Abstract
Caffeine (1,3,7-trimethylxanthine) is a secondary metabolite produced by certain plant species and an important component of coffee (Coffea arabica and Coffea canephora) and tea (Camellia sinensis). Here we describe the structures of two S-adenosyl-l-methionine-dependent N-methyltransferases that mediate caffeine biosynthesis in C. canephora ‘robusta’, xanthosine (XR) methyltransferase (XMT), and 1,7-dimethylxanthine methyltransferase (DXMT). Both were cocrystallized with the demethylated cofactor, S-adenosyl-l-cysteine, and substrate, either xanthosine or theobromine. Our structures reveal several elements that appear critical for substrate selectivity. Serine-316 in XMT appears central to the recognition of XR. Likewise, a change from glutamine-161 in XMT to histidine-160 in DXMT is likely to have catalytic consequences. A phenylalanine-266 to isoleucine-266 change in DXMT is also likely to be crucial for the discrimination between mono and dimethyl transferases in coffee. These key residues are probably functionally important and will guide future studies with implications for the biosynthesis of caffeine and its derivatives in plants.
Caffeine is a plant alkaloid known to have sensory and stimulatory effects when consumed in beverages such as coffee (Coffea arabica and Coffea canephora) and tea (Camellia sinensis). The pharmacological actions are thought to be mediated via blockade of the adenosine A1 and A2A receptors (Cauli and Morelli, 2005). The exact biological role of caffeine and related purine alkaloids of plants is still unclear, although there are now two main hypotheses. It has been proposed that caffeine protects young leaves and fruit from predators (chemical defense theory [Frischknecht, 1985; Hollingsworth et al., 2002]) and that the caffeine released by the seed coat prevents germination of other seeds (allelopathic or autotoxic theory [Friedman and Waller, 1983]). In agreement with these proposals, it is known that caffeine is accumulated in both the seeds and young leaves of coffee plants (Ashihara and Suzuki, 2004). Strong supporting evidence for the chemical defense theory has recently been obtained by Uefuji et al. (2005), who demonstrated that leaves of transgenic tobacco (Nicotiana tabacum) plants engineered to produce caffeine were less susceptible to insect feeding versus control leaves that did not contain caffeine.
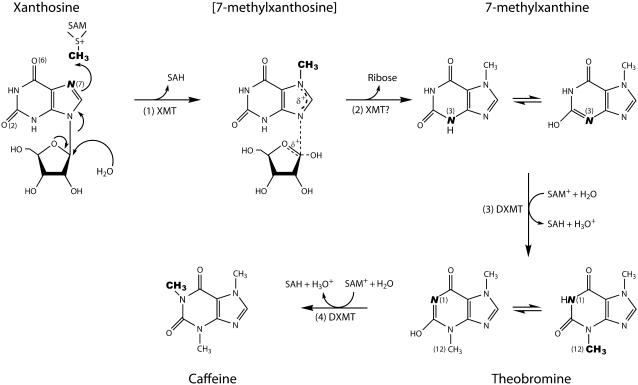
The first N-methyltransferase from the caffeine biosynthesis pathway to be biochemically characterized was cloned from young tea leaves and designated tea caffeine synthase1 (TCS1; Kato et al., 2000). Subsequently, an orthologous recombinant N-methyltransferase was cloned from the leaf cDNA of coffee and biochemically characterized as 7-methylxanthine (7mX) methyltransferase1 (MXMT1; Ogawa et al., 2001). A further group of homologous recombinant N-methyltransferases were then cloned from leaf and endosperm cDNA of coffee, biochemically characterized, and designated coffee 7-methylxanthosine synthase1 (XRS1; Mizuno et al., 2003a), coffee caffeine synthase1 (CCS1; Mizuno et al., 2003b), and coffee theobromine (Tb) synthase (CTS1 and CTS2; Mizuno et al., 2003b). Three S-adenosyl-l-methionine (SAM)-dependent N-methyltransferases were also cloned from immature fruit cDNA of coffee and biochemically characterized (Uefuji et al., 2003). These have been designated xanthosine (XR) methyltransferase (XMT), MXMT2 or Tb synthase, and 3,7-dimethylxanthine methyltransferase (DXMT or caffeine synthase). More recently, a Tb synthase (PCS1) from Camellia plants was characterized (Yoneyama et al., 2006). Thus, after many years of biochemical study, the core pathway has largely been clarified and occurs via three SAM-dependent N-methylation steps and a Rib removal step (Fig. 1), the Rib removal step being carried out by a currently unidentified 7-methylxanthosine (7mXR) nucleosidase.
Figure 1.
Caffeine biosynthetic pathway in C. canephora ‘robusta’. The first (1), third (3), and fourth (4) steps are N-methylations and the second (2) step is cleavage of Rib. Water is depicted as the proton acceptor for nitrogens accepting the reactive methyl group from SAM to produce an oxonium ion (H3O+). Italic letters highlight the atom undergoing methylation and the transferred groups are highlighted in bold.
The N-methyltransferases from the caffeine biosynthetic pathway have high protein sequence homology (>80% identity), but exhibit remarkable substrate selectivity (Kato and Mizuno, 2004). They also belong to the wider motif B′ methyltransferases specific to plants (Kato and Mizuno, 2004). This important family includes enzymes involved in the biosynthesis of small and volatile methyl esters that are proposed to act as interplant signaling molecules in plant defense (Zubieta et al., 2003). Only one member of this family, salicylic acid O-methyltransferase (SAMT), has been structurally characterized so far (Zubieta et al., 2003). In an effort to understand the subtle substrate selectivity within the family of N-methyltransferases from the caffeine biosynthetic pathway, as well as between members of the larger motif B′-containing family of methyltransferase proteins, we decided to undertake structural and biochemical characterization of XMT and DXMT from Coffea canephora ‘robusta’. Here we describe the structures of XMT and DXMT complexed with S-adenosyl-l-Cys (SAH), the demethylated product, and their respective substrates, either XR or Tb. These structures are compared to the SAMT structure complexed with salicylic acid and SAH (Zubieta et al., 2003). The structural results presented here offer several insights into the substrate specificity from this family of SAM-dependent methyltransferases.
RESULTS
Enzymatic Activity
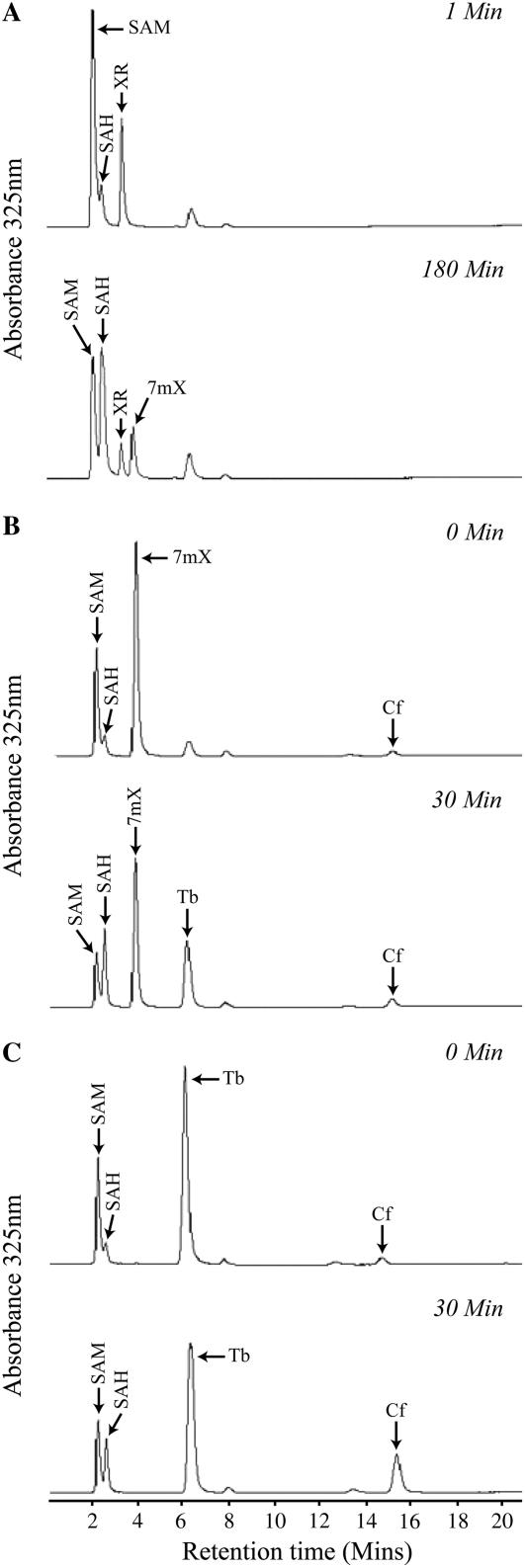
XMT and DXMT were expressed in Escherichia coli and highly purified for both biochemical and structural studies. Biochemical analysis of the highly purified XMT used for crystallization shows that this preparation catalyzes the addition of a methyl group to the N7 of XR and generates 7mX instead of 7mXR (Fig. 2A). This unexpected observation is dependent on SAM because initial assays of recombinant XMT were unsuccessful due to the presence of SAH, which was added for crystallization trials. The XMT activity was restored by diafiltration into the reaction buffer containing 3 mm SAM. XMT showed no activity toward either 7mX or Tb (data not shown). DXMT is capable of converting 7mX to Tb (Fig. 2B), as well as converting Tb to caffeine (Fig. 2C), and exhibits no detectable activity toward XR (data not shown).
Figure 2.
HPLC elution profiles for methyltransferase reactions of XMT and DXMT. A, XMT reaction activity with XR as substrate, with a sample taken at t = 1 min (top) and t = 180 min (bottom). B, DXMT reaction with 7mX as substrate, with a sample taken at t = 0 min before recombinant enzyme was added (top) and t = 30 min (bottom). C, DXMT reaction with Tb as substrate, with a sample taken at t = 0 min before recombinant enzyme was added (top) and t = 30 min (bottom).
Structure Determination and Refinement
DXMT could be solved by molecular replacement using SAMT as a starting model. However, the subsequent refinement was problematic and the structure was eventually solved by the single anomalous diffraction method with selenomethionine (SeMet)-incorporated DXMT. The final model comprises 348 of a possible 384 residues, one SAH, two Tb molecules in different orientations, and 146 water molecules. The disordered residues comprise the N- and C-terminal residues (two and five residues, respectively) and some surface loops (residues 12–15, 82–91, 169–174, and 303–311). The DXMT dimer observed in solution (McCarthy et al., 2007) is preserved in the crystal lattice, where each monomer is related by a crystallographic 2-fold axis along B. XMT was solved by molecular replacement using a model derived from the DXMT structure with flexible loops or insertions deleted. The final model comprises four XMT molecules in the asymmetric unit and 499 water molecules. Each of the XMT molecules contained 344 of a possible 372 residues, one SAH, and one XR. The 13 N-terminal residues have very weak electron density, but were excluded, and there was no discernible density for two surface loops (residues 85–89 and 303–306) or the five C-terminal residues of each molecule. The four molecules in the asymmetric unit form two dimer pairs. All the crystallographic information is summarized in Table I. The R and Rfree values for both DXMT and XMT are larger than those for comparable structures found in the Protein Data Bank and this is mainly due to the generous cutoff criteria used for determination of the high-resolution limit.
Table I.
Data collection, phasing, and refinement statistics
Values in parentheses are for the outermost resolution shell. 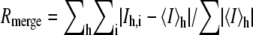 calculated for the whole dataset. Figure of merit
calculated for the whole dataset. Figure of merit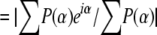 , where P(α) is the phase probability distribution and α is the phase.
, where P(α) is the phase probability distribution and α is the phase. 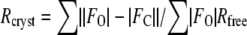 was calculated as for
was calculated as for 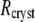 , with 5% of the data omitted from the structural refinement.
, with 5% of the data omitted from the structural refinement.
| Crystal | DXMT (Se-Met) | DXMT | XMT |
|---|---|---|---|
| Space group | C2221 | C2221 | P21 |
| Cell dimensions a, b, c (Å) | 49.9, 106.1, 141.7, 90, 90, 90 | 50.2, 105.6, 140.9, 90, 90, 90 | 57.6, 119.8, 116.4, 90, 102.2, 90 |
| Wavelength (Å) | 0.9793 | 0.9393 | 0.9393 |
| Resolution (Å; outer shell) | 30 − 2.66 (2.8 − 2.66) | 30 − 2.0 (2.1 − 2.0) | 30 − 2.2 (2.3 − 2.2) |
| Completeness (%; final shell) | 97.2 (81.7) | 97.2 (90.2) | 96.7 (93.4) |
| Rmerge (final shell) | 12.1 (39.9) | 5.3 (50.2) | 7.8 (44.3) |
| <I/σ(I)> (final shell) | 17.9 (3.6) | 15.6 (3.2) | 13.1 (3.0) |
| Unique reflections | 20,304 | 25,059 | 75,624 |
| Total reflections | 226,661 | 120,487 | 350,898 |
| FOM (centric/acentric) | 0.11/0.41 | ||
| Rcryst (final shell) | 0.221 | 0.23 | |
| Rfree | 0.276 | 0.284 | |
| rmsd, bonds/angles | 0.012 Å/1.6° | 0.015 Å/1.7° |
Overall Structure
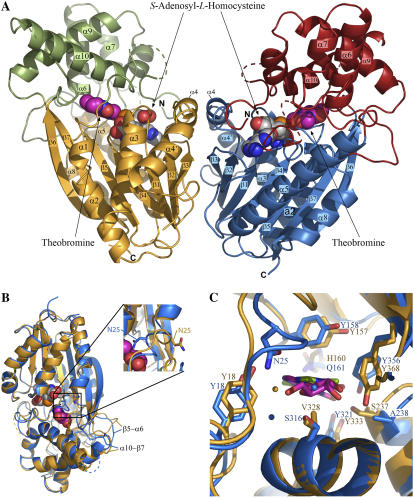
The XMT and DXMT structures are nearly identical and can be superimposed with a root-mean-square derivation (rmsd) of 1.0 Å for 331 Cα atoms from a possible 356. XMT and DXMT consist of two domains, the core SAM-dependent methyltransferase domain (residues 23–162, 190–227, 266–295, and 367–379 for DXMT) and an α-helical cap domain (residues 1–22, 163–189, 228–265, and 296–366 for DXMT; Figs. 3A and 4). In fact, their overall structure is nearly identical to that found in the recently solved SAMT, with a Cα rmsd of 1.6 Å between SAMT and XMT for 307 residues and a Cα rmsd of 1.7 Å between SAMT and DXMT for 306 residues. The major structural differences between all three of these structures occur in loop regions at or proximal to the active site (Fig. 3, B and C). The first involves the deletion of one amino acid between residues Ser-22 to Leu-28 in DXMT (Ser-22 to Ile-29 in XMT), resulting in a structural difference for this region between DXMT, XMT, and SAMT. The second occurs in the loop region between β5 and α6, where there is a structural difference between all three methyltransferases (XMT, DXMT, and SAMT) and includes an extra four residues in SAMT (Fig. 4). The third difference occurs in the loop region between β6 and α9, which corresponds to a large insertion in XMT and DXMT (Fig. 4), of which Asp-303 to Asp-311 and Ile-303 to Asp-305 are disordered in DXMT and XMT, respectively. The final structural difference occurs in the loop region between α10 and β7.
Figure 3.
Structure of DXMT and superposition of XMT and DXMT. A, Ribbon diagram of DXMT dimer structure, with SAH in gray and Tb in magenta. The secondary structure is labeled with the SAM-dependent methyltransferase domain in gold or blue and the α-helical cap domain in green or red. B, Superposition of XMT (blue) and DXMT (gold). C, Superposition of the XMT (blue) and DXMT (gold) active sites with important substrate-interacting residues highlighted. Tb is in gray and XR is in green.
Figure 4.
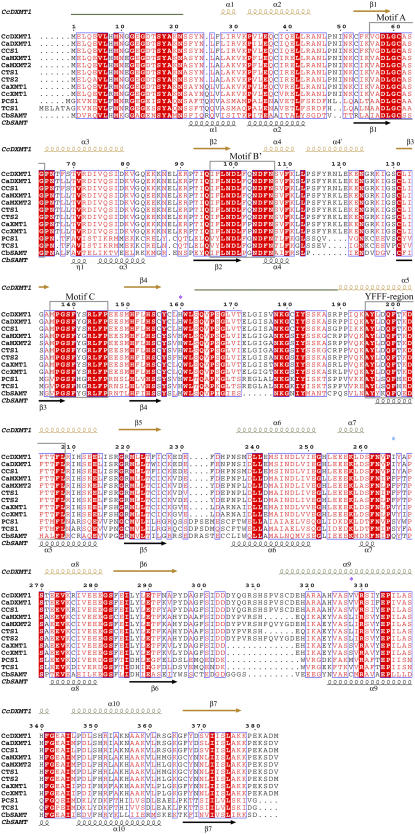
Multiple sequence alignment of representative N-methyltransferases and SAMT. Secondary structure plots are given for DXMT (above) and SAMT (below). The core SAM-binding domain is colored in orange, whereas the helical capping domain is colored in green. The SAM-binding motifs (A, B′, and C) and the conserved YFFF region are shown by gray boxes. Purple asterisks mark the crucial residues for XMTs versus DXMTs and a cyan asterisk marks the crucial residue in 7mX versus Tb recognition. Accession numbers are as follows: CaDXMT1, AB084125; CaMXMT1, AB084794; CaMXMT2, AB084126; CaXMT1, AB048793; CTS1, AB034700; CTS2, AB054841; CcDXMT1, DQ422955 (this study); CcXMT1, DQ422954 (this study); CbSAMT, AAF00108; PCS1, AB207817; and TCS1, AB031280.
The biochemically characterized members of this family exist as dimers in solution (Zubieta et al., 2003). The dimerization interface involves residues from α4 and β3 (Fig. 3A), and leaves each of the monomer active sites independent. Dimerization buries a total of 1,080 Å2 and 1,146 Å2 of solvent-accessible surface for each monomer of XMT and DXMT, respectively. This represents 7% and 8% of the total surface area for each monomer of XMT and DXMT, respectively, which is at the low end of dimer interfaces analyzed (Jones and Thornton, 1995) and similar to that observed in SAMT (Zubieta et al., 2003). Much of the interface is hydrophobic and sequence conserved between XMT, DXMT, and SAMT, with the side chains of Phe-102, Phe-106, Phe-110, Leu-132, and Met-136 being buried between the monomers in DXMT. A number of identical hydrogen bonds are also observed, such as those between the sequence-conserved side chains Asp-105 and Asn-107 in DXMT and the two main chain hydrogen bonds made by Ala-135 in DXMT.
SAH/SAM-Binding Site
SAH is bound in a similar position and conformation in XMT, DXMT, and SAMT to other SAM-dependent methyltransferases (Fig. 3A). SAH/SAM binding is mediated through extensive hydrogen bonding and van der Waals interactions. Two hydrogen-bonding interactions to the adenine ring of the SAH come from the highly conserved motif C (Fig. 4). The first one is between the exocyclic amino group (N6) and the hydroxyl group of Ser-139 in DXMT and the second one occurs between N1 and the backbone amide of Phe-140 in DXMT. There is also an additional water-mediated hydrogen bond between the exocyclic amino group (N6), N7, and the carboxyl group of Leu-162 in DXMT. The adenine ring is sandwiched between the hydrophobic side chains of Leu-101 in DXMT from motif B′ (Fig. 4) and Phe-140 in DXMT from motif C (Fig. 4). The adenine ring lies coplanar with the phenyl ring in a π-stacking interaction on one face, whereas the Leu caps the hydrophobic face on the opposite side. The Rib hydroxyls of SAH form hydrogen-bonding interactions with strictly conserved Asp-100 in DXMT from motif B′ (Fig. 4). There is also an additional sulfhydryl hydrogen bond between the Cys-158 in DXMT (Cys-159 in XMT) and the Rib O4′ of SAH in XMT and DXMT.
The direct hydrogen-bonding interactions between the amino tail of SAH and the backbone carboxyl residues of Gly-60 from motif A (Fig. 4) and Cys-156 in DXMT are conserved in all three structures. XMT and DXMT also make water-mediated interactions between the amino tail of SAH, the protein main chain atoms of Leu-59, Gly-60, and Cys-156, and the side chains of Asp-58 and Thr-70 in DXMT. The carboxyl tail of SAH forms a hydrogen-bonding interaction with the side chain of Tyr-18 in all three methyltransferases. An additional hydrogen bond (3.2 Å) is observed in DXMT between the carboxyl tail and Asn-66. Interestingly, this sequence-conserved Asn has moved away from the SAH carboxyl tail to a distance of approximately 4.2 Å in both XMT and SAMT. Additional water-mediated hydrogen bonds to the carboxyl tail are also observed in both XMT and DXMT.
Substrate-Binding Site
XMT was cocrystallized in the presence of XR and it is well defined in the structure (Supplemental Fig. S1). DXMT was cocrystallized in the presence of Tb and exists in two conformations (Supplemental Fig. S1), one mimicking 7mX binding. The substrate-binding site of XMT and DXMT is located in a similar position to that found in SAMT (Fig. 5, A–D). The substrate is properly positioned in the active site for methylation in all the methyltransferases from this family through both hydrogen bonding and van der Waals interactions. Only some of these interactions are sequence conserved and there are many differences important for both substrate recognition and catalysis.
Figure 5.
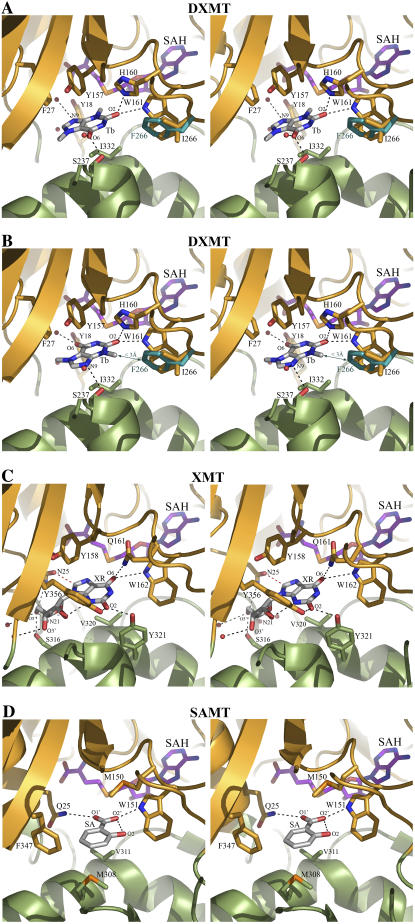
Stereo view of the ligand-binding sites. A, Tb bound to DXMT in the 7mX-like conformation. B, Tb bound to DXMT and either an Ile or Phe at position 266. C, XR-binding site in XMT. D, Salicylic acid (SA)-binding site in SAMT.
XR makes a total of nine hydrogen bonds to the XMT protein, six with the purine ring and three with the Rib moiety (Fig. 5C). The O5′ hydroxyl group from the Rib moiety of XR makes hydrogen-bonding interactions to the backbone carboxyl groups of Asn-21 and Ser-22, the hydroxyl group of Ser-316, and the amide group of Asn-25. The O5′ hydroxyl group also makes an additional water-mediated hydrogen bond to the backbone carboxyl groups of Asn-21 and Ser-316. This water also makes a hydrogen bond to the O4′ Rib atom in addition to a direct hydrogen bond to the Asn-25. The final XR Rib interaction is a water-mediated hydrogen bond between the O3′ hydroxyl group and the carboxyl group of Tyr-297.
In SAMT, the carboxylate moiety of salicylic acid is precisely positioned via hydrogen-bonding interactions with the sequence-conserved Trp-151 (Fig. 5D), which is predicted to be important for substrate recognition in this family of methyltransferases (Zubieta et al., 2003). A strictly conserved Trp is again involved in hydrogen-bonding interactions with the substrate in both XMT and DXMT, in each case making a hydrogen bond to a carboxylate moiety (Fig. 5, A–C). Trp-162 forms a hydrogen bond to the O6 carboxylate of the XR purine ring in XMT and Trp-161 forms a hydrogen bond with the O2 carboxylate of either 7mX or Tb in DXMT. The amide of the Gln-161 side chain in XMT forms an additional hydrogen bond to the carboxylate O6 of XR (Fig. 5C). This residue is replaced by His-60 in DXMT, which forms a similar hydrogen bond to the O2 of Tb or 7mX (Fig. 5, A and B). Interestingly, this position is occupied by Met-150 in SAMT, which acts as a clamp for one face of the benzyl ring of salicylic acid in SAMT (Zubieta et al., 2003; Fig. 5D).
The largest structural difference between all the methyltransferases occurs in the loop between β5 and α6. This loop contributes to an important hydrogen-bonding interaction in DXMT substrate binding, where the O6 carboxylate group of 7mX-like Tb forms a direct hydrogen bond with the Ser-237 hydroxyl group (Fig. 5A) and a water-mediated hydrogen bond with its amide backbone. However, in the DXMT binding of Tb, Ser-237 forms these hydrogen bonds with N9, whereas the O6 carboxylate group of Tb forms a direct and water-mediated hydrogen bond with the Tyr-18 hydroxyl group (Fig. 5B). Ser-237 is mutated to an Ala in XMT, resulting in a loss of hydrogen-bonding interaction and a loop movement away from the binding site of approximately 1.6 Å relative to DXMT (Fig. 3B). In SAMT, this loop moves by approximately 3.6 Å relative to DXMT and forms part of the salicylic hydrophobic binding pocket.
In XMT, the O2 carboxylate group of XR forms a hydrogen bond with the hydroxyl groups of both Tyr-321 and Tyr-356 (Fig. 5C). Interestingly, although Tyr-333 in DXMT is structurally equivalent to Tyr-321 in XMT, this sequence-conserved Tyr is too far from either the 7mX or Tb in DXMT to form a hydrogen bond with the substrate (Fig. 5, A and B). The Tyr-356 of XMT is the sequence-conserved Tyr-368 in DXMT, which adopts a different conformation due to the movement of the β5-α6 loop (Fig. 3, B and C).
These hydrogen-bonding interactions are supplemented by hydrophobic interactions between the protein and the hydrophobic faces of the purine ring. XR is involved in hydrophobic interaction with Met-9, Tyr-18, Tyr-24, Tyr-158, Ile-227, Val-320, Tyr-321, and Tyr-356 in XMT. Val-320 caps one face of the purine ring, whereas Ile-227 and Tyr-158 abut the opposite face, with the purine ring perpendicular to the phenyl ring of Tyr-158 in a π-stacking interaction (Fig. 5C). Tb forms hydrophobic interactions with Met-9, Leu-26, Phe-27, Tyr-157, Ile-226, Met-238, Ile-266, Val-328, Ile-332, Tyr-333, and Tyr-368 in DXMT. In particular, Ile-332 forms a hydrophobic interaction with one face, whereas Phe-27 and Tyr-157 abut the opposite face, with the purine ring again perpendicular to the sequence-conserved Tyr-157 in a π-stacking orientation (Fig. 5, A and B).
DISCUSSION
The work presented here details the structural analysis of two very closely related enzymes involved in caffeine biosynthesis in coffee. These proteins are part of the general motif B′ family of methyltransferases, which transfer the activated methyl group of SAM to different plant secondary metabolites. Biochemical studies on the purified proteins (Fig. 2) confirmed that one is an XMT and the other is a DXMT, as predicted from the analysis of their primary sequence (McCarthy et al., 2007). The structural studies presented here have identified a number of important loops or amino acid changes that are necessary for alterations in substrate recognition and/or catalysis.
N Terminus
The N-terminal part of the protein is flexible, facilitating the entry of the substrates and cofactor into the active site and the exit of the reaction products. XMT has an extra Gln residue between Asn-25 and Leu-26 (as numbered in DXMT; Fig. 4), allowing important hydrogen-bonding changes to occur. The Asn-25 of XMT makes extensive hydrogen-bonding interactions with the Rib moiety of the XR substrate (Fig. 5C). This agrees with the proposal that a shorter side chain is necessary to accommodate the larger substrates of the N-methyltransferases (Zubieta et al., 2003). The carboxylate group of Asn-25 in XMT is only 2.9 Å away from C8, indicating that it may also form an aromatic hydrogen bond to XR (Fig. 5C). In contrast to XMT, Asn-25 of DXMT is not involved in hydrogen-bonding interactions with either 7mX or Tb and actually points away from the active site (Fig. 3B). The space vacated by the Asn-25 movement is occupied by four well-ordered water molecules, which form an extensive hydrogen-bonding network between DXMT and 7mX or Tb. The additional space also allows for the closer approach of Tyr-18 to the substrate in DXMT, enabling it to hydrogen bond with the O6 carboxyl group in Tb (Fig. 5B). Tyr-18 is strictly conserved in the motif B′ methyltransferases and its hydroxyl group is in close proximity to the positively charged S atom of SAM. A similarly positioned Tyr-21 in an unrelated N-methyltransferase, Gly N-methyltransferase, has been implicated in its methyl transfer mechanism (Takata et al., 2003) and hence a catalytic role for this residue in motif B′ methyltransferases appears likely.
Substrate Discrimination between XMT and DXMT
Two elements important for substrate discrimination between XMT and DXMT can be identified. The first involves Ser-316 in XMT, which forms a hydrogen bond with the O5′ hydroxyl group from the Rib moiety of XR (Fig. 5C). Ser-316 corresponds to a Val in DXMT and to a Cys in SAMT (Fig. 4). A Val at this position would disrupt the hydrogen bond and introduce a steric clash with the Rib moiety of XR (Fig. 3C). Ser-316 is therefore likely to be crucial for XR substrate specificity in XMT. The second occurs in the loop connecting α10 to β7 (Fig. 3B), where large conformational differences are observed between the three methyltransferases in this family. The Tyr-356 of XMT adopts a different conformation from the corresponding sequence-conserved Tyr-368 of DXMT (Fig. 3C). These differences result in the Tyr-356 of XMT hydrogen bonding with the O2 carboxyl group of XR (Fig. 5C), whereas the Tyr-368 in DXMT adopts a conformation close to the Rib site and probably distorts the site enough to eliminate its potential to accept a Rib (Fig. 3C). The alternate conformations of these Tyr side chains are mainly due to movement of the β5-α6 loop away from the substrate-binding site in XMT. This is probably due to the sequence divergence observed between XMT and DXMT (Fig. 4) and specifically where a change from Pro-235 in DXMT to Arg-236 in XMT allows the β5-α6 loop in each structure to adopt a different conformation.
Another difference between the XMT and DXMT substrate-binding sites occurs at position Tyr-321 in XMT, which forms a hydrogen bond with the O2 carboxyl group of XR (Fig. 5C). In DXMT, the structurally conserved Tyr-333 is too far from any potential hydrogen-bonding partners and does not directly contribute to substrate binding. Tyr-333 instead forms a hydrogen-bonding interaction with Ser-237, orienting it for optimal hydrogen-bonding interactions with 7mX or Tb in DXMT. Ala-312 occupies this position in SAMT (Fig. 4) and is necessary to accommodate the large Trp-226, which forms part of the salicylate-binding site in SAMT (Zubieta et al., 2003).
Substrate Specificity between MXMTs and DXMTs
Coffee plants contain both MXMTs (or CTS1/2) and DXMTs (Kato and Mizuno, 2004). It is still unclear why both enzymes are required because the DXMTs can readily catalyze both methyl transfers (Fig. 2; Uefuji et al., 2003). However, whereas multiple isoforms of the MXMTs are expressed in various tissue types in coffee plants, the DXMTs are predominantly expressed in immature fruit (Uefuji et al., 2003). This observation may have important consequences for caffeine production in the immature fruit. There are 15 residues within 5 Å of Tb in DXMT. Only three of these are of significance when comparing the sequences of MXMT with DXMT because eight are identical and four involve very conservative changes. The first involves the substitution of Phe-27 in DXMT by an Ala in MXMT (Fig. 4). There is no obvious reason from our DXMT structure why this substitution would favor the 7mX substrate unless there is a conformational change. The next involves substitution of Ser-237 in DXMT for Pro in MXMT (Fig. 4). This would disrupt the hydrogen bond to either the O6 carboxyl group of 7mX or the N9 of Tb (Fig. 5, A and B), so it is unclear how this change would discriminate between 7mX and Tb. The last involves the substitution of Ile-266 in DXMT by a Phe in the MXMTs (Fig. 4). A Phe at this position in the MXMTs would result in a steric clash with a methyl group on the N3 position, precluding the binding of Tb and its subsequent methylation (Fig. 5B). The N1 position in 7mX is not methylated, allowing it to bind and methylation to occur in the MXMTs. However, it should be noted that the sequences of a MXMT from Camellia plants (PCS1; Yoneyama et al., 2006) and a DXMT from tea (TCS1; Kato et al., 2000) vary quite significantly in these regions (Fig. 4). So, while this Ile-to-Phe substitution is likely to be important for substrate discrimination in coffee, it is unlikely to account for the substrate selectivity observed between all the MXMT and DXMT proteins.
Dual Activity for XMT
A highly purified preparation of XMT was not active on 7mX or Tb as anticipated. Surprisingly, the only clearly detectable product from the methylation of XR was 7mX (Fig. 2A). A similar activity was reported for a crude preparation of XMT from coffee (Uefuji et al., 2003) and the 7mX nucleosidase activity was reportedly due to a nonspecific E. coli purine-nucleoside phosphorylase (PNP; Uefuji et al., 2003). However, it is highly unlikely that PNP would uniquely act on 7mXR. The structure and mechanism of PNP is well established (Koellner et al., 2002; Bennett et al., 2003) and XR, with its free N7 atom, would be a far better substrate. The reaction is dependent on SAM because no product peaks were observed in XMT assays when high concentrations of SAH were still present and we believe that the methyl transfer and nucleoside cleavage may indeed be coupled (Fig. 1). This also explains why transgenic tobacco plants expressing XMT, MXMT, and DXMT are able to produce caffeine (Ogita et al., 2005; Uefuji et al., 2005). Similar reactions and mechanisms involving the formation of an oxocarbenium intermediate are observed in the N-ribohydrolases (Degano et al., 1998; Lee et al., 2005) and phosphorylases (Bennett et al., 2003). A plausible mechanism for XMT Rib cleavage would commence with the methylation of N7, resulting in a partial positive charge on the purine ring moiety. This positive charge would induce the flow of electrons from the Rib moiety, resulting in the carbenium intermediate. A nucleophilic attack by water on the oxocarbenium intermediate could then occur because the Rib moiety is partially solvent exposed and oxocarbenium ions are an extremely reactive species in aqueous solution (Amyes and Jencks, 1989). It is also possible that Tyr-18 maybe capable of activating water via a charge-dipole interaction with the SAM carboxylate. From the XMT structure, there are two other residues, Tyr-24 and Asn-25, which may also be important for nucleosidase activity. The carboxyl group of the Asn-25 side chain in XMT is within 2.9 Å of the C8 atom in XR and may be involved in stabilizing a positive charge on the purine ring. Any lengthening of the C1′-N9 bond and subsequent movement of the Rib ring would allow the aromatic ring of Tyr-24 to form a stabilizing π-cation interaction with the Rib carbenium ion.
Catalytic Residues Involved in N-Methyltransferase Activity
It is very clear that DXMT can methylate both 7mX and Tb (Fig. 2B). There are no residues located within the transmethylation pocket that could act as a general acid/base for the methyl transfer reaction, as observed in SAMT (Zubieta et al., 2003). The substrates for the O-methyltransferases are expected to be fully or predominantly deprotonated at cellular pH values and should only require their correct positioning for methylation to occur (Zubieta et al., 2003). However, the conserved Tyr-18 noted earlier could form a charge-dipole interaction with the positively charged S atom of SAM, facilitating the methyl transfer reaction in this family of methyltransferases, as observed in Gly N-methyltransferases (Takata et al., 2003). The N-methyltransferases may also require some additional help for methylation because nitrogen is not as electronegative as oxygen. The His-160 side chain in DXMT forms a hydrogen bond with O2 in both 7mX and Tb substrate binding (Fig. 5, A and B). Both 7mX and Tb can exist as enolate ions (Fig. 1) and it is quite likely that the His-160 in DXMT could stabilize the enolate ion, making either N1 or N3 more electronegative for subsequent methylation. His-160 is conserved in all the MXMTs and DXMTs, but not in the XMTs or SAMT (Fig. 4), where its position would not aid in catalysis. The importance of this amino acid sequence in the substrate specificity of N-methyltransferases was previously postulated (Ogawa et al., 2001; Mizuno et al., 2003b), and here we also predict its potential catalytic importance.
CONCLUSION
The structures of XMT and DXMT presented here identify a number of key residues involved in substrate recognition and catalysis and will aid in the annotation of the many uncharacterized N-methyltransferase sequences available in GenBank. The subtle differences required for substrate recognition in the important motif B′ family of methyltransferases could even be exploited to produce new compounds for the pharmaceutical industry. Our results also suggest the plausibility of engineering a single protein capable of producing caffeine from XR. Such a protein could facilitate the production of herbivore-resistant crops that are more ecologically friendly.
MATERIALS AND METHODS
Purification, Crystallization, and Data Collection
Native XMT and DXMT were purified and crystallized as previously described (McCarthy et al., 2007). SeMet DXMT protein was produced via inhibition of the Met metabolism pathway (Doublie, 1997) and prepared using the same expression and purification protocols as the native DXMT (McCarthy et al., 2007). The SeMet DXMT crystallized in similar conditions, 23% to 28% polyethylene glycol 3350, 200 mm Li2SO4, 100 mm Tris-HCl, pH 8.5 to 8.7, containing 2 mm dithiothreitol (DTT), 2 mm SAH, and 2 mm Tb, to the native DXMT with plate-like morphology in 1 to 3 d at 20°C. The crystals were then flash frozen at 100 K after transferring them to identical crystallization conditions containing 38% polyethylene glycol 3350. These crystals were orthorhombic, space group C2221, with one molecule in the asymmetric unit and a 2.0 Å dataset was collected. A highly redundant dataset to 2.66 Å was collected from a SeMet DXMT crystal at the peak of the SeMet signal, as measured by x-ray fluorescence. XMT in complex with SAH and XR crystallized in space group P21 with four molecules per asymmetric unit and a 2.2 Å dataset was collected. All x-ray data were collected on beamline ID14-4 at the European Synchrotron Radiation Facility. All x-ray data were integrated and scaled using the XDS suite (Kabsch, 1993) and a summary of the data statistics is given in Table I.
Structure Determination and Refinement
Six SeMet sites were located on the basis of their anomalous differences using SHELXD (Uson and Sheldrick, 1999). The sites were refined and experimental phases to 2.66 Å were calculated using the single anomalous dispersion procedure in SHARP and further improved with the density modification package SOLOMON in SHARP (de La Fortelle and Bricogne, 1997). These phases were input into ARP/wARP (Perrakis et al., 1999) and resulted in a fragmented polyalanine model of 281 residues. Subsequent rounds of model building followed by phased refinement using the SHARP phases in REFMAC (Murshudov et al., 1997) allowed the majority of the model to be built. XMT was solved by molecular replacement using PHASER (McCoy et al., 2005) and the DXMT structure as a search model.
The crystal structure refinements of DXMT and XMT were performed using REFMAC (Murshudov et al., 1997), with a randomly chosen subset of 5% of reflections for the calculation of the free R factor. All the crystallographic information is summarized in Table I. A well-ordered SAH was easily modeled into both structures in the early stages of refinement. A Tb molecule in two orientations was modeled into DXMT (Supplemental Fig. S1, A and B) and a well-ordered XR molecule was easily modeled into XMT (Supplemental Fig. S1C). Ordered water molecules were added at locations where there was |FO| − |FC| density greater than 3σ above the mean and within hydrogen-bonding contact to a neighboring molecule. Model building was carried out with Coot (Emsley and Cowtan, 2004) and the stereochemical quality of the protein molecules was validated with PROCHECK (Laskowski et al., 1993). Restraints for XR were generated by using the Dundee PRODRG server (Schuttelkopf and van Aalten, 2004). Sequence alignments were done with ClustalW (Thompson et al., 1994) and ESPript (Gouet et al., 1999) and the figures were prepared with PYMOL (DeLano, 2002).
Determination of Enzymatic Activity
Reactions with purified recombinant DXMT protein were set up as follows: 30 μL of 10 mm SAM, 30 μL of 10× reaction buffer (500 mm Tris-HCl, pH 7.9, 10 mm MgCl2, 100 mm NaCl, 1 mm DTT), and 60 μL of 5 mm substrates (XR, 7mX, and Tb) were made up to a final volume of 285 μL with water. A 100-μL aliquot was taken as a control reaction and then 15 μL of purified protein (21 μg in 1× reaction buffer) were added to the remaining reaction. Both the control and enzyme reactions were incubated at 37°C. Forty-five- or 50-μL samples were taken at various times and added to 200 μL HPLC buffer A (92% water, 8% acetonitrile, 0.1% phosphoric acid, pH 2.2) to stop the reaction. Sixty micrograms of purified XMT were first diafiltered into reaction buffer (50 mm Tris-HCl, pH 7.9, 1 mm MgCl2, 10 mm NaCl, 0.1 mm DTT, and 3 mm SAM) to give a final volume of 1 mL. Then, 240-μL aliquots of XMT in reaction buffer were added to 60 μL of 5 mm solutions of XR, 7mX, or Tb. A 50-μL control sample was immediately taken from each of the reactions and added to 200 μL HPLC buffer A. The reactions were then placed at 37°C and 50-μL samples were taken at various times and added to 200 μL HPLC buffer A to stop the reactions.
The samples were then passed through a 0.2-μm filter and were analyzed by HPLC using a Waters reverse-phase C18 column (4 μm, 4.6 × 250 mm) and a gradient of 8% to 50% acetonitrile. Solvent A was 91.9% milliQ water, 8% CH3CN, and 0.1% H3PO4; solvent B was 49.9% milliQ water, 50% CH3CN, and 0.1% H3PO4. Solvents were sparged with 30% helium and the flow rate was 1 mL/min. The gradient was as follows: at 0 min, 98% A/2% B; at 5 min, 92% A/8% B; at 25 min, 50% A/50% B; at 30 min, 30% A/70% B; at 35 min, 30% A/70% B; then from 37 to 45 min, 98% A/2% B. Detection was done using a Waters photodiode array detector.
The nucleotide sequences reported in this article have been deposited in the DDBJ/GenBank/EBI Data Bank with accession numbers DQ422954 (XMT) and DQ422955 (DXMT). The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rscb.org) with the accession codes 2EG5 (XMT) and 2EFJ (DXMT).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Electron density maps for bound XR and Tb molecules.
Supplementary Material
Acknowledgments
We thank G. Cheminade for carrying out the HPLC analysis, the European Molecular Biology Laboratory/European Synchrotron Radiation Facility Joint Structural Biology Group for access to ID14-4, R. Ravelli for help with data collection and structural solution, and V. Petiard, R. Ravelli, and S. Cusack for their support during this work. Finally, we are grateful to C. Petosa and E. Mitchell for critically reading this manuscript.
This article is dedicated to the memory of Nicholas P. Chopey.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Andrew A. McCarthy (andrewmc@embl.fr).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Amyes TL, Jencks WP (1989) Lifetimes of oxocarbenium ions in aqueous solution from common ion inhibition of the solvolysis of α-azido ethers by added azide ion. J Am Chem Soc 111 7888–7900 [Google Scholar]
- Ashihara H, Suzuki T (2004) Distribution and biosynthesis of caffeine in plants. Front Biosci 9 1864–1876 [DOI] [PubMed] [Google Scholar]
- Bennett EM, Li C, Allan PW, Parker WB, Ealick SE (2003) Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase. J Biol Chem 278 47110–47118 [DOI] [PubMed] [Google Scholar]
- Cauli O, Morelli M (2005) Caffeine and the dopaminergic system. Behav Pharmacol 16 63–77 [DOI] [PubMed] [Google Scholar]
- de La Fortelle E, Bricogne G (1997) Heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. In CW Carter Jr, RM Sweet, eds, Methods of Enzymology, Vol 276. Academic Press, New York, pp 472–494 [DOI] [PubMed]
- Degano M, Almo SC, Sacchettini JC, Schramm VL (1998) Trypanosomal nucleoside hydrolase: a novel mechanism from the structure with a transition-state inhibitor. Biochemistry 37 6277–6285 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) Pymol. DeLano Scientific, Palo Alto, CA
- Doublie S (1997) Preparation of selenomethionyl proteins for phase determination. In CW Carter, RM Sweet, eds, Methods of Enzymololgy, Vol 276. Academic Press, New York, pp 523–530
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60 2126–2132 [DOI] [PubMed] [Google Scholar]
- Friedman J, Waller GR (1983) Caffeine hazards and their prevention in germinating seeds of coffee. J Chem Ecol 9 1099–1106 [DOI] [PubMed] [Google Scholar]
- Frischknecht PM (1985) Purine alkaloid formation in buds and developing leaflets of Coffea arabica: expression of an optimal defense strategy? Phytochemistry 3 613–616 [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15 305–308 [DOI] [PubMed] [Google Scholar]
- Hollingsworth RG, Armstrong JW, Campbell E (2002) Caffeine as a repellent for slugs and snails. Nature 417 915–916 [DOI] [PubMed] [Google Scholar]
- Jones S, Thornton JM (1995) Protein-protein interactions: a review of protein dimer structures. Prog Biophys Mol Biol 63 31–65 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26 795–800 [Google Scholar]
- Kato M, Mizuno K (2004) Caffeine synthase and related methyltransferases in plants. Front Biosci 9 1833–1842 [DOI] [PubMed] [Google Scholar]
- Kato M, Mizuno K, Crozier A, Fujimura T, Ashihara H (2000) Caffeine synthase gene from tea leaves. Nature 406 956–957 [DOI] [PubMed] [Google Scholar]
- Koellner G, Bzowska A, Wielgus-Kutrowska B, Luic M, Steiner T, Saenger W, Stepinski J (2002) Open and closed conformation of the E. coli purine nucleoside phosphorylase active center and implications for the catalytic mechanism. J Mol Biol 315 351–371 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26 283–291 [Google Scholar]
- Lee JE, Smith GD, Horvatin C, Huang DJ, Cornell KA, Riscoe MK, Howell PL (2005) Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis. J Mol Biol 352 559–574 [DOI] [PubMed] [Google Scholar]
- McCarthy AA, Biget L, Lin C, Petiard V, Tanksley SD, McCarthy JG (2007) Cloning, expression, crystallisation and preliminary x-ray analysis of the XMT and DXMT N-methyltransferases from Coffea canephora (robusta). Acta Crystallograph Sect F Struct Biol Cryst Commun 63 304–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr D 61 458–464 [DOI] [PubMed] [Google Scholar]
- Mizuno K, Kato M, Irino F, Yoneyama N, Fujimura T, Ashihara H (2003. a) The first committed step reaction of caffeine biosynthesis: 7-methylxanthosine synthase is closely homologous to caffeine synthases in coffee (Coffea arabica L.). FEBS Lett 547 56–60 [DOI] [PubMed] [Google Scholar]
- Mizuno K, Okuda A, Kato M, Yoneyama N, Tanaka H, Ashihara H, Fujimura T (2003. b) Isolation of a new dual-functional caffeine synthase gene encoding an enzyme for the conversion of 7-methylxanthine to caffeine from coffee (Coffea arabica L.). FEBS Lett 534 75–81 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53 240–255 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Herai Y, Koizumi N, Kusano T, Sano H (2001) 7-Methylxanthine methyltransferase of coffee plants: gene isolation and enzymatic properties. J Biol Chem 276 8213–8218 [DOI] [PubMed] [Google Scholar]
- Ogita S, Uefuji H, Morelli M, Sano H (2005) Metabolic engineering of caffeine production. Plant Biotechnol 22 461–468 [Google Scholar]
- Perrakis A, Morris R, Lamzin VS (1999) Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6 458–463 [DOI] [PubMed] [Google Scholar]
- Schuttelkopf AW, van Aalten DM (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D 60 1355–1363 [DOI] [PubMed] [Google Scholar]
- Takata Y, Huang Y, Komoto J, Yamada T, Konishi K, Ogawa H, Gomi T, Fujioka M, Takusagawa F (2003) Catalytic mechanism of glycine N-methyltransferase. Biochemistry 42 8394–8402 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uefuji H, Shinjiro O, Yamaguchi Y, Koizumi N, Sano H (2003) Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the caffeine biosynthetic pathway in coffee plants. Plant Physiol 132 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uefuji H, Tatsumi Y, Morimoto M, Kaothien-Nakayama P, Ogita S, Sano H (2005) Caffeine production in tobacco plants by simultaneous expression of three coffee N-methyltransferases and its potential as a pest repellant. Plant Mol Biol 59 221–227 [DOI] [PubMed] [Google Scholar]
- Uson I, Sheldrick GM (1999) Advances in direct methods for protein crystallography. Curr Opin Struct Biol 9 643–648 [DOI] [PubMed] [Google Scholar]
- Yoneyama N, Morimoto H, Ye CX, Ashihara H, Mizuno K, Kato M (2006) Substrate specificity of N-methyltransferase involved in purine alkaloids synthesis is dependent upon one amino acid residue of the enzyme. Mol Genet Genomics 275 125–135 [DOI] [PubMed] [Google Scholar]
- Zubieta C, Ross JR, Koscheski P, Yang Y, Pichersky E, Noel JP (2003) Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 15 1704–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.