Seeds are essential for flowering plant reproduction because they protect, nourish, and contain the developing embryo that represents the next sporophytic generation. In addition, seeds contain energy resources that sustain the young sporophyte during germination before photosynthesis begins. In legumes, food reserves stored in embryonic cotyledons make seeds important as a food source for both human and animal consumption. For example, soybean (Glycine max) is now one of the most important seed crops in the world (Wilcox, 2004). Research on legume seed development has led to direct applications, such as seeds with more nutrients (Kinney, 1998; Wang et al., 2003; Krishnan, 2005), reduced allergens (Herman et al., 2003), and novel constituents, such as edible vaccines (Moravec et al., 2007). In the current genomic era, it is now possible to begin to understand what genes are required to make a legume seed and how regulatory networks are interconnected in legume genomes to program seed formation. In the future, this information should permit novel approaches to breed and engineer legume seeds with new agronomic traits and, most importantly, help provide a sustainable food supply for a growing human population. This Update outlines how our laboratories have been using legumes and functional genomics to identify genes that program legume seed development.
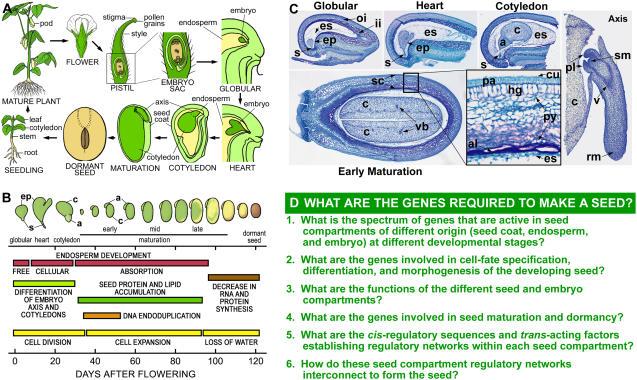
Seed development is triggered by a novel double-fertilization process that leads to the differentiation of the embryo, endosperm, and seed coat, which are the major compartments of the seed (Fig. 1, A–C; Goldberg et al., 1994; Miller et al., 1999; Gehring et al., 2004; Laux et al., 2004; Moise et al., 2005). These compartments have different origins and play distinct roles in seed formation. The maternally derived seed coat differentiates from the ovule integuments that surround the embryo sac and plays a major role in protecting the embryo and transferring nutrients from the maternal plant to the developing embryo (Fig. 1, A and C; Murray, 1987; Borisjuk et al., 2004; Moise et al., 2005). By contrast, the embryo and endosperm are direct descendents of the fertilized egg and central cell, respectively. The endosperm proliferates to occupy most of the postfertilization embryo sac and nourishes the embryo early in development (Gehring et al., 2004). In many flowering plants, such as legumes, the endosperm is absorbed by the embryo during development and is not present in the mature seed (Fig. 1, A–C; Goldberg et al., 1994). After fertilization, the zygote divides asymmetrically, giving rise to a small apical cell that develops into the embryo proper and a large basal cell that forms the suspensor. The suspensor is a terminally differentiated structure that supports and nourishes the embryo proper and degenerates later in development (Yeung and Meinke, 1993). The embryo proper, on the other hand, represents the new sporophytic generation and contains the shoot and root meristems that are responsible for generating organ systems of the mature plant after seed germination (Fig. 1C; Goldberg et al., 1994; Laux et al., 2004).
Figure 1.
Soybean seed development. A, Cartoon depicting soybean life cycle. B, Schematic representation of soybean seed development. Embryo morphologies and developmental events were adapted and modified from Goldberg et al. (1989). C, Paraffin transverse 10-μm sections of soybean globular, heart, cotyledon, and early maturation seeds. Inset contains a magnified view (40×) of the seed coat. Axis longitudinal section was obtained from an early maturation seed. D, Major unanswered questions in seed development. a, Axis; al, aleurone; c, cotyledon; cu, cuticle; ep, embryo proper; es, endosperm; hg, hourglass cells; ii, inner integument; oi, outer integument; pa, palisade layer; pl, plumule; py, parenchyma; rm, root meristem; s, suspensor; sc, seed coat; sm, shoot meristem; v, vascular tissues; vb, vascular bundle.
MAJOR QUESTIONS REMAIN UNANSWERED IN SEED DEVELOPMENT
Many developmental and physiological events occur within each seed compartment during development (Fig. 1B) and are programmed, in part, by the activity of different genes (Goldberg et al., 1989, 1994; Stangeland et al., 2003; Gehring et al., 2004; Haughn and Chaudhury, 2005). Seed development, therefore, is the result of a mosaic of distinct gene expression programs occurring in parallel in different seed compartments (e.g. embryo, endosperm, seed coat) as well as within specific regions and tissues (e.g. embryo proper, suspensor, epidermis). What these programs are and how they are integrated into unique regulatory networks within the plant genome remain major unanswered questions (Fig. 1D). Specifically, it is not yet known what genes in different seed compartments play important roles in cell fate specification, differentiation, and morphogenesis during early seed and embryo development. Molecular identification and characterization of these genes will help identify regulatory networks that program and coordinate the development of each seed compartment. In addition, it is not known what the functions are of many genes that are expressed in different seed compartments. Identifying the function of compartment-specific genes should provide new insight into their roles in seed development. At present, new genomic resources allow seed biologists to use global gene expression profiling and comparative genomics to answer many questions that only a short time ago seemed out of reach. These questions, and others (Fig. 1D), are challenging the field of seed biology, and their answers should provide new insights into the process of seed development and lead to improved seeds for human and animal consumption.
LEGUMES ARE AN EXCELLENT MODEL SYSTEM TO STUDY SEED DEVELOPMENT
Legumes represent one of the largest and most diverse families of flowering plants, with approximately 20,000 species classified (Doyle and Luckow, 2003). There are three subfamilies in legumes and the largest, Papilionoideae, contains most of the model species in which different aspects of plant biology have been studied. The most common legume models are peanut (Arachis hypogaea), Lotus (Lotus japonicus), Medicago (Medicago truncatula), soybean (Glycine max), scarlet runner bean (SRB; Phaseolus coccineus), common bean (Phaseolus vulgaris), pea (Pisum sativum), and broad bean (Vicia faba). The latter five species have been used historically to study seed and embryo development (Goldberg et al., 1989; Johnson et al., 1994; Coste et al., 2001; Weterings et al., 2001; Weber et al., 2005).
Several features make legumes an excellent model system to study seed and embryo development. For example, many legumes, such as soybean and peanut, are food crops of major economic importance. The mature seeds of these legumes are rich in proteins, carbohydrates, and oils, and accumulate to high nutritional value. These stored seed food reserves make legumes, such as soybean, the second most important crop for human nutrition and animal feed (Rubel et al., 1972; Duranti and Gius, 1997; Graham and Vance, 2003). One advantage of using crop models to study seed biology is to be able to modify traits of agronomic importance, such as improved seed nutritional composition, reduced allergen levels, or increased seed number and size (Kinney, 1998; Herman et al., 2003; Wang et al., 2003; Gupta et al., 2006). In addition, legume seed biology has been studied for more than 150 years using descriptive, physiological, biochemical, molecular, and genetic approaches (see below). These studies have provided a solid intellectual framework for using legume models to study and dissect seed development in our current genomic era. The recent development of genomic tools, such as genome sequences, ESTs, oligonucleotide and cDNA microarrays, and comprehensive databases, such as the Legume Information System (http://www.comparative-legumes.org), make legumes an excellent model to study seed development at a global scale (VandenBosch and Stacey, 2003; Gepts et al., 2005; Gonzales et al., 2005). These genomic tools allow comparative genomic analyses in closely related species (Zhu et al., 2005) and should facilitate the identification and investigation of genes important for seed development.
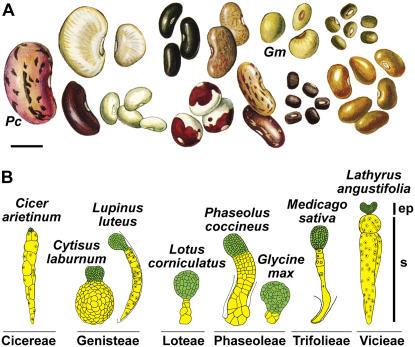
One of the most fascinating characteristics of legumes is that collectively they produce a large range of seed sizes (Fig. 2A). For example, some legume seeds are giants and are excellent models for developmental studies, particularly during early stages of seed development. The large size of SRB globular-stage seed and embryo allows manipulation and isolation of embryonic regions, such as the embryo proper and suspensor, using hand-dissection techniques (Walbot et al., 1972; Sussex et al., 1973; Weterings et al., 2001). Due to their size, large quantities of cells and tissues from these SRB embryonic regions can be obtained, facilitating molecular and biochemical studies. Manipulation of seeds and embryos at early stages of development is difficult in other model plant species with smaller seeds, such as Arabidopsis (Arabidopsis thaliana), making many legumes particularly useful to study early developmental seed biology.
Figure 2.
Diversity of legume seed size and embryo morphology. A, Legume seed shape and size variation. Scale bar = 1 cm. Adapted and modified from Vaughan et al. (1997). B, Variation in embryo-proper (ep) and suspensor (s) morphologies within and between legume tribes. Embryos are not drawn to scale. Images were adapted and modified from Martin (1914), Lersten (1983), and Chamberlin et al. (1994). Pc, P. coccineus; Gm, G. max.
A second novel feature of legumes is that their embryos show a wide range of morphological forms (Fig. 2B). For example, two closely related species, soybean and SRB, have morphologically distinct suspensors. The soybean suspensor is small, consisting of a few cells, whereas the SRB suspensor is much larger and contains several hundred cells (Fig. 2B). The variety in size and shape of legume seeds and embryos makes them excellent models for comparative morphological studies using a functional genomics approach. This strategy can lead to a better understanding of the function, evolution, and diversity of legume seeds and their corresponding compartments.
LEGUMES HAVE BEEN USED TO STUDY SEED DEVELOPMENT FOR MORE THAN 150 YEARS
Historically, legumes have been used to address important questions of seed and embryo development. In fact, early work with legumes contributed to the development of major ideas in biology. For example, during the early 1800s, Matthias Schleiden used several legumes, including Medicago and Vicia, to investigate the endosperm and describe the process of seed development (Schleiden and Vogel, 1838, 1842). These studies contributed to his role in establishing the cell theory. In the mid-1800s, Gregor Mendel used peas to study the inheritance of phenotypic variation, including seed color and shape, leading to his Laws of Inheritance and the establishment of modern-day genetics (Mendel, 1865).
From the late 1800s to the middle of the 1900s, legumes were used to describe the processes of seed and embryo development, including the cellular events that occur before and after fertilization, early embryo cell cleavages, and endosperm differentiation. For example, Guignard's compendium of more than 40 legume species described the rich diversity of legume embryo and suspensor morphologies (Fig. 2B; Guignard, 1882). These studies, and others, contributed to our overall understanding of seed and embryo development at the descriptive level (Martin, 1914; Brown, 1917; Cooper, 1938).
Studies on legume seed formation transitioned from descriptive anatomy to experiments at the molecular, biochemical, and physiological levels during the 1970s (Dure, 1975), although ultrastructural and histochemical studies of legume seed development continued (Johansson and Walles, 1993; Nishizawa et al., 1994; Duval et al., 1995). Work in many legumes provided some of the earliest measurements of RNA, DNA, carbohydrates, lipids, and protein levels in seeds (Rubel et al., 1972; Clutter et al., 1974; Hill and Breidenbach, 1974; Davies, 1976; Pattee et al., 1981; Singh et al., 1981; Adams et al., 1982; Dhillon and Miksche, 1983). These studies provided new insights into the processes by which food reserves accumulate and are stored in seeds, as well as demonstrating that genome endoreduplication processes occur in specific seed compartments (e.g. cotyledon, suspensor). In addition, legumes such as SRB were used to dissect and manipulate the embryo proper and suspensor experimentally to study the role of growth hormones (e.g. GA) in early embryo development (Sussex et al., 1973; Cionini et al., 1976; Alpi et al., 1979).
During this same period, our laboratory used RNA-excess DNA-RNA hybridization experiments to show that approximately 14,000 to 18,000 diverse mRNAs are present in soybean embryos at different developmental stages (Goldberg et al., 1981b, 1989). We also demonstrated that most diverse mRNA species are present throughout seed development, but that small numbers of mRNAs, including those encoding storage proteins, are regulated quantitatively at specific developmental stages (Goldberg et al., 1981a). Seed protein genes were chosen as models to investigate gene regulation during legume seed development because they encode superprevalent mRNAs that could be easily identified and isolated and because of their importance as a food source for human and animal consumption.
Research on genes active in legume seed development exploded during the late 1970s and 1980s when it became possible to clone and study individual mRNAs and genes and reinsert them into plants using newly developed transformation techniques (Bevan et al., 1983; Estrella-Herrera et al., 1983; Fraley et al., 1983). In addition, Murai et al. (1983) demonstrated that the common bean phaseolin seed storage protein gene could be transferred to sunflower (Helianthus annuus) cells and expressed. This sunbean experiment showed that gene cloning and Agrobacterium tumefaciens transformation techniques could be combined to transfer foreign genes into plant cells and study their function. Research in several laboratories with legume seed protein genes, such as β-conglycinin, glycinin, Kunitz trypsin inhibitor, and lectin, showed that their mRNA accumulation patterns are regulated temporally and spatially (Goldberg et al., 1981a, 1983; Meinke et al., 1981; Rerie et al., 1992) and controlled by both transcriptional and posttranscriptional processes (Evans et al., 1984; Beach et al., 1985; Chappell and Chrispeels, 1986; Walling et al., 1986). Subsequent work determined that cis-regulatory sequences flanking legume seed protein genes could confer embryo-specific expression patterns in heterologous plants, such as tobacco (Nicotiana tabacum) and petunia (Petunia hybrida; Chen et al., 1986; Okamuro et al., 1986; Jofuku et al., 1987; Higgins et al., 1988; Naito et al., 1988; Baumlein et al., 1992; Wohlfarth et al., 1998; Chandrasekharan et al., 2003). This work provided insights into the mechanisms controlling gene activity during seed development and showed that the cis-regulatory sequences and transcription factors controlling legume seed protein gene expression are highly conserved in other plant species.
At present, the remarkable development of new genomic resources makes it possible to study legume gene expression during seed and embryo development at a global level. Currently, Medicago, Lotus, and soybean cDNA and oligonucleotide microarrays are available (Endo et al., 2002; Vodkin et al., 2004; Firnhaber et al., 2005). An increasing collection of legume seed transcriptome and proteomic data (Thibaud-Nissen et al., 2003; Firnhaber et al., 2005; Hajduch et al., 2005; Buitink et al., 2006; Dhaubhadel et al., 2007), cDNA library collections, and public EST databases (Journet et al., 2002; Shoemaker et al., 2002; Asamizu et al., 2004; Firnhaber et al., 2005; Ramirez et al., 2005) have helped to provide a global view of gene activity at specific seed developmental stages. Sequences of the Medicago, Lotus, soybean, common bean, and peanut genomes (Broughton et al., 2003; Gepts et al., 2005; Young et al., 2005; Jackson et al., 2006) should provide an invaluable resource for identifying and characterizing genes that play critical roles during seed and embryo development in the near future.
A NOVEL STRATEGY TO DISSECT REGULATORY NETWORKS PROGRAMMING EARLY SEED AND EMBRYO DEVELOPMENT
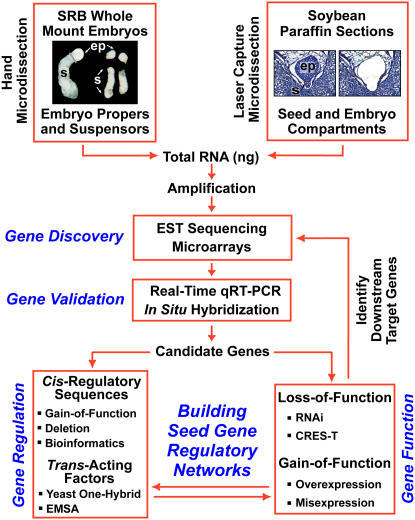
Our laboratory has developed a genomics strategy to begin to identify the regulatory networks that program legume seed and embryo development (Fig. 3). One aspect of this strategy is to use the giant SRB embryo, pioneered by Sussex and colleagues (Walbot et al., 1972; Sussex et al., 1973), as an entry point to dissect the molecular events that occur during early embryogenesis (Fig. 4). More recently, we incorporated soybean into our strategy (Fig. 5) because it contrasts with SRB in terms of early embryo morphology (Fig. 2B) and because our laboratory has worked on soybean embryo development for more than 25 years (Goldberg et al., 1989). Recent development of soybean and SRB genomic resources allows both of these legumes to be used as excellent systems to identify genes that play important roles in the differentiation of unique seed and embryo compartments and to build compartment-specific regulatory networks that are crucial for seed formation (Figs. 4–6).
Figure 3.
Genomics strategy for identifying legume seed gene regulatory networks.
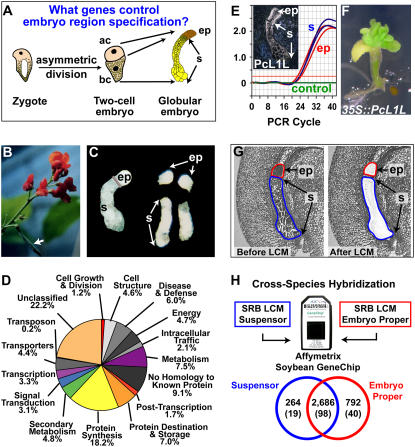
Figure 4.
Using SRB as a genomics engine to uncover genes active early in embryogenesis. A, Model for the specification of the embryo proper and suspensor, adapted and modified from Weterings et al. (2001). ac, Apical cell; bc, basal cell; ep, embryo proper; s, suspensor. B, SRB plant with a pod indicated by the arrow. C, Hand-dissected SRB globular-stage embryos before and after separating the embryo proper and suspensor. D, Functional category distribution of SRB suspensor ESTs. E, Real-time qRT-PCR validation of PcL1L mRNA accumulation pattern. Inset, In situ hybridization of SRB globular-stage embryo mRNA using a PcL1L antisense probe. In situ data were taken from Kwong et al. (2003). F, PcL1L overexpression in transgenic Arabidopsis seedlings. G, SRB globular-stage seed paraffin sections before and after capturing the embryo proper and suspensor by LCM. H, Cross-species hybridization of SRB embryo-proper and suspensor mRNA captured in G with an Affymetrix Soybean GeneChip containing 37,593 soybean probe sets. Only SRB sequences with a high similarity to soybean EST sequences on the GeneChip will hybridize. Therefore, the proportion of heterologous SRB suspensor and embryo-proper transcripts detected on the GeneChip is lower than that detected for homologous soybean RNAs (Fig. 5C). Venn diagram of transcripts detected in the suspensor and embryo proper is shown. Numbers in parentheses refer to the number of transcription factor gene transcripts detected. Data are available at http://estdb.biology.ucla.edu/seed.
Figure 5.
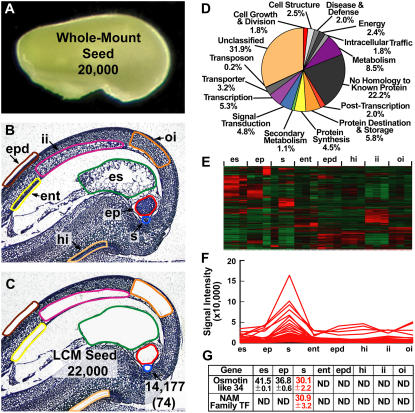
Using LCM and transcriptional profiling to identify genes required to make a soybean seed. A, Globular-stage soybean seed showing the approximate number of transcripts detected in the entire seed using the Affymetrix Soybean GeneChip (Whole-Mount Seed). B and C, Globular-stage soybean seed paraffin sections before (B) and after (C) capturing the highlighted seed compartments by LCM. The approximate total number of diverse transcripts detected collectively from LCM seed compartments is shown (LCM Seed) in addition to the total number of transcripts detected in the suspensor (arrow). The number in parentheses refers to suspensor transcripts not detected in other seed compartments at the level of the GeneChip (i.e. suspensor-specific transcripts). Raw data were deposited in the Gene Expression Omnibus (GEO) as data series GSE6414 (http://www.ncbi.nlm.nih.gov/geo) and can also be accessed at http://estdb.biology.ucla.edu/seed. D, Functional category distribution of soybean suspensor transcripts detected by GeneChip analysis. E, Unsupervised hierarchical clustering of the top 2,000 most varying transcripts detected in all globular-stage seed compartments using dChip version 1.3 (Li and Wong, 2001). F, Supervised cluster analysis of suspensor developmentally regulated transcripts. G, Real-time qRT-PCR validation of suspensor-specific transcripts. Values represent the mean and sd of the threshold cycle (Ct) for two biological replicates with two technical replicates each. Ct values were adjusted to an 18S rRNA internal control. One Ct cycle represents a 2-fold difference in RNA prevalence. Lower Ct values indicate higher RNA levels. ent, Endothelium; ep, embryo proper; epd, epidermis; es, endosperm; hi, hilum; ii, inner integument; oi, outer integument; s, suspensor.
Figure 6.
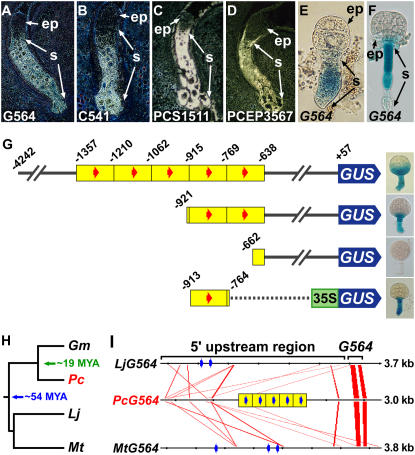
Identifying DNA sequences important for suspensor transcription. A to D, In situ hybridization of SRB globular-stage embryos using antisense probes from G564 (A), C541 (B), PCS1511 (C), and PCEP3567 (D) cDNAs. G564 and C541 in situ hybridization data were taken from Weterings et al. (2001). E and F, GUS enzymatic activity in transgenic tobacco (E) and Arabidopsis (F) embryos carrying a G564∷GUS chimeric gene. G, G564 5′-deletion and gain-of-function analyses in transgenic tobacco plants. The number indicates position relative to the transcription start site (+1). Yellow blocks indicate approximately 150-bp tandem duplications. Red arrows indicate a 10-bp motif (GAAAAGC/TGAA) identified to be conserved in the upstream sequences of G564 and C541 (Weterings et al., 2001). Deletion data were taken from Weterings et al. (2001). The −913 to −764 gain-of-function construct was made by fusing this G564 upstream region with a cauliflower mosaic virus 35S∷GUS chimeric gene and transforming tobacco plants according to Koltunow et al. (1990). H, Legume phylogenetic tree, including SRB, soybean, Lotus, and Medicago. Soybean and Lotus/Medicago diverged from SRB approximately 19 and 54 million years ago, respectively (Lavin et al., 2005). I, Conserved regions among legume G564 genes. Red lines indicate closely related sequences displayed by FamilyRelationsII (20-bp window size, 75% similarity or greater; Brown et al., 2005). Blue arrows indicate motifs identified by MEME (20-bp window size; Bailey and Elkan, 1994) to be significantly enriched in the G564 upstream regions. ep, Embryo proper; Gm, G. max; Lj, L. japonicus; Mt, M. truncatula; MYA, million years ago; Pc, P. coccineus; s, suspensor.
USING SRB AS A GENOMICS ENGINE TO DISSECT EARLY EMBRYOGENESIS
Our laboratory has utilized SRB as a model system (Weterings et al., 2001) to identify genes and regulatory networks programming early embryo developmental events using a genomics approach (Figs. 3 and 4). We have focused on the question of how the embryo-proper and suspensor regions are specified from the apical and basal cells of a two-cell embryo, respectively (Fig. 4A; Weterings et al., 2001). SRB is unique in this regard because of its giant seed and large embryo (Fig. 2A), permitting hand dissection of embryo-proper and suspensor regions at early developmental stages (Fig. 4, B and C; Walbot et al., 1972; Sussex et al., 1973; Weterings et al., 2001). We prepared cDNA libraries from hand-dissected embryo-proper and suspensor regions of SRB globular-stage embryos (Fig. 3; Weterings et al., 2001) and ESTs were sequenced to determine what genes are active in the embryo proper and suspensor. Our SRB early embryo EST database is available at both http://estdb.biology.ucla.edu/PcEST and GenBank (accession nos. CA896559–CA916678). Figure 4D shows the distribution of >16,000 SRB suspensor ESTs grouped by functional categories, illustrating the large diversity of genes that are active in a terminally differentiated suspensor. Surprisingly, more than 300 suspensor transcription factor ESTs were identified that are distributed into a variety of transcription factor gene families (A.Q. Bui, B.H. Le, and R.B. Goldberg, unpublished data), providing a glimpse into the spectrum of regulatory genes active in one region of a legume embryo shortly after fertilization. What roles these transcription factors play in suspensor differentiation and function remain to be determined.
As part of our strategy, we carried out real-time quantitative reverse transcription (qRT)-PCR and in situ hybridization experiments (Fig. 3) to quantify and localize the accumulation of SRB mRNAs during early embryo development. One advantage of using giant SRB embryos is that qRT-PCR can be used to quantitate mRNA levels in different regions of a single embryo. For example, PcL1L mRNA uncovered in our EST studies is localized throughout the globular-stage SRB embryo (Fig. 4E, inset; Kwong et al., 2003). PcL1L is a relative of the Arabidopsis LEAFY COTYLEDON1 (LEC1) CAAT-box-binding transcription factor gene that is a critical regulator of embryo development (Lotan et al., 1998). Real-time qRT-PCR studies on regions dissected from a single SRB embryo at the globular stage show similar PcL1L mRNA prevalences in the embryo-proper and suspensor regions (Fig. 4E). Remarkably, Arabidopsis plants transformed with a full-length PcL1L cDNA under the control of the cauliflower mosaic virus 35S gene promoter (R.W. Kwong, J. Pelletier, and J.J. Harada, unpublished data) formed ectopic embryo-like structures on seedlings (Fig. 4F), a phenotype similar to that obtained with a 35S∷AtLEC1 chimeric gene (Lotan et al., 1998). These results suggest that PcL1L is an important regulator of SRB embryo development, illustrating the power of the SRB system as a gene discovery engine to uncover regulatory genes that play essential roles in seed development.
USING SOYBEAN TO IDENTIFY GENES REQUIRED TO MAKE A SEED
Even though the giant SRB embryo is a novel system for studying the specification of embryo regions early in development, this legume does have some limitations for global studies of seed formation. For example, although we have produced an EST dataset for SRB globular-stage embryo regions (http://estdb.biology.ucla.edu/PcEST), few other genomic resources are available. In addition, transformation procedures have not yet been developed for SRB. Finally, because SRB is not a major food crop, it is unlikely that a genome project will be carried out to sequence the SRB genome. To complement our use of SRB as a gene discovery engine and overcome these deficiencies, we decided to go back to the future (Goldberg et al., 1989) and use soybean to dissect genes important for making a seed. At the present time, a large number of genomic resources have been developed for soybean, including microarrays, EST databases, and genome sequences (Shoemaker et al., 2002; Vodkin et al., 2004; Jackson et al., 2006). In addition, transformation procedures are well established for soybean, making it feasible to address questions of gene function (Ko et al., 2006; Olhoft et al., 2006). Finally, the unique morphological differences between soybean and SRB embryos (e.g. suspensor size and shape) should permit structure-function questions of legume embryo diversity to be studied (Fig. 2B).
Although we have been able to take advantage of the giant SRB embryo to dissect by hand embryo-proper and suspensor regions (Figs. 3 and 4C), this approach is time consuming and not practical with legumes that have smaller embryos, such as soybean (Fig. 2B). One way to overcome the limitations of hand dissection and to be able to isolate different regions from any legume seed and embryo regardless of size (Fig. 2) is to make use of spectacular progress in laser capture microdissection (LCM) technology (Day et al., 2005; Nelson et al., 2006). LCM technology makes it possible to study gene activity in the entire seed because any seed compartment, region, or tissue can be isolated easily throughout development (Fig. 5). LCM technology has been used successfully in rice (Oryza sativa), maize (Zea mays), tobacco, soybean, and Arabidopsis in which a variety of plant tissues and cell types have been isolated and studied, including those in an early Arabidopsis embryo (Asano et al., 2002; Kerk et al., 2003; Nakazono et al., 2003; Casson et al., 2005; Klink et al., 2005; Sanders et al., 2005; Spencer et al., 2007).
We have been using LCM with soybean seeds to identify all the genes required to make a seed (Figs. 3 and 5). In combination with GeneChip technology, we can investigate the global gene activity profiles in different compartments of the entire seed. For example, we used LCM to isolate the endosperm, suspensor, embryo proper, endothelium, inner integument, outer integument, epidermis, and hilum from a globular-stage soybean seed (Fig. 5, B and C). We hybridized RNAs isolated from each of these seed regions, as well as from intact globular-stage seeds (Fig. 5, A–C), with soybean Affymetrix GeneChips (J.A. Wagmaister, X. Wang, A.Q. Bui, B.H. Le, and R.B. Goldberg, unpublished data). We then compared the spectrum of diverse transcripts present in the entire globular-stage soybean seed (Fig. 5A) to those obtained with the eight laser-captured seed regions (Fig. 5, B and C). These data are available at http://estdb.biology.ucla.edu/seed as part of our National Science Foundation (NSF) Plant Genome Research Project.
Approximately 20,000 diverse transcripts were found to be present in the whole-mount globular-stage soybean seed (Fig. 5A), a value close to that which we obtained more than a quarter of a century ago using Rot curve hybridization technology (Goldberg et al., 1981b, 1989). A smaller number of diverse mRNAs were found to be present in individual soybean globular-stage seed regions. For example, approximately 14,000 diverse transcripts were detected in the suspensor, including those that encode about 700 transcription factors (Fig. 5C). These values are comparable to those obtained with each of the other globular-stage seed regions (i.e. 14,000–17,000 diverse transcripts and 600–800 transcription factor mRNAs; J.A. Wagmaister, X. Wang, A.Q. Bui, and R.B. Goldberg, unpublished data). Soybean suspensor transcripts are distributed into functional categories (Fig. 5D) similar to what we observed from the analysis of giant SRB suspensor ESTs (Fig. 4D). These findings indicate that there is a large diversity of biological functions in the small soybean suspensors (Fig. 5D) and that there does not appear to be any apparent differences in the functional groupings of soybean and SRB suspensor mRNAs, despite large differences in size and morphology (Fig. 2B).
We estimated independently that there are approximately 22,000 diverse transcripts present in a soybean globular-stage seed (Fig. 5, B and C) by taking the union of each individual seed mRNA set captured by LCM (Fig. 5C) in close agreement with the result obtained with intact globular-stage seeds (Fig. 5A). These data indicate that (1) at least 20,000 to 22,000 diverse mRNAs are required to make a globular-stage soybean seed; (2) the majority of the diverse mRNAs present in each globular-stage seed region are shared with other regions; and (3) there are small sets of seed region-specific mRNAs.
We used hierarchical clustering to determine whether sets of mRNAs that are shared between different soybean globular-stage seed regions are coregulated at a quantitative level (Fig. 5, E and F). Our analyses identified groups of shared coregulated mRNAs that accumulate at a higher level in a particular seed region (Fig. 5E). For example, approximately 65 suspensor transcripts accumulate at a 2-fold or higher level in the suspensor compared with other seed regions (Fig. 5F).
Finally, comparison of diverse mRNAs present in each region of a soybean globular-stage seed identified small sets of region-specific transcripts. For example, 74 mRNAs were detected in the suspensor that are undetectable in other seed regions at the level of the GeneChip (Fig. 5C; J.A. Wagmaister, X. Wang, A.Q. Bui, B.H. Le, and R.B. Goldberg, unpublished data). Real-time qRT-PCR experiments validated the GeneChip specificity of two of these suspensor transcripts—an osmotin-like 34 mRNA and a mRNA encoding a member of the NAM transcription factor family (Fig. 5G; C. Cheng, J.A. Wagmaister, A.Q. Bui, and R.B. Goldberg, unpublished data). Taken together, this example illustrates that it is possible to combine LCM and GeneChip technologies to profile the spectrum of mRNAs that are present in any legume seed compartment and region throughout development (Fig. 3). The challenge will be to identify which mRNAs play a critical role in the differentiation of each seed region and how their corresponding genes are organized into regulatory networks in the soybean genome (Fig. 3).
USING COMPARATIVE GENOMICS TO IDENTIFY SEED mRNAS IN DIVERSE LEGUME SPECIES
Legumes exhibit a wide range of diversity in seed size and embryo morphology (Fig. 2), providing an outstanding opportunity to use LCM and functional genomics to compare the mRNA sets present in the different legume seeds. For example, it should be possible to isolate and compare the RNA sets present in legume suspensors that vary greatly in size and form (e.g. SRB, Medicago, Lotus, and soybean; Fig. 2B) and address the question of what role, if any, variation in suspensor size plays in legume embryo development. Although it is unlikely that GeneChips will be constructed containing diverse mRNAs for each legume species, it is possible to take advantage of the close relationship between legumes at the DNA and RNA levels to use cross-species hybridization approaches to compare gene activity within the seed regions of any legume. Cross-species hybridization approaches have been applied successfully in plants, animals, and fungi where species have diverged from a common ancestor more than 75 million years ago, such as pig and human (Chismar et al., 2002; Moody et al., 2002; Adjaye et al., 2004; Becher et al., 2004; Ji et al., 2004; Wang et al., 2004; Chalmers et al., 2005; Nowrousian et al., 2005). This evolutionary distance is greater than that separating SRB, Medicago, Lotus, and soybean, which have been shown to diverge from a common ancestor approximately 54 million years ago (Fig. 6; Lavin et al., 2005).
To test this approach, we carried out cross-species hybridization using laser-captured SRB suspensors and embryo-proper RNAs hybridized with soybean GeneChips (Fig. 4, G and H). Our results indicated that most diverse SRB embryo mRNAs that are conserved enough to be detected by the soybean GeneChip are shared by SRB embryo-proper and suspensor regions (Fig. 4H). By contrast, small sets of mRNAs, including those encoding transcription factors, are specific to each region of the globular-stage SRB embryo at the level of the GeneChip (Fig. 4H). The results from these LCM cross-species hybridization studies complement those generated by sequencing hand-dissected SRB embryo-proper and suspensor ESTs (Figs. 3 and 4D). In addition, they identified 1,000 new embryo proper- and suspensor-specific mRNAs, including those encoding approximately 60 transcription factors that might play key roles in embryo region specification during early embryogenesis (Fig. 4A). Our data indicate that cross-species hybridization using mRNAs from diverse legumes can be successful in identifying genes active during seed development at a global level. Coupling LCM with cross-species hybridization using available legume GeneChips and microarrays (e.g. soybean) should provide an entry point for identifying genes that play important roles in the development of model legume seeds, such as soybean, Medicago, and Lotus, as well as in those of nonmodel legumes where few genomic resources are available (Fig. 2B).
IDENTIFYING REGULATORY NETWORKS REQUIRED TO PROGRAM A LEGUME SEED
Using the genomics strategy that we developed to study the early stages of legume seed and embryo development (Fig. 3), we identified genes, including those encoding transcription factors, that are active specifically in the embryo proper and suspensor of SRB and soybean globular-stage embryos (Figs. 4, D and H, and 5, D and G). We also identified genes that are active specifically in other compartments of the seed (e.g. endosperm, integuments, hilum; Fig. 5C). What DNA sequences and transcription factors regulate compartment-specific genes within a seed and how compartment-specific genes are organized into regulatory networks within a plant genome are important questions of seed biology (Fig. 1D).
As a first step to uncover regulatory networks that operate within legume seeds, we used in situ hybridization to identify mRNAs from our EST database that accumulate specifically in the SRB suspensor (Figs. 3 and 4). For example, G564, C541, PCS1511, and PCEP3567 mRNAs accumulate at a high level in the suspensor of SRB globular-stage embryos (Fig. 6, A–D; Weterings et al., 2001; A.Q. Bui, Y. Bi, and R.B. Goldberg, unpublished data). G564 and C541 mRNAs encode proteins with unknown functions (Weterings et al., 2001). By contrast, PCS1511 and PCEP3567 mRNAs encode GA 3β-hydroxylase and a homeodomain transcription factor related to WOX9 (Haecker et al., 2004), respectively. Other mRNAs, such as those encoding additional enzymes in the GA biosynthetic pathway (e.g. ent-kaurene synthase, ent-kaurene oxidase, GA 20-oxidase), show a similar accumulation pattern in SRB globular-stage embryos (A.Q. Bui, Y. Bi, and R.B. Goldberg, unpublished data), suggesting that their corresponding genes might be organized into a suspensor regulatory network. To begin dissecting suspensor regulatory networks, our laboratory analyzed in detail the G564 gene 5′-upstream region (Fig. 3; Weterings et al., 2001).
We showed that approximately 4.2 kb of the G564 upstream region activates transcription in the suspensor of transgenic tobacco globular-stage embryos, demonstrating that suspensor-specific expression is controlled primarily at the transcriptional level (Fig. 6E; Weterings et al., 2001). In addition, the G564 upstream region also activates suspensor transcription in transgenic Arabidopsis embryos (Fig. 6F; X. Wang, T. Kawashima, and R.B. Goldberg, unpublished data), suggesting that the machinery regulating suspensor-specific transcription is conserved in flowering plants. G564 5′-deletion and gain-of-function analyses identified regions important for suspensor transcription (Fig. 6G; Weterings et al., 2001). The G564 upstream region possesses five approximately 150-bp tandem duplications (Fig. 6G). Each duplication is capable of activating suspensor transcription, indicating that cis-regulatory sequences within each duplicated fragment are sufficient to direct transcription in the suspensor (Weterings et al., 2001; T. Kawashima, Y. Bi, and R.B. Goldberg, unpublished data). Computational analysis uncovered a conserved 10-bp sequence (GAAAAGC/TGAA) in the upstream regions of the SRB G564 and C541 genes (Fig. 6G, red arrows; Weterings et al., 2001), suggesting that this motif might play an important role in regulating suspensor-specific transcription during early embryogenesis. If so, the 10-bp motif might be conserved in the upstream regions of other SRB suspensor-specific genes and their orthologs in closely related legumes.
The spectacular increase in legume genome sequences enables comparative approaches to be used to identify conserved cis-regulatory sequences among related legume species. For example, we uncovered G564 orthologs in soybean, Lotus, and Medicago (Fig. 6, H and I; T. Kawashima and R.B. Goldberg, unpublished data). Soybean separated from SRB approximately 19 million years ago and from Lotus and Medicago approximately 54 million years ago (Fig. 6H; Lavin et al., 2005). Results obtained from two different computational analyses uncovered short conserved regions between the 5′-upstream DNA sequences of G564 genes in SRB, Lotus, and Medicago (Fig. 6I). The first approach used FamilyRelationsII to identify blocks of similar DNA sequences shared between G564 upstream regions (Fig. 6I, red lines; Brown et al., 2005). In addition, this program showed that G564 structure is conserved (two exons and one intron) in these three legumes (Fig. 6I), suggesting strongly that the G564 genes are orthologous. Blocks of similar DNA sequences in the upstream regions of orthologous genes have been shown to contain cis-regulatory sequences (Yuh et al., 2002). The closely related sequence blocks found in the legume G564 upstream regions might also contain cis-regulatory sequences important for suspensor-specific transcription. We also used Multiple Em for Motif Elicitation (MEME; Bailey and Elkan, 1994) to identify sequences significantly enriched in the G564 upstream regions (Fig. 6I, blue arrows; T. Kawashima and R.B. Goldberg, unpublished data). Significantly, regions identified by MEME in SRB include the conserved 10-bp motif sequence (Weterings et al., 2001). Whether the 10-bp motif is an important suspensor cis-regulatory sequence and what trans-acting factors regulate transcription in the suspensor remain to be determined.
One or more of the suspensor-specific transcription factors that we identified using EST and LCM-GeneChip analyses (Figs. 4 and 5) might interact with the conserved 10-bp motif and other cis-regulatory sequences to control transcription in the suspensor of SRB and other legumes. Similarly, transcription factors specific to other seed compartments (Fig. 5) might play an important role in controlling transcription in different parts of the seed. To date, the molecular mechanisms by which region-specific transcription factors are interconnected to form seed regulatory networks remain unknown. Studying the function of region-specific transcription factors is essential for understanding the importance of these proteins in seed development and for uncovering downstream target genes to construct seed gene regulatory networks (Fig. 3).
Advances in soybean transformation procedures (Ko et al., 2006; Olhoft et al., 2006) have made it possible to use loss-of-function and gain-of-function strategies to study gene function directly in soybean (Fig. 3). Although T-DNA is used commonly to generate loss-of-function alleles in Arabidopsis (Alonso et al., 2003), this technique might not be appropriate in soybean for several reasons. The soybean genome is 8 times larger than that of Arabidopsis (Arumuganathan and Earle, 1991) and contains a majority of repetitive sequences (Goldberg, 1978), requiring a large effort to generate T-DNA insertions at a saturation level. Because soybean transformation procedures are not as efficient as the seed transformation technique used in Arabidopsis (Clough and Bent, 1998), it would be challenging to produce large numbers of independent transgenic lines. In addition, soybean is a polyploid (Shoemaker et al., 1996; Hymowitz, 2004) and the presence of homeologous genes may complicate the interpretation of knockout results due to gene redundancy. A more productive approach is to utilize RNA interference (RNAi) knockdown strategies to study gene functions that have proven useful in a variety of eukaryotes, including soybean (Fig. 3; Subramanian et al., 2005; Amore and Davidson, 2006; Nunes et al., 2006). For example, Herman et al. (2003) used RNAi in soybean to eliminate allergenic proteins from soybean seeds. The advantage of RNAi is that it can be used to target specific genes and has the potential to knock down sets of closely related genes (Miki et al., 2005; Kaur et al., 2006). This approach, and an analogous one using chimeric repressors to knock down related genes (Fig. 3, CRES-T; Hiratsu et al., 2003), should be feasible for studying the functions of transcription factors identified by LCM and GeneChip analysis in different compartments of a soybean seed, including those that are redundant in the soybean genome (Fig. 3). The sequence of the soybean genome (Jackson et al., 2006) combined with RNAi studies should make it possible to identify downstream target genes that are regulated by region-specific transcription factors at the global level, facilitating identification of seed and embryo regulatory networks (Fig. 3).
FUTURE PERSPECTIVES
The study of legume seed development has become exciting due to the availability of new genomic resources and sophisticated techniques, such as LCM and RNA profiling using GeneChip arrays. We have identified genes that are unique to a particular seed compartment and that are coregulated within the context of the soybean globular-stage seed (Fig. 5, C, E, and F). The completion of the soybean genome sequence (Jackson et al., 2006) should allow us to identify conserved motifs among the upstream regions of these unique and coregulated genes, thus facilitating the identification of compartment-specific cis-regulatory sequences that connect seed genes into regulatory networks. In addition to the soybean genome sequence, other legume genome sequences will be available soon (Broughton et al., 2003; Gepts et al., 2005; Young et al., 2005). Genome sequences from diverse legume species will provide an invaluable resource for comparative analysis to identify conserved cis-regulatory sequences that connect seed genes into regulatory networks, an approach that has been successful in other eukaryotes, such as the sea urchin (Yuh et al., 2002; Bolouri and Davidson, 2003). Comparative analysis between legume genome sequences, combined with the GeneChip and EST data obtained from seed and embryo compartments of SRB and soybean (Figs. 4 and 5) and other plants (e.g. Arabidopsis; Casson et al., 2005), should facilitate the discovery of genes essential for seed and embryo development, including those important for specific legume traits, such as seed size and embryo morphology (Fig. 2). In addition, comparative analysis of legume genomes with other nonlegume genomes, such as Arabidopsis, rice, and poplar (Populus spp.), will advance the discovery of genes important for seed development in flowering plants (Graham et al., 2004; Zhu et al., 2005). Finally, once a soybean whole-genome GeneChip becomes available, the entire mRNA profiles of all seed and embryo compartments can be determined, completing the identification of seed- and embryo-specific genes. Taken together, remarkable advances in genomic resources will allow us to answer questions regarding seed and embryo development (Fig. 1D) that were not possible only a few years ago. It is now becoming realistic in this genomic era to understand what genes and regulatory networks are required to make a legume seed.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers CA896559 to CA916678 (ESTs) and AF325187 (G564).
Acknowledgments
We are grateful to all the members of our laboratory, past and present, who have helped to establish soybean and SRB as powerful systems to investigate seed development. We particularly acknowledge Dr. Koen Weterings, Dr. Yuping Bi, and Dr. Xingjun Wang for contributing to many of the experiments summarized in this Update. In addition, we thank Ms. Chen Cheng for carrying out the real-time qRT-PCR experiment presented in Figure 5G. We would also like to acknowledge Ian Sussex, Roger Beachy, Tim Hall, Maarten Chrispeels, Niels Nielsen, Lila Vodkin, Don Boulter, T.J. Higgins, Klaus Muntz, and Uli Wobus whose laboratories helped to provide a foundation for understanding gene activity during legume seed development.
This work was supported by the National Science Foundation Plant Genome Program (grant no. DBI–0501720), the Department of Energy (grant no. DE–FG03–97ER20263), and Ceres Inc. T.K. is a recipient of a Nakajima Foundation predoctoral fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Robert B. Goldberg (bobg@ucla.edu).
References
- Adams CA, Norby SW, Rinne RW (1982) Protein modification and utilization of starch in soybean (Glycine max (L.) Merr.) seed saturation. J Exp Bot 33 279–287 [Google Scholar]
- Adjaye J, Herwig R, Herrmann D, Wruck W, Benkahla A, Brink TC, Nowak M, Carnwath JW, Hultschig C, Niemann H, et al (2004) Cross-species hybridisation of human and bovine orthologous genes on high density cDNA microarrays. BMC Genomics 5 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Alpi A, Lorenzi R, Cionini PG, Bennici A, Damato F (1979) Identification of gibberellin A1 in the embryo suspensor of Phaseolus coccineus. Planta 147 225–228 [DOI] [PubMed] [Google Scholar]
- Amore G, Davidson EH (2006) cis-Regulatory control of cyclophilin, a member of the ETS-DRI skeletogenic gene battery in the sea urchin embryo. Dev Biol 293 555–564 [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9 208–218 [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S (2004) Characteristics of the Lotus japonicus gene repertoire deduced from large-scale expressed sequence tag (EST) analysis. Plant Mol Biol 54 405–414 [DOI] [PubMed] [Google Scholar]
- Asano T, Masumura T, Kusano H, Kikuchi S, Kurita A, Shimada H, Kadowaki K (2002) Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: toward comprehensive analysis of the genes expressed in the rice phloem. Plant J 32 401–408 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2 28–36 [PubMed] [Google Scholar]
- Baumlein H, Nagy I, Villarroel R, Inze D, Wobus U (1992) Cis-analysis of a seed protein gene promoter—the conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. Plant J 2 233–239 [PubMed] [Google Scholar]
- Beach LR, Spencer D, Randall PJ, Higgins TJV (1985) Transcriptional and post-transcriptional regulation of storage protein gene-expression in sulfur-deficient pea-seeds. Nucleic Acids Res 13 999–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher M, Talke IN, Krall L, Kramer U (2004) Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J 37 251–268 [DOI] [PubMed] [Google Scholar]
- Bevan MW, Flavell RB, Chilton M (1983) A chimeric antibiotic resistance gene as a selectable marker for plant cell transformation. Nature 304 184–187 [PubMed] [Google Scholar]
- Bolouri H, Davidson EH (2003) Transcriptional regulatory cascades in development: initial rates, not steady state, determine network kinetics. Proc Natl Acad Sci USA 100 9371–9376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisjuk L, Rolletschek H, Radchuk R, Weschke W, Wobus U, Weber H (2004) Seed development and differentiation: a role for metabolic regulation. Plant Biol 6 375–386 [DOI] [PubMed] [Google Scholar]
- Broughton WJ, Hernandez G, Blair M, Beebe S, Gepts P, Vanderleyden J (2003) Beans (Phaseolus spp.)—model food legumes. Plant Soil 252 55–128 [Google Scholar]
- Brown CT, Xie Y, Davidson EH, Cameron RA (2005) Paircomp, FamilyRelationsII and Cartwheel: tools for interspecific sequence comparison. BMC Bioinformatics 6 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MM (1917) The development of the embryo-sac and of the embryo in Phaseolus vulgaris. Bull Torrey Bot Club 44 535–544 [Google Scholar]
- Buitink J, Leger JJ, Guisle I, Vu BL, Wuilleme S, Lamirault G, Le Bars A, Le Meur N, Becker A, Kuester H, et al (2006) Transcriptome profiling uncovers metabolic and regulatory processes occurring during the transition from desiccation-sensitive to desiccation-tolerant stages in Medicago truncatula seeds. Plant J 47 735–750 [DOI] [PubMed] [Google Scholar]
- Casson S, Spencer M, Walker K, Lindsey K (2005) Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis. Plant J 42 111–123 [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Goldstone K, Smith JC, Gilchrist M, Amaya E, Papalopulu N (2005) A Xenopus tropicalis oligonucleotide microarray works across species using RNA from Xenopus laevis. Mech Dev 122 355–363 [DOI] [PubMed] [Google Scholar]
- Chamberlin MA, Horner HT, Palmer RG (1994) Early endosperm, embryo, and ovule development in Glycine max (L.) Merr. Int J Plant Sci 155 421–436 [Google Scholar]
- Chandrasekharan MB, Bishop KJ, Hall TC (2003) Module-specific regulation of the beta-phaseolin promoter during embryogenesis. Plant J 33 853–866 [DOI] [PubMed] [Google Scholar]
- Chappell J, Chrispeels MJ (1986) Transcriptional and posttranscriptional control of phaseolin and phytohemagglutinin gene expression in developing cotyledons of Phaseolus vulgaris. Plant Physiol 81 50–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZL, Schuler MA, Beachy RN (1986) Functional analysis of regulatory elements in a plant embryo-specific gene. Proc Natl Acad Sci USA 83 8560–8564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chismar JD, Mondala T, Fox HS, Roberts E, Langford D, Masliah E, Salomon DR, Head SR (2002) Analysis of result variability from high-density oligonucleotide arrays comparing same-species and cross-species hybridizations. Biotechniques 33 516–518, 520, 522, 524 [DOI] [PubMed] [Google Scholar]
- Cionini PG, Bennici A, Alpi A, Damato F (1976) Suspensor, gibberellin and in vitro development of Phaseolus coccineus embryos. Planta 131 115–117 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Clutter M, Brady T, Walbot V, Sussex I (1974) Macromolecular synthesis during plant embryogeny—cellular rates of RNA synthesis in diploid and polytene cells in bean embryos. J Cell Biol 63 1097–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DC (1938) Embryology of Pisum sativum. Bot Gaz 100 123–132 [Google Scholar]
- Coste F, Ney B, Crozat Y (2001) Seed development and seed physiological quality of field grown beans (Phaseolus vulgaris L.). Seed Sci Technol 29 121–136 [Google Scholar]
- Davies DR (1976) DNA and RNA contents in relation to cell and seed weight in Pisum sativum. Plant Sci Lett 7 17–25 [Google Scholar]
- Day RC, Grossniklaus U, Macknight RC (2005) Be more specific! Laser-assisted microdissection of plant cells. Trends Plant Sci 10 397–406 [DOI] [PubMed] [Google Scholar]
- Dhaubhadel S, Gijzen M, Moy P, Farhangkhoee M (2007) Transcriptome analysis reveals a critical role of CHS7 and CHS8 genes for isoflavonoid synthesis in soybean seeds. Plant Physiol 143 326–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon SS, Miksche JP (1983) DNA, RNA, protein and heterochromatin changes during embryo development and germination of soybean (Glycine max L.). Histochem J 15 21–37 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Luckow MA (2003) The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol 131 900–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duranti M, Gius C (1997) Legume seeds: protein content and nutritional value. Field Crops Res 53 31–45 [Google Scholar]
- Dure LS (1975) Seed formation. Annu Rev Plant Physiol Plant Mol Biol 26 259–278 [Google Scholar]
- Duval M, Pepin R, Job C, Derpierre C, Douce R, Job D (1995) Ultrastructural localization of the major biotinylated protein from Pisum sativum seeds. J Exp Bot 46 1783–1786 [Google Scholar]
- Endo M, Matsubara H, Kokubun T, Masuko H, Takahata Y, Tsuchiya T, Fukuda H, Demura T, Watanabe M (2002) The advantages of cDNA microarray as an effective tool for identification of reproductive organ-specific genes in a model legume, Lotus japonicus. FEBS Lett 514 229–237 [DOI] [PubMed] [Google Scholar]
- Estrella-Herrera L, Depicker A, Van Montagu M, Schell J (1983) Expression of chimeric genes transferred into plant cells using a Ti-plasmid-derived vector. Nature 303 209–213 [PubMed] [Google Scholar]
- Evans LS, Dimitriadis L, Hinkley DA (1984) Seed protein quantities of field-grown soybeans exposed to simulated acidic rain. New Phytol 97 71–76 [Google Scholar]
- Firnhaber C, Puhler A, Kuster H (2005) EST sequencing and time course microarray hybridizations identify more than 700 Medicago truncatula genes with developmental expression regulation in flowers and pods. Planta 222 269–283 [DOI] [PubMed] [Google Scholar]
- Fraley RT, Rogers SG, Horsch RB, Sanders PR, Flick JS, Adams SP, Bittner ML, Brand LA, Fink CL, Fry JS, et al (1983) Expression of bacterial genes in plant cells. Proc Natl Acad Sci USA 80 4803–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Choi Y, Fischer RL (2004) Imprinting and seed development. Plant Cell 16 S203–S213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepts P, Beavis WD, Brummer EC, Shoemaker RC, Stalker HT, Weeden NF, Young ND (2005) Legumes as a model plant family. Genomics for food and feed report of the Cross-Legume Advances Through Genomics Conference. Plant Physiol 137 1228–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB (1978) DNA sequence organization in the soybean plant. Biochem Genet 16 45–68 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Barker SJ, Perez-Grau L (1989) Regulation of gene expression during plant embryogenesis. Cell 56 149–160 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Depaiva G, Yadegari R (1994) Plant embryogenesis—zygote to seed. Science 266 605–614 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Hoschek G, Ditta GS, Breidenbach RW (1981. a) Developmental regulation of cloned superabundant embryo mRNAs in soybean. Dev Biol 83 218–231 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Hoschek G, Tam SH, Ditta GS, Breidenbach RW (1981. b) Abundance, diversity, and regulation of mRNA sequence sets in soybean embryogenesis. Dev Biol 83 201–217 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Hoschek G, Vodkin LO (1983) An insertion sequence blocks the expression of a soybean lectin gene. Cell 33 465–475 [DOI] [PubMed] [Google Scholar]
- Gonzales MD, Archuleta E, Farmer A, Gajendran K, Grant D, Shoemaker R, Beavis WD, Waugh ME (2005) The Legume Information System (LIS): an integrated information resource for comparative legume biology. Nucleic Acids Res 33 D660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MA, Silverstein KAT, Cannon SB, VandenBosch KA (2004) Computational identification and characterization of novel genes from legumes. Plant Physiol 135 1179–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PH, Vance CP (2003) Legumes: importance and constraints to greater use. Plant Physiol 131 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guignard L (1882) Recherches anatomiques et physiologiques sur des legumineuses. PhD thesis. University of Paris, Paris
- Gupta PK, Rustgi S, Kumar N (2006) Genetic and molecular basis of grain size and grain number and its relevance to grain productivity in higher plants. Genome 49 565–571 [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131 657–668 [DOI] [PubMed] [Google Scholar]
- Hajduch M, Ganapathy A, Stein JW, Thelen JJ (2005) A systematic proteomic study of seed filling in soybean. Establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol 137 1397–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn G, Chaudhury A (2005) Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci 10 472–477 [DOI] [PubMed] [Google Scholar]
- Herman EM, Helm RM, Jung R, Kinney AJ (2003) Genetic modification removes an immunodominant allergen from soybean. Plant Physiol 132 36–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins TJV, Newbigin EJ, Spencer D, Llewellyn DJ, Craig S (1988) The sequence of a pea vicilin gene and its expression in transgenic tobacco plants. Plant Mol Biol 11 683–695 [DOI] [PubMed] [Google Scholar]
- Hill JE, Breidenbach RW (1974) Proteins of soybean seeds II. Accumulation of major protein components during seed development and maturation. Plant Physiol 53 747–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34 733–739 [DOI] [PubMed] [Google Scholar]
- Hymowitz T (2004) Speciation and cytogenetics. In HR Boerma, J Specht, eds, Soybeans: Improvement, Production, and Uses, Ed 3. American Society of Agronomy, Madison, WI, pp 97–129
- Jackson SA, Rokhsar D, Stacey G, Shoemaker RC, Schmutz J, Grimwood J (2006) Toward a reference sequencing of the soybean genome: a multiagency effort. Crop Sci 46 S55–S61 [Google Scholar]
- Ji W, Zhou W, Gregg K, Yu N, Davis S, Davis S (2004) A method for cross-species gene expression analysis with high-density oligonucleotide arrays. Nucleic Acids Res 32 e93 [DOI] [PMC free article] [PubMed]
- Jofuku KD, Okamuro JK, Goldberg RB (1987) Interaction of an embryo DNA binding protein with a soybean lectin gene upstream region. Nature 328 734–737 [DOI] [PubMed] [Google Scholar]
- Johansson M, Walles B (1993) Functional-anatomy of the ovule in broad bean, Vicia faba L. 2. Ultrastructural development up to early embryogenesis. Int J Plant Sci 154 535–549 [Google Scholar]
- Johnson S, Liu CM, Hedley CL, Wang TL (1994) An analysis of seed development in Pisum sativum XVIII. The isolation of mutants defective in embryo development. J Exp Bot 45 1503–1511 [Google Scholar]
- Journet EP, van Tuinen D, Gouzy J, Crespeau H, Carreau V, Farmer MJ, Niebel A, Schiex T, Jaillon O, Chatagnier O, et al (2002) Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Res 30 5579–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J, Sebastian J, Siddiqi I (2006) The Arabidopsis-mei2-like genes play a role in meiosis and vegetative growth in Arabidopsis. Plant Cell 18 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk NM, Ceserani T, Tausta SL, Sussex IM, Nelson TM (2003) Laser capture microdissection of cells from plant tissues. Plant Physiol 132 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney AJ (1998) Plants as industrial chemical factories—new oils from genetically engineered soybeans. Fett Lipid 100 173–176 [Google Scholar]
- Klink VP, Alkharouf N, MacDonald M, Matthews B (2005) Laser capture microdissection (LCM) and expression analyses of Glycine max (soybean) syncytium containing root regions formed by the plant pathogen Heterodera glycines (soybean cyst nematode). Plant Mol Biol 59 965–979 [DOI] [PubMed] [Google Scholar]
- Ko TS, Korban SS, Somers DA (2006) Soybean (Glycine max) transformation using immature cotyledon explants. Methods Mol Biol 343 397–405 [DOI] [PubMed] [Google Scholar]
- Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB (1990) Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2 1201–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HB (2005) Engineering soybean for enhanced sulfur amino acid content. Crop Sci 45 454–461 [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Wurschum T, Breuninger H (2004) Genetic regulation of embryonic pattern formation. Plant Cell (Suppl) 16 S190–S202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol 54 575–594 [DOI] [PubMed] [Google Scholar]
- Lersten NR (1983) Suspensors in Leguminosae. Bot Rev 49 233–257 [Google Scholar]
- Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93 1195–1205 [DOI] [PubMed] [Google Scholar]
- Martin JN (1914) Comparative morphology of some Leguminosae contributions from the Hull Botanical Laboratory. Bot Gaz 58 154–167 [Google Scholar]
- Meinke DW, Chen J, Beachy RN (1981) Expression of storage-protein genes during soybean seed development. Planta 153 130–139 [DOI] [PubMed] [Google Scholar]
- Mendel G (1865) Versuche über Plflanzenhybriden. In Verhandlungen des naturforschenden Vereines, Vol Bd. IV für das Jahr. Abhandlungen, Brünn, Czech Republic, pp 3–47
- Miki D, Itoh R, Shimamoto K (2005) RNA silencing of single and multiple members in a gene family of rice. Plant Physiol 138 1903–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SS, Bowman LAA, Gijzen M, Miki BLA (1999) Early development of the seed coat of soybean (Glycine max). Ann Bot (Lond) 84 297–304 [Google Scholar]
- Moise JA, Han S, Gudynaite-Savitch L, Johnson DA, Miki BLA (2005) Seed coats: structure, development, composition, and biotechnology. In Vitro Cell Dev Biol Plant 41 620–644 [Google Scholar]
- Moody DE, Zou Z, McIntyre L (2002) Cross-species hybridisation of pig RNA to human nylon microarrays. BMC Genomics 3 27 [DOI] [PMC free article] [PubMed]
- Moravec T, Schmidt MA, Herman EM, Woodford-Thomas T (2007) Production of Escherichia coli heat labile toxin (LT) B subunit in soybean seed and analysis of its immunogenicity as an oral vaccine. Vaccine 25 1647–1657 [DOI] [PubMed] [Google Scholar]
- Murai N, Sutton DW, Murray MG, Slightom JL, Merlo DJ, Reichert NA, Senguptagopalan C, Stock CA, Barker RF, Kemp JD, et al (1983) Phaseolin gene from bean is expressed after transfer to sunflower via tumor-inducing plasmid vectors. Science 222 476–482 [DOI] [PubMed] [Google Scholar]
- Murray DR (1987) Nutritive role of seed coats in developing legume seeds. Am J Bot 74 1122–1137 [Google Scholar]
- Naito S, Dube PH, Beachy RN (1988) Differential expression of conglycinin alpha-subunit and beta-subunit genes in transgenic plants. Plant Mol Biol 11 109–123 [DOI] [PubMed] [Google Scholar]
- Nakazono M, Qiu F, Borsuk LA, Schnable PS (2003) Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell 15 583–59612615934 [Google Scholar]
- Nelson T, Tausta SL, Gandotra N, Liu T (2006) Laser microdissection of plant tissue: What you see is what you get. Annu Rev Plant Biol 57 181–201 [DOI] [PubMed] [Google Scholar]
- Nishizawa NK, Mori S, Watanabe Y, Hirano H (1994) Ultrastructural-localization of the basic 7S globulin in soybean (Glycine Max) cotyledons. Plant Cell Physiol 35 1079–1085 [Google Scholar]
- Nowrousian M, Ringelberg C, Dunlap JC, Loros JJ, Kuck U (2005) Cross-species microarray hybridization to identify developmentally regulated genes in the filamentous fungus Sordaria macrospora. Mol Genet Genomics 273 137–149 [DOI] [PubMed] [Google Scholar]
- Nunes ACS, Vianna GR, Cuneo F, Amaya-Farfan J, de Capdeville G, Rech EL, Aragao FJL (2006) RNAi-mediated silencing of the myo-inositol-1-phosphate synthase gene (GmMIPS1) in transgenic soybean inhibited seed development and reduced phytate content. Planta 224 125–132 [DOI] [PubMed] [Google Scholar]
- Okamuro JK, Jofuku KD, Goldberg RB (1986) Soybean seed lectin gene and flanking nonseed protein genes are developmentally regulated in transformed tobacco plants. Proc Natl Acad Sci USA 83 8240–8244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olhoft PM, Donovan CM, Somers DA (2006) Soybean (Glycine max) transformation using mature cotyledonary node explants. Methods Mol Biol 343 385–396 [DOI] [PubMed] [Google Scholar]
- Pattee HE, Young CT, Giesbrecht FG (1981) Seed size and storage effects on carbohydrates of peanuts. J Agric Food Chem 29 800–802 [Google Scholar]
- Ramirez M, Graham MA, Blanco-Lopez L, Silvente S, Medrano-Soto A, Blair MW, Hernandez G, Vance CP, Lara M (2005) Sequencing and analysis of common bean ESTs. Building a foundation for functional genomics. Plant Physiol 137 1211–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie WG, Newbigin EJ, Higgins TJV (1992) Genes encoding seed globulins in legumes. In IM Morrison, ed, Advances in Plant Cell Biochemistry and Biotechnology: A Research Annual, Vol 1. Jai Press, Greenwich, CT, pp 53–104
- Rubel A, Rinne RW, Canvin DT (1972) Protein, oil, and fatty-acid in developing soybean seeds. Crop Sci 12 739–741 [Google Scholar]
- Sanders PM, Bui AQ, Le BH, Goldberg RB (2005) Differentiation and degeneration of cells that play a major role in tobacco anther dehiscence. Sex Plant Reprod 17 219–241 [Google Scholar]
- Schleiden MJ, Vogel T (1838) Über das Albumen, insbesondere der Leguminosen, Vol 19: Pt I. Verhandlungen der Kaiserlich Leopoldinisch-Carolinischen Deutschen Akademie der Naturforscher, Bonn
- Schleiden MJ, Vogel T (1842) Über das Albumen, insbesondere der Leguminosen, Vol 19: Pt II. Verhandlungen der Kaiserlich Leopoldinisch-Carolinischen Deutschen Akademie der Naturforscher, Breslau, Poland
- Shoemaker R, Keim P, Vodkin L, Retzel E, Clifton SW, Waterston R, Smoller D, Coryell V, Khanna A, Erpelding J, et al (2002) A compilation of soybean ESTs: generation and analysis. Genome 45 329–338 [DOI] [PubMed] [Google Scholar]
- Shoemaker R, Polzin K, Labate J, Specht J, Brummer EC, Olson T, Young N, Concibido V, Wilcox J, Tamulonis JP, (1996) Genome duplication in soybean (Glycine subgenus soja). Genetics 144 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U, Jambunathan R, Saxena NP (1981) Changes in carbohydrates, amino-acids and proteins in developing seed of chickpea. Phytochemistry 20 373–378 [Google Scholar]
- Spencer MWB, Casson SA, Lindsey K (2007) Transcriptional profiling of the Arabidopsis embryo. Plant Physiol 143 924–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangeland B, Salehian Z, Aalen R, Mandal A, Olsen OA (2003) Isolation of GUS marker lines for genes expressed in Arabidopsis endosperm, embryo and maternal tissues. J Exp Bot 54 279–290 [DOI] [PubMed] [Google Scholar]
- Subramanian S, Graham MY, Yu O, Graham TL (2005) RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and to enhanced susceptibility to Phytophthora sojae. Plant Physiol 137 1345–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussex I, Clutter M, Walbot V, Brady T (1973) Biosynthetic activity of suspensor of Phaseolus coccineus. Caryologia 25 261–272 [Google Scholar]
- Thibaud-Nissen F, Shealy RT, Khanna A, Vodkin LO (2003) Clustering of microarray data reveals transcript patterns associated with somatic embryogenesis in soybean. Plant Physiol 132 118–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBosch KA, Stacey G (2003) Summaries of legume genomics projects from around the globe. Community resources for crops and models. Plant Physiol 131 840–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan JG, Geissler C, Nicholson B, Nicholson B (1997) The New Oxford Book of Food Plants. Oxford University Press, New York
- Vodkin LO, Khanna A, Shealy R, Clough SJ, Gonzalez DO, Philip R, Zabala G, Thibaud-Nissen F, Sidarous M, Stromvik MV, et al (2004) Microarrays for global expression constructed with a low redundancy set of 27,500 sequenced cDNAs representing an array of developmental stages and physiological conditions of the soybean plant. BMC Genomics 5 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V, Clutter M, Sussex IM (1972) Reproductive development and embryogeny in Phaseolus. Phytomorphology 22 59–68 [Google Scholar]
- Walling L, Drews GN, Goldberg RB (1986) Transcriptional and post-transcriptional regulation of soybean seed protein mRNA levels. Proc Natl Acad Sci USA 83 2123–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TL, Domoney C, Hedley CL, Casey R, Grusak MA (2003) Can we improve the nutritional quality of legume seeds? Plant Physiol 131 886–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Dooley TP, Curto EV, Davis RL, VandeBerg JL (2004) Cross-species application of cDNA microarrays to profile gene expression using UV-induced melanoma in Monodelphis domestica as the model system. Genomics 83 588–599 [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U (2005) Molecular physiology of legume seed development. Annu Rev Plant Biol 56 253–279 [DOI] [PubMed] [Google Scholar]
- Weterings K, Apuya NR, Bi Y, Fischer RL, Harada JJ, Goldberg RB (2001) Regional localization of suspensor mRNAs during early embryo development. Plant Cell 13 2409–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox JR (2004) World distribution and trade of soybean. In RH Boerma, JE Specht, eds, Soybeans: Improvement, Production, and Uses, Ed 3. American Society of Agronomy, Madison, WI, pp 1–14
- Wohlfarth T, Braun H, Kirik V, Kolle K, Czihal A, Tewes A, Luerssen H, Misera S, Shutov A, Baumlein H (1998) Regulation and evolution of seed globulin genes. J Plant Physiol 152 600–606 [Google Scholar]
- Yeung EC, Meinke DW (1993) Embryogenesis in angiosperms—development of the suspensor. Plant Cell 5 1371–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Cannon SB, Sato S, Kim D, Cook DR, Town CD, Roe BA, Tabata S (2005) Sequencing the genespaces of Medicago truncatula and Lotus japonicus. Plant Physiol 137 1174–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuh CH, Brown CT, Livi CB, Rowen L, Clarke PJ, Davidson EH (2002) Patchy interspecific sequence similarities efficiently identify positive cis-regulatory elements in the sea urchin. Dev Biol 246 148–161 [DOI] [PubMed] [Google Scholar]
- Zhu H, Choi HK, Cook DR, Shoemaker RC (2005) Bridging model and crop legumes through comparative genomics. Plant Physiol 137 1189–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]