectively, shows that the catalytic efficiency (kcat/Km) of the reaction is reduced approximately 2.4-fold in the deletion mutant (Table I) and indicates that the C-terminal amino acids are important for efficient catalysis in Arabidopsis PDX2.
Biochemical Characterization of Arabidopsis PLP Synthase
Previously, we reported that Arabidopsis PDX1.1 and PDX1.3 can catalyze PLP formation using either of the pentose phosphate sugars, R5P or Ru5P, and either of the triose phosphate sugars, G3P or DHAP, in the presence of an external ammonium source (Tambasco-Studart et al., 2005). Now, with the availability of a functional recombinant PDX2, we can provide a more detailed analysis of PDX1 activity in the presence of its in vivo partner enzyme, PDX2. For an accurate determination of PLP synthase activity, PDX1 was first depleted of the copurifying pentose sugar (see below) as recently described in Raschle et al. (2007). With Gln as the N source, PLP synthase activity of PDX1 was strictly observed only in the presence of PDX2. Under these conditions, the activity of PDX1 displayed no preference for the triose sugar used but clearly preferred the pentose to the pentulose (Fig. 4A). As our earlier analysis of PLP synthesis using ammonium sulfate as the N source was biased by the presence of the copurifying pentose sugar, we reanalyzed PLP synthesis under conditions where no pentose sugar was bound to PDX1. Again, there was a preference for the pentose sugar (data not shown), but the rate with DHAP as the triose substrate was significantly decreased. Interestingly, if PDX2 was added, the rate was restored to that observed with G3P (Fig. 4B). The activities observed with either PDX1.1 and PDX1.3 were very similar, whereas no activity was observed with PDX1.2 under any of the conditions used (Fig. 4C), corroborating our earlier analysis (Tambasco-Studart et al., 2005) that PDX1.2 is not functional in PLP biosynthesis.
Figure 4.
Enzymatic formation of PLP. A, Initial rates of activity observed in the presence of R5P (0.5 mm) and either dl-G3P (1 mm) or DHAP (0.5 mm; solid and dashed line, respectively), or Ru5P (0.5 mm) and either dl-G3P (1 mm) or DHAP (0.5 mm; dotted and dashed/dotted line, respectively). All assays shown were carried out in the presence of 20 mm Gln and 10 μm each of PDX1.1 and PDX2. B, Initial rates observed as a function of the triose sugar in the presence of ammonium sulfate as the N source; R5P and dl-G3P in the presence or absence of PDX2 (solid and dashed line, respectively) or R5P and DHAP (dotted and dashed/dotted line, respectively). PDX1.1 was present under all conditions shown. C, Comparison of PLP synthase activity employing all three PDX1 homologs in the presence of PDX2; PDX1.1 (solid line), PDX1.2 (dotted line), and PDX1.3 (dashed line). The assays were performed using R5P, dl-G3P, and Gln. The concentrations of each component in B and C were as described in A.
Evidence for Coupling between PDX1 and PDX2
The glutaminase and synthase active sites are remote from each other in B. subtilis PLP synthase (Strohmeier et al., 2006). Thus, akin to all other characterized Gln amidotransferases, it is assumed that the ammonia produced at the PDX2 active site travels through a tunnel to the PDX1 active site where it is incorporated into the pyridine ring. For this process to work efficiently and to prevent the futile hydrolysis of Gln, the catalytic activities of the two active sites should be coordinated. We investigated this in both the plant and bacterial PLP synthase. The fractional ratio for the rate of PLP synthesis to Gln hydrolysis can be taken as a measure of channeling efficiency between the two active sites (Bera et al., 2000). Channeling is more pronounced (approximately 7-fold) with the Arabidopsis proteins than with the B. subtilis proteins (Table II). This is predominantly manifested in the lower rate of glutaminase activity of the Arabidopsis proteins (Table II) and implies that the coupling of the activities is stronger between the two plant proteins. Furthermore, we observed that as for the bacterial proteins (Raschle et al., 2005), the substrates of PDX1, either alone or in combination, have no effect on the glutaminase activity of PDX2 (data not shown), indicating that the coupling effect may be inherent to the protein anatomy.
Table II.
Coupling efficiency of PLP synthase
PLP formation and Gln hydrolysis were measured, and the ratio between the two was used to estimate the coupling efficiency between PDX1 and PDX2.
| Species | PLPa | Glutaminase | Couplingb |
|---|---|---|---|
| nmol min−1 | nmolmin−1 | ||
| Arabidopsis PDX1.1 wild type | 0.13 ± 0.008 | 0.37 ± 0.050 | 0.35 |
| Arabidopsis PDX1.1 L30A | 0.11 ± 0.020 | 1.14 ± 0.020 | 0.09 |
| B. subtilis | 0.15 ± 0.006 | 3.43 ± 0.190 | 0.05 |
Employing 10 μm each of PDX1 and PDX2 with R5P and DL-G3P as substrates.
Ratio between the respective reaction rates of PLP synthesis and glutaminase.
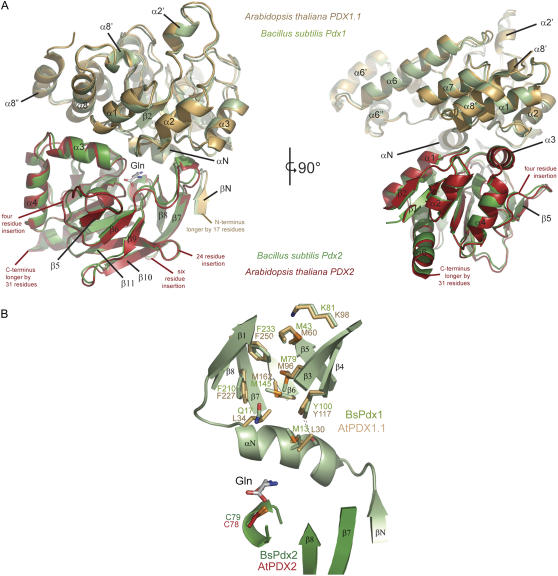
With the elucidation of the structure of the bacterial PLP synthase complex, a proposal was put forward for the route of ammonia transfer between the glutaminase and synthase active sites (Strohmeier et al., 2006; Zein et al., 2006). Specifically, in B. subtilis PLP synthase, a series of four Met residues (M13, M145, M79, and M43) that line the putative tunnel have been implicated as having the conformational plasticity to mediate ammonia transfer (Strohmeier et al., 2006). While three of these Met residues are tightly conserved (M145, M79, and M43 in BsPdx1), the one at the interface of Pdx1 and Pdx2 (M13 in BsPdx1) is not (see “Discussion”; Fig. 7B). In Arabidopsis PDX2, the equivalent of M13 in BsPdx1 is replaced by a Leu (L30), a residue that would be expected to display considerable less flexibility. In this context, we were prompted to investigate whether L30 has a role in the enhanced coupling observed with the Arabidopsis proteins compared to the bacterial ones. Indeed, the L30A mutant of PDX1.1 had a substantially reduced coupling efficiency (approximately 4-fold) approaching that observed with the B. subtilis proteins; the effects are predominantly manifested by an increase in the rate of glutaminase activity (Table II).
Figure 7.
Modeling the three-dimensional structure of the domains of PLP synthase from Arabidopsis. A, A ribbon representation of PLP synthase from B. subtilis (PDB code 2NV2) is shown in green and is overlaid by a model of PDX1.1 (gold) and PDX2 (red) from Arabidopsis. The two figures show views turned by 90° around a vertical axis in the paper plane. Sequence insertions in the plant enzymes compared to the bacterial enzyme have not been modeled but are indicated in the figure. Visible secondary structure elements have been labeled for reference. B, The part of PLP synthase depicted is proposed to be the route of ammonia transfer. Residues involved in the B. subtilis protein are shown in ball and stick representation (green) overlaid by the equivalent residues of Arabidopsis PDX1.1 (gold). A number of residues are seen to be different in PDX1.1, notably M13 in BsPdx1 is replaced by a Leu.
Evidence That the Mechanism of Action of the Plant PLP Synthase Is Similar to That of Its Prokaryotic Counterparts
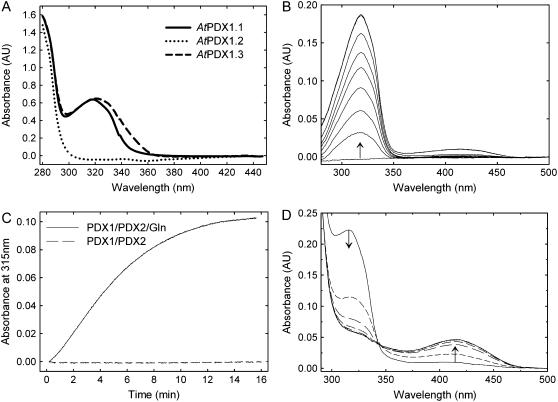
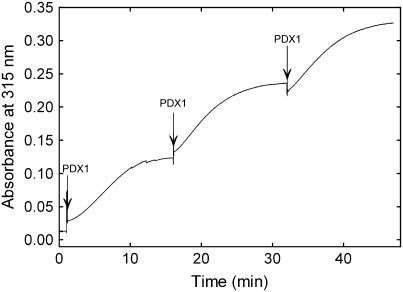
We have recently identified a covalently bound reaction intermediate during PLP biosynthesis by B. subtilis, which is chromophoric, with a characteristic absorbance maximum at 315 nm (Raschle et al., 2007). It is observed with freshly isolated protein and can be reconstituted by addition of the pent(ul)ose phosphate substrate to PDX1 in the presence of PDX2 and Gln (Raschle et al., 2007). Here, we observed that the freshly isolated plant proteins, PDX1.1 and PDX1.3, display an absorbance maximum in this region, whereas PDX1.2 does not (Fig. 5A). Furthermore, the chromophoric adduct could be reconstituted with the functional plant PDX1s in a fashion identical to that described for the bacterial protein (Fig. 5B). Formation of the chromophore was absolutely dependent on Gln hydrolysis by the PDX2 subunit (Fig. 5C). The addition of either of the triose phosphate substrates, G3P or DHAP, resulted in its conversion to PLP, demonstrating the catalytic competence of the intermediate (Fig. 5D). This data implies that the mechanism of action of PLP synthase is conserved across taxa. We also observed that, similar to the B. subtilis protein (Raschle et al., 2007), formation of the chromophore is a function of the concentration of the PDX1 protein (Fig. 6). While PDX2 and Gln are requisite for chromophore formation by PDX1, only a catalytic amount was necessary, i.e. it did not have to be added again with the subsequent PDX1 additions. This implies that PDX2 is released from the adduct form of PDX1, making it available to activate the newly added free PDX1.
Figure 5.
Characteristics of the chromophoric reaction intermediate. A, UV-visible absorbance spectra of freshly isolated PDX1.1, PDX1.2, and PDX1.3, respectively, in 50 mm Tris-Cl, pH 8.0. B, Reconstitution of the observed chromophoric adduct employing free PDX1.1 (20 μm), PDX2 (20 μm), Gln (10 mm), and R5P (0.2 mm) in Tris-Cl, pH 8.0. The pentose sugar was added at time 0, and the spectra were acquired after 1.5, 4.5, 6.0, 7.5, 9.0, 12.0, 25.5, and 27 min. C, Formation of the chromophoric adduct in the presence (solid line) and absence (dashed line) of Gln (10 mm) employing PDX1.1 (10 μm), PDX2 (10 μm), and R5P (0.3 mm) in Tris buffer, pH 8.0. D, Demonstration of the catalytic competence of the chromophoric adduct; after reconstitution of the latter as described in B (taken as time 0), dl-G3P (0.3 mm) was added, and spectra were acquired at 3, 6, 9, 12, 15, 18, 19.5, and 21 min. The direction of the arrows indicates either the decrease or increase in absorbance observed.
Figure 6.
Formation of the chromophoric adduct as a function of the concentration of PDX1. PDX1.1 was added at the time intervals indicated by arrows (10 μm at each time point) to a preincubation mix of PDX2 (10 μm), Gln (10 mm), and R5P (0.3 mm) in 50 mm Tris-Cl, pH 8.0.
DISCUSSION
In contrast to what had been tacitly assumed, the de novo pathway of vitamin B6 biosynthesis in plants proceeds via a DXP-independent pathway (Tambasco-Studart et al., 2005). This pathway involves the PDX1 and PDX2 proteins, both of which are essential for plant survival, as the knockout of either one results in an arrest of embryogenesis at the globular stage (Tambasco-Studart et al., 2005; Titiz et al., 2006). The functionality of PDX2 in the biosynthesis of vitamin B6, both in vivo and in vitro, has been addressed in detail in this study.
The ability to rescue pdx2 seeds by supplementation with vitamin B6 establishes the role of PDX2 in the biosynthesis of this vitamin. Furthermore, the spatial and temporal expression of PDX2 roughly correlates with that observed for the functional PDX1 proteins, PDX1.1 and PDX1.3 (Tambasco-Studart et al., 2005; Titiz et al., 2006), i.e. it is found in all organs examined, is expressed in a similar fashion throughout the developmental cycle, and is light inducible (Titiz et al., 2006). In particular, there is a remarkable similarity at the protein level. At the transcript level, a comparison of their abundance indicates an overall tendency in the order PDX1.3 > PDX1.1 > PDX2. In some cases, PDX2 is more predominant than either PDX1.1 or PDX1.3, e.g. roots and etiolated seedlings (Supplemental Fig. S3). Data provided by Genevestigator (Zimmermann et al., 2004) support a high correlation between PDX2, PDX1.1, and PDX1.3 but very weak correlation with PDX1.2 (Supplemental Fig. S4). In contrast to certain other metabolic mutations that can be successfully rescued in a relatively straightforward manner by addition of the missing metabolite, e.g. the biotin defective mutant bio2 (Patton et al., 1998), we did not observe the rescue of mutant pdx2/pdx2 embryos in PDX2/pdx2 plants by providing vitamin B6. Rather, it was necessary to isolate the mutant seeds and continuously supply them with an excess of the vitamin to allow growth. This appears to indicate that there is no transport of vitamin B6 in the silique of Arabidopsis. If the mutant seed is removed from the silique, rescue is possible, implying that uptake can occur through the seed coat. While the homozygous pdx2 plants can be rescued, we have not been able to produce progeny from these plants.
In microorganisms, PDX1 and PDX2 have been clearly proven to function as a Gln amidotransferase, with both proteins required for the functionality of either domain, i.e. the synthase and glutaminase, respectively (Belitsky, 2004; Dong et al., 2004; Raschle et al., 2005; Gengenbacher et al., 2006). Our data now confirm this for plants as well. The plant and microbial enzymes share properties, e.g. glutaminase activation by PDX1 and dependency on PDX2 for formation of the PDX1 chromophoric adduct, but there are some notable differences. A key feature of the majority of Gln amidotransferases is the coordination of the glutaminase and synthase activities, thereby preventing the futile hydrolysis of Gln in the absence of product synthesis. In all Gln amidotransferases scrutinized thus far, but with the notable exception of PLP synthase (Strohmeier et al., 2006), it appears that coupling of glutaminase and synthase activities is mediated through interdomain signaling upon binding of the synthase substrate. It has been suggested that a specific interdomain salt bridge may be responsible for this coordination (Myers et al., 2005). As this has not been observed with PLP synthase (Strohmeier et al., 2006), a different mechanism must be anticipated. Formation of an ammonia channel that connects the physically separated active sites provides a mechanism to sequester ammonia from solvent and to deliver it to the PLP synthesis domain for nucleophilic attack on the carbon scaffolds. Specific residues that could control the delivery of ammonia to the PLP synthase active site, in addition to those that control glutaminase activation, could be envisaged as additional means to regulate coordination between the disparate active sites. As stated above, M13 in B. subtilis Pdx1, which is at the base of the predicted ammonia channel, is replaced by a Leu in Arabidopsis PDX1. Mutation of the latter residue severely disrupts the coupling of glutaminase and synthase activities observed with the plant proteins (Table II). Bacterial PLP synthase appears to only be approximately 15% as efficient as the plant PLP synthase in coupling the respective activities. Indeed, almost 95% of the ammonia derived from Gln hydrolysis was lost from the bacterial enzyme under our in vitro experimental conditions. An additional factor may be required to control coupling in vivo with the bacterial system. As a Leu would have different properties than a Met, the proposed mechanism of entry to the ammonia tunnel must be divergent in the Arabidopsis proteins. To test whether this residue is structurally compatible, we created a model of the Arabidopsis PDX1.1 and PDX2 proteins on the basis of the recently solved bacterial PLP synthase complex (Strohmeier et al., 2006). This model indicates that while the core of Arabidopsis PLP synthase would be predicted to be essentially the same as that of B. subtilis (Strohmeier et al., 2006; Fig. 7A), the entrance to the presumed ammonia tunnel must be different (Fig. 7B). This may be the reason for the observation of a less leaky tunnel in Arabidopsis.
As observed previously (Tambasco-Studart et al., 2005), PDX1.2 is not able to catalyze PLP biosynthesis in vitro, and, furthermore, we now show that it is unable to activate the glutaminase activity of PDX2. The structure of the PLP synthase complex has revealed that the N terminus of PDX1, in particular the unique helix αN, is instrumental not only for complex formation but also for activation of the glutaminase (Strohmeier et al., 2006). Thus, it is remarkable that despite the extreme conservation of PDX1 across taxa, the most divergent region between the amino acid sequences is in fact the N terminus (T.B. Fitzpatrick, unpublished data). In particular, key residues involved in the polar contacts that allow precise alignment of αN are not conserved in the plant proteins. This is suggestive of a variant mode of interaction between PDX1 and PDX2, and PDX2 activation in Arabidopsis, and is supported by the structural model (Fig. 7A). This statement is corroborated further by the fact that B. subtilis and P. falciparum PDX1/PDX2 chimeras do not attain full catalytic activity (Gengenbacher et al., 2006). Wagner et al. (2006) have shown that in contrast to Arabidopsis PDX1.1 and PDX1.3, PDX1.2 does not associate with PDX2. A comparison of the N termini of the Arabidopsis PDX1s reveals high conservation between PDX1.1 and PDX1.3, while PDX1.2 diverges strongly, and this may explain why it cannot associate with PDX2 (Supplemental Fig. S2B).
Another notable difference from the bacterial PLP synthase is that efficient utilization of DHAP, but not G3P, as a substrate by Arabidopsis PDX1 is dependent on the presence of PDX2. This indicates that PDX2 induces a conformational change in PDX1 such that it can utilize DHAP with an efficiency equal to that of G3P as the triose sugar in PLP biosynthesis. The exact mechanism behind this observation cannot be deciphered at present, as the binding site of the triose sugar is currently not known. Interestingly, the inability to use DHAP as a substrate in the absence of PDX2 has also been observed for the P. falciparum enzyme (Gengenbacher et al., 2006). The B. subtilis proteins do not show this dependence; instead, Pdx2 is necessary for utilization of the pentulose phosphate sugar, Ru5P, as a substrate (Raschle et al., 2005). This may reflect subtle differences in the active site for the isomerization of triose and pentose sugars and indicates a closer relationship between the plant and apicomplexan machinery, respectively.
MATERIALS AND METHODS
Plant Material
Wild-type plants of Arabidopsis (Arabidopsis thaliana) ecotype Columbia, the mutant line SALK_072168, as well as the rescued pdx2 plants, were grown on soil at 22°C under 16-h-light/8-h-dark cycles, with 100 μmol photons m−2 s−1. For the expression analyses, tissue from roots, stems, flowers, siliques, rosette leaves, and cauline leaves were collected from at least 10 plants. Cotyledons were collected 18 d after stratification from seedlings grown under the same conditions. For the developmental expression analysis, samples from rosette leaves were collected over a period of 2 to 11 weeks. When grown in sterile culture, Arabidopsis seeds were first surface sterilized, plated on Murashige and Skoog medium (Murashige and Skoog, 1962) containing 1% (w/v) Suc and 0.9% (w/v) agar, and kept for 4 d in the dark at 4°C after which the plates were kept in the conditions as described above for soil-grown plants. For light induction analysis, plates were first transferred to continuous dark at 20°C for 5 d. The etiolated seedlings were then transferred to light at an intensity of 150 μmol photons m−2 s−1, and whole plantlets were collected after 15 min and 1, 5, and 24 h of transfer to light.
Rescue of pdx2 Seeds
Immature pdx2 seeds, which have an albino phenotype and are arrested at the globular stage of embryo development (Tambasco-Studart et al., 2005), were isolated from PDX2/pdx2 plants and placed in an enriched-vitamin medium, composed of Murashige and Skoog salts, 3% Suc, 100 mg/L myo-inositol, 500 mg/L MES, 0.9% agar, 0.1 mg/L 1-naphthylacetic acid, 1 mg/L 6-benzylaminopurine, 1 mg/L biotin, 1 mg/L nicotinic acid, and 1 mg/L thiamine, adjusted to pH 5.7 with KOH. As a control, wild-type or PDX2/pdx2 immature seeds, which are green and are not arrested in embryo development, were cultured under the same conditions. Immature seeds cultured in the presence of vitamin B6 were on medium supplemented with 50 μm each of pyridoxol and pyridoxal. The immature seeds were placed on solid medium with additional bathing in liquid medium (composition as above) immediately after their isolation. This liquid medium was supplied at 3-d intervals over 21 d. Growth conditions were 22°C, 16 h light/8 h dark, and a light intensity of 35 μmol photons m−2 s−1. The teratomata rescued in the presence of vitamin B6 were transferred to soil after 50 d of growth on the enriched-vitamin medium and were watered daily with a solution of 1 mm pyridoxol and 1 mm pyridoxal.
Vitamin B6 Quantification
The total amount of B6 vitamins in the rescued pdx2 and wild-type (control) plants was estimated by employing a microbiological assay as described by Tambasco-Studart et al. (2005). Leaves (5 mg fresh weight) were harvested from the rescued plants that had been maintained without vitamin B6 supplementation for 28 d. The vitamin was extracted using 0.02 m H2SO4 as described by Kall (2003) with the following modifications: a mortar and pestle were used to homogenize the leaf material, the extract was adjusted to pH 5.2 with potassium hydroxide, centrifuged, and the supernatant analyzed. Tissue from at least three independent rescued plants was used.
Measurement of Chlorophyll Content
Chlorophyll was extracted from leaves with 100% acetone and determined spectrophotometrically from the absorbance at 646 and 663 nm according to Lichtenthaler (1987).
Isolation of RNA and Quantitative RT-PCR
Total RNA was extracted using the RNeasy mini kit (Qiagen AG) and treated with ribonuclease-free DNase (Qiagen), according to the manufacturer's instructions. First-strand cDNA synthesis was performed at 42°C for 60 min in a volume of 20 μL, containing 1 or 2 μg of total RNA, 20 pmol of oligo(dT), and 200 units of reverse transcriptase from the Advantage RT-for-PCR kit (Takara Bio Europe/CLONTECH). For transcript quantification, the equivalent of 50 ng of total RNA was employed. Quantitative analyses were performed using an ABI Prism 7700 instrument (Applied Biosystems) by fluorescence-based real-time PCR with the fluorescent dye SYBR Green. The PCR conditions were as follows: an initial denaturation at 95°C for 15 min, followed by 45 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Relative mRNA abundance was calculated using the comparative ΔCt method and normalized against the constitutively expressed actin-2 (At3g18780) or ubiquitin (At5g25760) gene. The primers used were: PDX2 forward, 5′-AGATGGGGAAACCTGTTTGG-3′; PDX2 reverse, 5′-CTGTATGTCTCTGGCCCACC-3′; actin-2 forward, 5′-ATTCTTGCTTCCCTCAGCAC-3′; actin-2 reverse, 5′-CCCCAGCTTTTTAAGCCTTT-3′. The control primers for the Arabidopsis ubiquitin gene were from Sigma-Aldrich.
Immunochemical Analyses
For western-blot analysis, proteins were extracted from frozen plant material by homogenization in liquid N using a mortar and pestle and 1 volume of 50 mm Tris-HCl, pH 7.5, containing 100 mm NaCl, 0.5% Triton X-100, 0.5 mm EDTA, and protease inhibitor cocktail (Sigma-Aldrich). Debris was removed by centrifugation at 10,000g for 15 min at 4°C. Total protein was quantified using a Coomassie protein assay kit (Socochim) and was subjected to 12.5% SDS-PAGE (30 μg/lane). Immunodetection was carried out essentially as described by Titiz et al. (2006), with the exception that the primary antibody used was that raised against the recombinant Arabidopsis PDX2 in rabbit (1:1,000 dilution).
Cloning and Expression of PDX2
PDX2 was amplified by PCR from isolated cDNA with the primers 5′-GGAATTCCATATGACCGTCGGAGTTTTAGCTTTGC-3′ (forward) and either 5′-CCGCTCGAGTTGAAATATAGGAAGATCAGGCTTAG-3′ or 5′-CCGCTCGAGGCTCCTTGCTCAATCTCTTTCGTCATC-3′ (reverse) to obtain sequences encoding the full-length protein and a truncated version lacking the C-terminal 30 amino acids, respectively. In both cases, the gene was cloned without the stop codon into the NdeI/XhoI restriction sites of pET21a (Novagen), such that the proteins were expressed with a C-terminal hexa-His affinity tag. The constructs, pET-AtPDX2H and pET-AtPDX2ΔC30H, were verified by sequence analysis after transformation into DH5α cells (Stratagene) and were transformed into Escherichia coli BL21 cells for expression. Expression of the proteins was induced by addition of isopropyl-1-thio-d-galactopyranoside to a final concentration of 0.25 mm and growth at 26°C for 6 h. The cells were then harvested by centrifugation and stored at −80°C until further purification.
Protein Extraction and Purification
In the case of PDX2 and PDX2ΔC30, cell paste from 2 L of culture was resuspended in 25 mL of lysis buffer (50 mm phosphate buffer, pH 8.0, containing 300 mm NaCl, 0.1 mm EDTA, 10% glycerol, 2 mm phenylmethylsulfonyl fluoride, and 10 mm imidazole). Lysozyme (1 mg/mL) was added and after 30 min on ice, the suspension was further lysed by sonication followed by centrifugation at 25,000g for 30 min at 4°C. The extracted protein was subjected to nickel-nitrilotriacetic acid chromatography according to the protocol supplied by the manufacturer (Qiagen). Isolated protein was promptly desalted using a PD10 column (Bio-Rad) into storage buffer (20 mm Tris-Cl pH 8.0, containing 10 mm NaCl and 0.1 mm EDTA). The purity of the isolated proteins was judged by SDS-PAGE and the identity confirmed by N-terminal sequencing. PDX1.1 to 1.3 were isolated in a fashion identical to that described previously (Tambasco-Studart et al., 2005).
Determination of Enzyme Activities
Glutaminase and PLP synthase activities of PDX2 and PDX1, respectively, were performed essentially as described by Raschle et al. (2005) with the exception that 2.5 units of Glu dehydrogenase was used in the glutaminase assay.
Coupling Efficiency
To calculate the coupling efficiency between the PLP synthase subunits, the ratio between the rate of PLP synthesis and Gln hydrolysis was determined. Reactions were carried out in the presence of R5P (0.5 mm), dl-G3P (1 mm), and l-Gln (10 mm) with 10 μm each of PDX1.1 and PDX2. Rates of Gln hydrolysis and PLP synthesis were monitored separately as described above.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number Q8LAD0.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Rate of Gln hydrolysis as a function of the concentration of Gln.
Supplemental Figure S2. Sequence analysis of PDX proteins.
Supplemental Figure S3. Absolute transcript abundance of PDX2 (black bars) compared to PDX1.1 (light gray bars) and PDX1.3 (dark gray bars).
Supplemental Figure S4. Gene Correlator analysis of the expression of PDX2 in relation to PDX1 (Zimmermann et al., 2004).
Supplementary Material
Acknowledgments
We are grateful to the SALK institute for supplying line 072168 for analysis.
This work was supported by the ETH Zurich (grant no. 0094/41–2703.5).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Teresa B. Fitzpatrick (tfitzpatrick@ethz.ch).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Belitsky BR (2004) Physical and enzymological interaction of Bacillus subtilis proteins required for de novo pyridoxal 5′-phosphate biosynthesis. J Bacteriol 186 1191–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera AK, Smith JL, Zalkin H (2000) Dual role for the glutamine phosphoribosylpyrophosphate amidotransferase ammonia channel: interdomain signaling and intermediate channeling. J Biol Chem 275 7975–7979 [DOI] [PubMed] [Google Scholar]
- Bilski P, Li MY, Ehrenshaft M, Daub ME, Chignell CF (2000) Vitamin B6 (pyridoxine) and its derivatives are efficient singlet oxygen quenchers and potential fungal antioxidants. Photochem Photobiol 71 129–134 [DOI] [PubMed] [Google Scholar]
- Burns KE, Xiang Y, Kinsland CL, McLafferty FW, Begley TP (2005) Reconstitution and biochemical characterization of a new pyridoxal-5′-phosphate biosynthetic pathway. J Am Chem Soc 127 3682–3683 [DOI] [PubMed] [Google Scholar]
- Cane DE, Du S, Robinson JK, Hsiung Y, Spenser ID (1999) Biosynthesis of vitamin B6: enzymatic conversion of 1-deoxy-D-xylulose-5-phosphate to pyridoxol phosphate. J Am Chem Soc 121 7722–7723 [Google Scholar]
- Chen H, Xiong L (2005) Pyridoxine is required for post-embryonic root development and tolerance to osmotic and oxidative stresses. Plant J 44 396–408 [DOI] [PubMed] [Google Scholar]
- Dong YX, Sueda S, Nikawa J, Kondo H (2004) Characterization of the products of the genes SNO1 and SNZ1 involved in pyridoxine synthesis in Saccharomyces cerevisiae. Eur J Biochem 271 745–752 [DOI] [PubMed] [Google Scholar]
- Drewke C, Leistner E (2001) Biosynthesis of vitamin B6 and structurally related derivatives. In G Litwack, ed, Vitamins and Hormones, Vol 63. Academic Press, San Diego, pp 121–155 [DOI] [PubMed]
- Ehrenshaft M, Bilski P, Li MY, Chignall CF, Daub ME (1999) A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc Natl Acad Sci USA 96 9374–9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengenbacher M, Fitzpatrick TB, Raschle T, Flicker K, Sinning I, Müller S, Macheroux P, Tews I, Kappes B (2006) Vitamin B6 biosynthesis by the malaria parasite Plasmodium falciparum: biochemical and structural insights. J Biol Chem 281 3633–3641 [DOI] [PubMed] [Google Scholar]
- Kall MA (2003) Determination of total vitamin B6 in foods by isocratic HPLC: a comparison with microbiological analysis. Food Chem 82 315–327 [Google Scholar]
- Laber B, Maurer W, Scharf S, Stepusin K, Schmidt FS (1999) Vitamin B6 biosynthesis: formation of pyridoxine 5′-phosphate from 4-(phosphohydroxy)-L-threonine and 1-deoxy-D-xylulose-5-phosphate by PdxA and PdxJ protein. FEBS Lett 449 45–48 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 18 350–382 [Google Scholar]
- Mittenhuber G (2001) Phylogenetic analyses and comparative genomics of vitamin B6 (pyridoxine) and pyridoxal phosphate biosynthesis pathways. J Mol Microbiol Biotechnol 3 1–20 [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth of bioassays with tobacco tissue culture. Physiol Plant 15 493–497 [Google Scholar]
- Myers RS, Amaro RE, Luthey-Schulten ZA, Davisson VJ (2005) Reaction coupling through interdomain contacts in imidazole glycerol phosphate synthase. Biochemistry 44 11974–11985 [DOI] [PubMed] [Google Scholar]
- Osmani AH, May GS, Osmani SA (1999) The extremely conserved pyroA gene of Aspergillus nidulans is required for pyridoxine synthesis and is required indirectly for resistance to photosensitizers. J Biol Chem 274 23565–23569 [DOI] [PubMed] [Google Scholar]
- Patton DA, Schetter AL, Franzmann LH, Nelson K, Ward ER, Meinke DW (1998) An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin biosynthesis. Plant Physiol 116 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle T, Amrhein N, Fitzpatrick TB (2005) On the two components of pyridoxal 5′-phosphate synthase from Bacillus subtilis. J Biol Chem 280 32291–32300 [DOI] [PubMed] [Google Scholar]
- Raschle T, Arigoni D, Brunisholz R, Amrhein N, Fitzpatrick TB (2007) Reaction mechanism of pyridoxal 5′-phosphate synthase: detection of an enzyme bound chromophoric intermediate. J Biol Chem 282 6098–6105 [DOI] [PubMed] [Google Scholar]
- Raushel FM, Thoden JB, Holden HM (1999) The amidotransferase family of enzymes: molecular machines for the production and delivery of ammonia. Biochemistry 38 7891–7899 [DOI] [PubMed] [Google Scholar]
- Spenser ID, Hill RE (1995) The biosynthesis of pyridoxine. Nat Prod Rep 12 555–565 [DOI] [PubMed] [Google Scholar]
- Strohmeier M, Raschle T, Mazurkiewicz J, Rippe K, Sinning I, Fitzpatrick TB, Tews I (2006) Structure of a bacterial pyridoxal 5′-phosphate synthase complex. Proc Natl Acad Sci USA 103 19284–19289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambasco-Studart M, Titiz O, Raschle T, Forster G, Amrhein N, Fitzpatrick TB (2005) Vitamin B6 biosynthesis in higher plants. Proc Natl Acad Sci USA 102 13687–13692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Tateno Y, Gojobori T (2005) Evolution of vitamin B6 (pyridoxine) metabolism by gain and loss of genes. Mol Biol Evol 22 243–250 [DOI] [PubMed] [Google Scholar]
- Titiz O, Tambasco-Studart M, Warzych E, Apel K, Amrhein N, Laloi C, Fitzpatrick TB (2006) PDX1 is essential for vitamin B6 biosynthesis, development and stress tolerance in Arabidopsis. Plant J 48 933–946 [DOI] [PubMed] [Google Scholar]
- Wagner S, Bernhardt A, Leuendorf JE, Drewke C, Lytovchenko A, Mujahed N, Gurgui C, Frommer WB, Leistner E, Fernie AR, et al (2006) Analysis of the Arabidopsis rsr4-1/pdx1-3 mutant reveals the critical function of the PDX1 protein family in metabolism, development, and vitamin B6 biosynthesis. Plant Cell 18 1722–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrenger C, Eschbach ML, Müller IB, Warnecke D, Walter RD (2005) Analysis of the vitamin B6 biosynthesis pathway in the human malaria parasite Plasmodium falciparum. J Biol Chem 280 5242–5248 [DOI] [PubMed] [Google Scholar]
- Zalkin H, Smith JL (1998) Enzymes utilizing glutamine as an amide donor. Adv Enzymol Relat Areas Mol Biol 72 87–144 [DOI] [PubMed] [Google Scholar]
- Zein F, Zhang Y, Kang YN, Burns K, Begley TP, Ealick SE (2006) Structural insights into the mechanism of the PLP synthase holoenzyme from Thermotoga maritima. Biochemistry 45 14609–14620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Henning L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.