Abstract
Phosphatidylserine (PS) decarboxylase is involved in the synthesis of the abundant phospholipid phosphatidylethanolamine (PE), particularly in mitochondria, in many organisms, including yeast (Saccharomyces cerevisiae) and animals. Arabidopsis (Arabidopsis thaliana) contains three genes with sequence similarity to PS decarboxylases, and the respective gene products were functionally characterized after heterologous expression in yeast and Escherichia coli. While the PSD1 protein localizes to mitochondria, PSD2 and PSD3 are found in the endomembrane system. To study the role of PSD genes in plant phospholipid metabolism, Arabidopsis insertional mutants for psd1, psd2, and psd3 were obtained. The single mutants were decreased in PS decarboxylase activity to various extents, but mutant plants showed no obvious growth or morphological phenotype. A triple mutant, psd1 psd2 psd3, was generated that was totally devoid of PS decarboxylase activity. While the phospholipid composition in whole leaves was unchanged, the PE content in isolated mitochondria of psd1 psd2 psd3 was decreased. Therefore, the predominant proportion of PE in Arabidopsis is synthesized by alternative pathways, but a significant amount of mitochondrial PE is derived from the PS decarboxylase reaction. These results imply that, similar to yeast and animal cells, a specific phospholipid transfer from the endoplasmic reticulum to mitochondria exists in plants.
In contrast to chloroplasts of higher plants where galactolipids are predominant, phospholipids (e.g. phosphatidylcholine and phosphatidylethanolamine [PE]) are the most abundant lipids in extraplastidial membranes. Phospholipid metabolism in plants involves a complex network of biosynthetic pathways, some of which are localized to different subcellular compartments. Two phospholipids, PE and phosphatidyl-Ser (PS), which can be interconverted in many organisms, are characterized by the presence of an ammonium moiety in their head group. PE represents the most abundant nonbilayer-forming lipid in extraplastidial membranes (Webb and Green, 1991), and its deficiency was shown to result in severe developmental defects in a number of organisms (yeast [Saccharomyces cerevisiae]: Trotter and Voelker, 1995; Birner et al., 2001; hamster ovary cells: Emoto and Umeda, 2000; Drosophila: Pavlidis et al., 1994; mouse: Steenbergen et al., 2005). The aminophospholipid PS is of very low abundance in most membranes, and it serves as a precursor for PE synthesis. PS in animal cells is confined to the inner leaflet of the plasma membrane, and this distinct distribution is maintained by specific transporters. Exposure of PS to the outer leaflet of the plasma membrane serves as signal for apoptosis in animal cells and results in phagocytosis of affected cells (Vance and Steenbergen, 2005). In plants, distinct pools of PS seem to exist that localize to different membranes. In contrast to other membrane lipids, the PS pool of the endoplasmic reticulum (ER) and of the plasma membranes was shown to be enriched in very long chain fatty acids (Bohn et al., 2001; Vincent et al., 2001).
PS metabolism in different organisms was recently reviewed by Vance and Steenbergen (2005). At least three different pathways contribute to the synthesis of PE in spinach leaves, i.e. decarboxylation of PS (PS decarboxylase), phosphoethanolamine transfer from CDP-ethanolamine to diacylglycerol (aminoalcoholphospho-transferase; Kennedy/nucleotide pathway), and exchange of the head group of PE with Ser (base exchange enzyme; Marshall and Kates, 1973). The Kennedy pathway is believed to be the major pathway for PE synthesis in plants. In agreement with this hypothesis, the precursor for PE synthesis via the Kennedy pathway, ethanolamine, was shown to be predominantly produced by direct decarboxylation of Ser (Rontein et al., 2001; Rontein et al., 2003a). Furthermore, a mutation in the gene encoding CTP:phosphorylethanolamine cytidylyltransferase, a key enzyme of the Kennedy pathway for PE synthesis, was shown to cause embryo lethality for Arabidopsis (Arabidopsis thaliana; Mizoi et al., 2006).
In castor bean endosperm, radioactive label from [14C]Ser was incorporated into PS and PE, suggesting that PS was synthesized via the PE-PS base exchange enzyme and that a fraction of PS was decarboxylated (Moore, 1975). The PS synthase isolated from wheat (Triticum aestivum) encodes an enzyme that synthesizes PS from CDP-diacylglycerol and Ser (Delhaize et al., 1999). However, other plant species, such as Arabidopsis, do not contain genes with sequence similarity to wheat PS synthase, and, therefore, plants seem to employ distinct routs for PS production.
The first molecular study on PS decarboxylases was the isolation of the temperature-sensitive Escherichia coli mutant EH150, which accumulates large amounts of PS when grown at 42°C (Hawrot and Kennedy, 1975, 1978). Isolation of the PSD protein and cloning of the respective gene revealed that the 35-kD PSD proenzyme is cleaved at a specific LGS254T motif, resulting in the release of the two subunits of the mature enzyme: the C-terminal, 7-kD α-subunit carrying an N-terminal pyruvoyl prosthetic group, and the N-terminal, 28-kD β-subunit (Li and Dowhan, 1988).
Two PS decarboxylase proteins were found in yeast: PSD1 localizes to the mitochondria (Clancey et al., 1993; Trotter et al., 1993) and PSD2 to the Golgi and tonoplast compartments (Trotter and Voelker, 1995; Trotter et al., 1995). PSD1 represents the major PS decarboxylase activity (for review, see Voelker, 1997). The double mutant of yeast shows severe PE deficiency because psd1 psd2 cells contain only approximately 2% of PE instead of approximately 25% in wild type (Trotter et al., 1995; Birner et al., 2001). PE deficiency and growth retardation can be rescued with ethanolamine that serves as precursor for PE synthesis via the Kennedy pathway.
Additional PS decarboxylases have been isolated from bacteria (Bacillus subtilis; Matsumoto et al., 1998) and from mammalian cells (Chinese hamster ovary cells; Kuge et al., 1991). In animals, PS decarboxylase was shown to localize to the inner mitochondrial membrane (Dennis and Kennedy, 1972; van Golde et al., 1974). Decarboxylation of ER-derived PS in the mitochondria is believed to largely contribute to PE synthesis not only in yeast but also in animal cells (Vance and Steenbergen, 2005). As a consequence, a block in mitochondrial PS decarboxylase activity causes embryonic lethality and changes in mitochondrial ultra-structure in mouse (Steenbergen et al., 2005).
The first molecular study on higher plant PS decarboxylases was the isolation and characterization of the PSD1 cDNAs from tomato (Solanum lycopersicum) and Arabidopsis (Rontein et al., 2003b). Plant PSD1 displays a high degree of sequence similarity with yeast PSD1, and localization studies confirmed that the plant enzyme is found in mitochondria. The Arabidopsis genome contains two additional, uncharacterized loci with sequence similarity to PS decarboxylases. To study the role of PS decarboxylase in plants, we functionally characterized these two putative PSD genes after heterologous expression in yeast and E. coli and determined their subcellular localization. Arabidopsis single mutant lines for all three PSD loci were identified in a reverse genetic approach, and a triple mutant was generated that was totally devoid of PS decarboxylase activity. From the characterization of this mutant, it became clear that in plants, PS decarboxylase contributes only to a minor proportion of PE production, but that a block in PS decarboxylase affects mitochondrial phospholipid composition.
RESULTS
Isolation and Functional Characterization of PS Decarboxylases from Arabidopsis
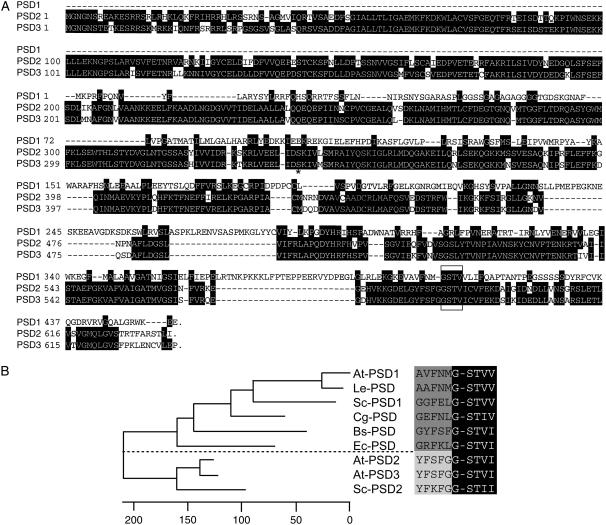
In many eukaryotic organisms, mitochondrial PS decarboxylase is involved in the synthesis of a major proportion of cellular PE. Mitochondrial PS decarboxylases were identified in yeast, Chinese hamster, tomato, and Arabidopsis (Kuge et al., 1991; Clancey et al., 1993; Rontein et al., 2003b; Fig. 1). Furthermore, bacterial PS decarboxylases with close sequence similarity to mitochondrial PSD forms were identified in E. coli and B. subtilis (Li and Dowhan, 1988; Matsumoto et al., 1998). Yeast contains a second PS decarboxylase, PSD2, that localizes to the Golgi/tonoplast and harbors a long N-terminal extension (Trotter et al., 1995). In addition to PSD1, the genomic sequence of Arabidopsis contains two further loci with sequence similarity to PS decarboxylases, At5g57190 and At4g25970 (Beisson et al., 2003), which were tentatively designated PSD2 and PSD3 (Fig. 1). The predicted amino acid sequences of these genes were very similar (amino acid identity of 76%) and more related to the yeast PSD2 protein than to the mitochondrial PSD1 proteins from plants or other organisms. Furthermore, similar to yeast PSD2, the two Arabidopsis proteins PSD2 and PSD3 contained long N-terminal extensions (Fig. 1).
Figure 1.
PS decarboxylases in Arabidopsis. A, Amino acid sequence comparison of Arabidopsis PSD1 (At4g16700), PSD2 (At5g57190), and PSD3 (At4g25970). Arabidopsis contains three genes with sequence similarities to PS decarboxylases. In contrast to the mitochondrial PS decarboxylase, PSD1, the PSD2 and PSD3 proteins contain an N-terminal extension of approximately 350 amino acids. A truncated form of PSD3 starting with the amino acid Ser-352 (indicated with an asterisk) was expressed in E. coli ( Fig. 2B). Identical amino acids are highlighted in black, and gaps are indicated with dashes. The conserved GSTV motif is marked with a box. B, Phylogenetic tree of PS decarboxylases. Amino acid sequences (the C-terminal 300 amino acids without N-terminal extensions) were compared using the ClustalW program of the Lasergene DNAstar software. Numbers indicate the nucleotide substitutions (×100). The sequences on the right depict the conserved sequence motif G-ST, at which the precursor protein is processed into the α and β polypeptides, constituting mature PS decarboxylase. The dashed line separates the two groups of PS decarboxylases containing the mitochondrial/bacterial and endomembrane forms. At-PSD1, AY189805 (At4g16700), Arabidopsis, and Le-PSD, AY093689, tomato, Rontein et al. (2003b); Sc-PSD1, L20973, yeast, Clancey et al. (1993); Cg-PSD, P27465, Chinese hamster, Kuge et al. (1991); Bs-PSD, P39822, B. subtilis, Matsumoto et al. (1998); Ec-PSD, J03916, E. coli, Li and Dowhan (1988); At-PSD2 (At5g57190) and At-PSD3, AV527283 (At4g25970), Arabidopsis, this study; Sc-PSD2, U19910, yeast, Trotter et al. (1995). The full-length coding sequences for At-PSD2 (At5g57190) and At-PSD3 (At4g25970) were deposited in GenBank with the accession numbers EF203902 and EF203901, respectively.
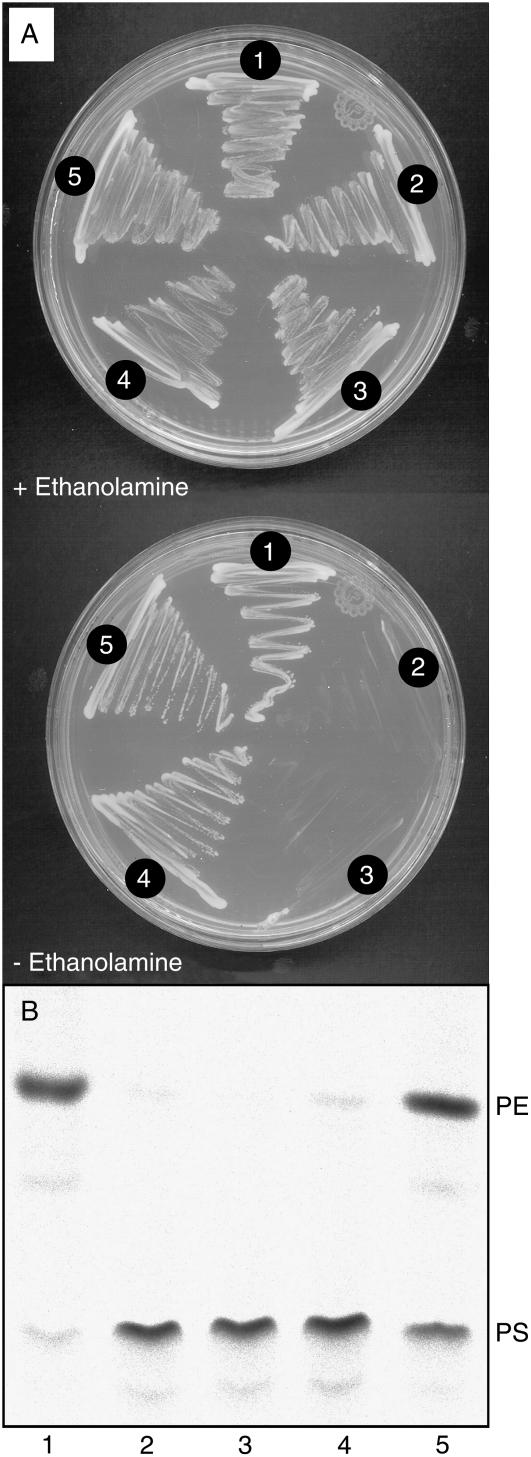
To functionally characterize the putative PS decarboxylases PSD2 and PSD3 from Arabidopsis, the respective cDNAs were heterologously expressed in psd mutants of yeast and E. coli. A yeast double mutant (psd1 psd2) deficient in both PSD genes is unable to synthesize PE via the PS decarboxylase pathway. This mutant is auxotroph for ethanolamine because PE can be synthesized from ethanolamine via the Kennedy pathway. Transformation of the yeast psd1 psd2 mutant with plasmids harboring the Arabidopsis PSD2 or PSD3 cDNAs restored growth on ethanolamine-free medium, indicating that these two sequences encode functional PS decarboxylases (Fig. 2A). An alternative strategy to study PS decarboxylase activity is the complementation of the E. coli EH150 mutant, which carries a temperature-sensitive mutation in its single psd gene. To address the question whether the C-terminal part of PSD3 carrying the PS decarboxylase sequence domain is sufficient to encode a functionally active enzyme, the entire PSD3 cDNA or its C-terminal part (starting at Ser-352; Fig. 1) was transferred into the E. coli mutant EH150. Proteins were isolated from the EH150 mutant carrying the Arabidopsis PSD3 constructs, and PS decarboxylase activity measured at 42°C with radioactively labeled PS. Figure 2B shows that PS decarboxylase activity in EH150 cells transformed with an empty vector or with the full-length Arabidopsis PSD3 cDNA was low as compared to cells harboring the N-terminally truncated form. Therefore, the C-terminal, PSD-like part of Arabidopsis PSD3 encodes a functionally active PS decarboxylase, and the long N terminal is not required for enzymatic activity but has an inhibitory effect when expressed in E. coli.
Figure 2.
Arabidopsis PSD2 and PSD3 encode functional PS decarboxylases. A, Arabidopsis PSD2 and PSD3 complement ethanolamine auxotrophy of the PS decarboxylase-deficient yeast psd1 psd2 double mutant. Wild type and psd1 psd2 double mutant yeast strains harboring different plasmids were grown in the presence or absence of ethanolamine, as indicated. 1, Wild-type yeast (strain SEY6210); 2, double mutant psd1 psd2 (RYY51); 3, psd1 psd2 transformed with empty vector; 4, psd1 psd2 transformed with Arabidopsis PSD3; 5, psd1 psd2 transformed with Arabidopsis PSD2. B, The full-length and the C-terminal domain of the PSD3 cDNA of Arabidopsis harbor PS decarboxylase activity. The temperature-sensitive E. coli strain EH150 deficient in PS decarboxylase activity was transformed with different Arabidopsis PSD3 cDNA constructs. After growth at 42°C, cells were harvested and PS decarboxylase activity measured with 14C-labeled PS. The figure shows the conversion of PS to PE after lipid separation by TLC and autoradiography. 1, E. coli XL1-Blue (control); 2, EH150; 3, EH150 carrying empty vector; 4, EH150 transformed with the full-length Arabidopsis PSD3 cDNA; and 4, EH150 carrying the C-terminal PS decarboxylase domain of Arabidopsis PSD3 (starting at Ser-352).
Subcellular Localization and Expression Pattern of PS Decarboxylases
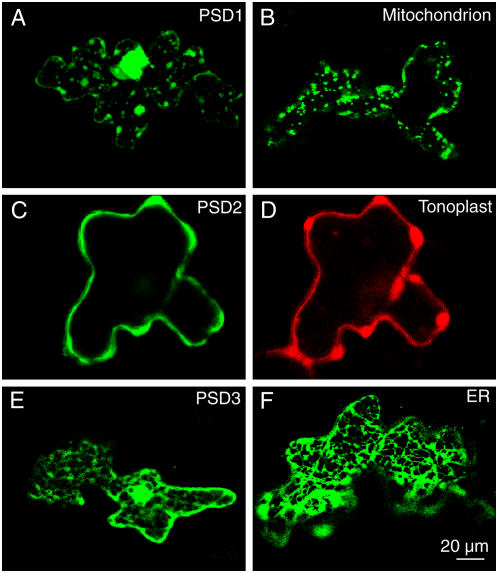
The fact that the two Arabidopsis PS decarboxylases PSD2 and PSD3 contain a long N-terminal extension might suggest that these two proteins do not localize to mitochondria. The full-length sequences of PSD2 and PSD3 were N-terminally fused to the sequence of the GFP and transiently expressed in Arabidopsis leaves. As a control, a GFP fusion construct of the mitochondrial PS decarboxylase (PSD1) from Arabidopsis was employed. Analysis by confocal microscopy and comparison to control constructs revealed that the PSD1-GFP fusion protein accumulated in mitochondria in agreement with previous results (Fig. 3, A and B; Rontein et al., 2003b). In contrast, the localization pattern of PSD2-GFP and PSD3-GPF fusion proteins was different. For PSD2-GFP, a colocalization experiment was done of a red fluorescent protein (DsRed) fusion with a tonoplast control protein. The green and red fluorescence of the two proteins was largely overlapping, indicating that PSD2 localizes to the tonoplast (Fig. 3, C and D). Furthermore, comparison of the fluorescence of the PSD3-GFP construct with that of a GFP fusion with an ER control protein revealed that PSD3 most likely localizes to the ER (Fig. 3, E and F). Therefore, the two PS decarboxylases PSD2 and PSD3 were localized to endomembrane fraction, presumably to the tonoplast and ER, respectively.
Figure 3.
Subcellular localization of Arabidopsis PS decarboxylases. N-terminal fusion constructs of full-length PSD proteins with GFP were transferred into Arabidopsis leaves by bombardment and subcellular localization analyzed by confocal fluorescence microscopy. A, PSD1; B, mitochondrial control (pre-101-GFP); C, PSD2; D, tonoplast control (KCO1-GFP); E, PSD3; F, ER control (KDEL-GFP). Bar = 20 μm.
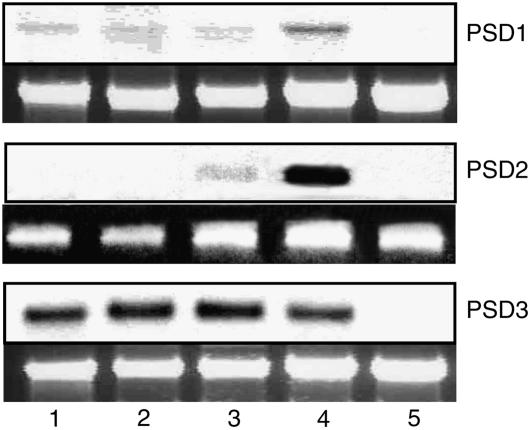
Expression of PS decarboxylase genes in different plant organs was analyzed by northern hybridization and semiquantitative PCR analysis. In agreement with previous results (Rontein et al., 2003b), the mitochondrial PSD1 was found to be expressed in roots and to a lesser extent in stems and leaves (Fig. 4A). PSD1 expression was highest in flowers but very low in siliques. PSD3 expression was comparable in roots, stems, leaves, and flowers, but very low in siliques (Fig. 4C). Because of the high sequence similarity with PSD3 and the low expression level, it was not possible to detect PSD2 expression by northern hybridization (data not shown). Therefore, a specific PSD2 primer pair was designed for semiquantitative reverse transcription (RT)-PCR to distinguish between PSD2 and PSD3 mRNA expression. As shown in Figure 4B, PSD2 expression was highest in flowers and lower in leaves. Expression in roots, shoots, and siliques was very low when measured by RT-PCR.
Figure 4.
Expression of PSD1, PSD2, and PSD3 in different Arabidopsis tissues. Expression of PS decarboxylase was determined with mRNA isolated from different plant tissues using northern hybridization (PSD1 and PSD3) or RT-PCR (PSD2). The top segments for PSD1 and PSD2 show the hybridization signals, and the bottom segments show a photo of the 26 ribosomal RNA bands stained with ethidium bromide. Because of its low expression, PSD2 mRNA was determined by semiquantitative RT-PCR. The top segment shows the band for the PSD2 RT-PCR product in agarose gel electrophoresis and the bottom segment for ubiquitin (control). 1, Roots; 2, stems; 3, leaves; 4, flowers; and 5, siliques.
Isolation of Arabidopsis Mutants Defective in PS Decarboxylase Activity
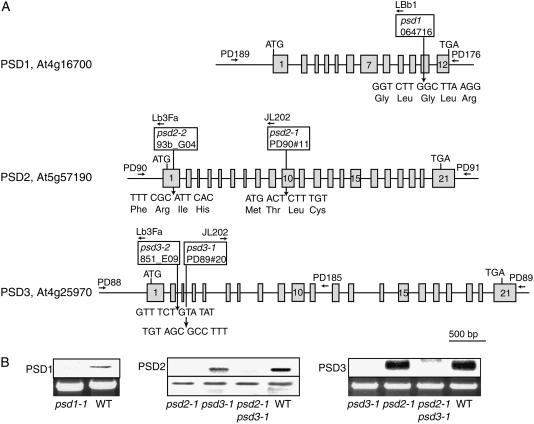
To study the role of PS decarboxylase in plant lipid metabolism, a reverse genetic approach was chosen. To this end, T-DNA insertion mutant lines for the three PSD genes were selected from different Arabidopsis mutant populations (Fig. 5A). One mutant line, psd1, was found in the SALK population. Screening of the Madison T-DNA insertion population resulted in the identification of two mutant lines, psd2-1 and psd3-1. Two additional mutant alleles, psd2-2 and psd2-3, were obtained from Syngenta. Sequencing of PCR fragments revealed that the insertions in all mutant plants are localized in exons or introns between the start and stop codons (Fig. 5A). Homozygous lines for all mutant alleles were identified by PCR, segregation analysis, and Southern-blot hybridization using genomic PSD fragments as probes (data not shown). PSD mRNA abundance in psd mutants was analyzed to unravel whether there still was residual expression (Fig. 5B). For psd1, northern hybridization was done with total RNA from flowers because PSD1 expression was high in flowers (see Fig. 4). PSD1 expression in the psd1 mutant was not detectable, in contrast to wild type, where a clear band was identified. Northern hybridization of leaf RNA revealed that one band corresponding to PSD3 was detectable in wild type, but this band was absent from the psd3-1 mutant (Fig. 5B). PSD2 expression was analyzed by RT-PCR of leaf RNA and PCR fragments separated by agarose gel electrophoresis. A band for PSD2 was observed in wild type, but this band was absent from psd2-1. In conclusion, the mRNA of the respective PSD genes was shown to be absent from the psd1, psd2-1, and psd3-1 mutant lines, indicating that these insertion lines represent null alleles.
Figure 5.
PS decarboxylase mutants of Arabidopsis. A, Localization of T-DNA insertions in psd1, psd2, and psd3 mutants. Positions of insertions are indicated relative to the exon/intron structures of PSD genes. Exons are depicted by boxes and numbered from 5′ to 3′. Arrows show positions of oligonucleotides used for PCR analysis. For PSD1, one insertional mutant (psd1) was identified in the SALK collection. Two mutant alleles were found for PSD2, psd2-1 and psd2-2, from the Madison collection and from Syngenta, respectively. The psd3-1 and psd3-2 mutant alleles were derived from the Madison and from Syngenta, respectively. B, Expression of PSD genes in Arabidopsis psd1, psd2-1, and psd3-1 mutants. Expression of PSD1 and PSD3 were recorded by northern hybridization of total RNA isolated from flowers or leaves, respectively. The top segment shows the hybridization signal, and the bottom segment shows a photo of the 26S rRNA band stained with ethidium bromide (control). Because of its low expression, PSD2 was determined by semiquantitative RT-PCR. The top and bottom segments show the RT-PCR band for PSD2 and ubiquitin, respectively.
Growth and morphology of the different mutant lines was indistinguishable from the respective wild-type ecotypes, indicating that single mutations in PSD genes have no effect on overall plant physiology. To address the question whether the different PS decarboxylase forms are functionally redundant, double mutants were generated by crosses between the psd2-1 and psd3-1 as well as the psd2-2 and psd3-2 mutant lines. PSD2 and PSD3 share a high degree of sequence similarity, and the two proteins localize to the endomembrane system, suggesting that they might have redundant functions. However, similar to the psd single mutants, growth and morphology of the two double mutant lines, psd2-1 psd3-1 and psd2-2 psd3-2, were very similar to wild type. To obtain plants carrying T-DNA insertions in all three PS decarboxylase genes, triple mutant lines (psd1 psd2-1 psd3-1 and psd1 psd2-2 psd3-2) were generated by crossing the double mutant lines psd2-1 psd3-1 or psd2-2 psd3-2 with the psd1 single mutant. Growth of the two lines psd1 psd2-1 psd3-1 and psd1 psd2-2 psd3-2 was not different from wild type (data not shown).
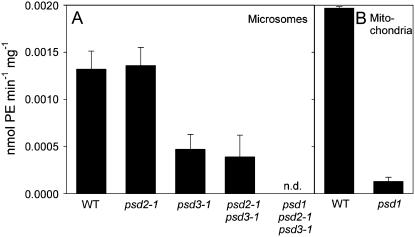
PS decarboxylase activity was measured in microsomal membranes isolated from leaves. The enzyme activities in wild type, psd2-1, and psd2-2 were very similar, indicating that the PSD2 enzyme is responsible for only a minor fraction of the total PS decarboxylase activity in leaves (Fig. 6A; data not shown). This is in agreement with the low expression level of PSD2 (Fig. 4; data not shown). Microsomal PSD activities in psd3-1, psd3-2, and the psd2-1 psd3-1 double mutant were decreased to about one-third of wild-type activity (Fig. 6A; data not shown). Therefore, PSD3 represents the major PS decarboxylase activity in Arabidopsis leaves. PSD activity in microsomal fractions of the psd1 psd2-1 psd3-1 triple mutant was below detection limit. The residual amount of PSD activity found in the psd2-1 psd3-1 double mutant is derived from PSD1, suggesting that the microsomal fraction contained some mitochondrial membranes. The complete absence of PSD activity from the psd1 psd2-1 psd3-1 triple mutant is in agreement with the assumption that this mutant plant carries three null mutations in the PSD genes and that Arabidopsis contains no other membrane-associated protein capable of decarboxylating PS. Mitochondria were isolated from dark-grown seedlings and employed for PSD activity assays (Fig. 6B). PSD activity in mitochondria of the psd1 mutant was decreased to less than 10% of wild-type activity. The very low activity in psd1 mitochondria presumably is derived from PSD2 and PSD3 activities still associated with the mitochondrial preparation.
Figure 6.
PS decarboxylase activity in psd mutants. Plant protein was incubated with [14C]PS, and after lipid extraction [14C]PE production was quantified after TLC and autoradiography. A, PS decarboxylase activity in microsomal fractions. B, PS decarboxylase activity in mitochondria.
Phospholipid Composition in PS Decarboxylase-Deficient Mutants
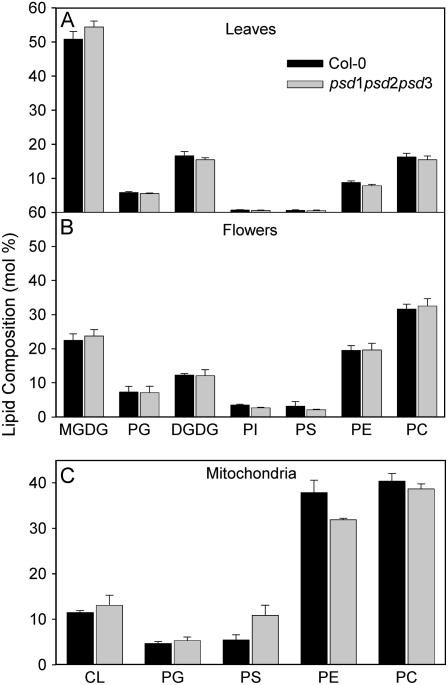
PS decarboxylase activity was shown to be required for the synthesis of a large proportion of PE in yeast and animals. Therefore, a block in PSD gene expression in Arabidopsis mutants was expected to affect the PE content, particularly in mitochondria, possibly accompanied with an increase of the substrate, PS. Total lipids were isolated from leaves of wild type and the single, double, and triple mutant lines (psd1, psd2-1, psd3-1, psd2-1 psd3-1, and psd1 psd2-1 psd3-1) and after separation via two-dimensional thin-layer chromatography (TLC), quantified by gas chromatography (GC) of fatty acid methyl esters. Lipid contents were unchanged in all lines analyzed (Fig. 7A). Therefore, partial or total deficiency in PSD activity in Arabidopsis mutants has no measurable effect on PE synthesis in leaves. The contents of phospholipids in leaves are considerably low because leaves are rich in galactolipids. Therefore, lipid analysis of nongreen plant organs (e.g. flowers) that contain higher proportions of phospholipids might reveal small differences in phospholipid composition that would not be detectable in leaves. Membrane lipid composition of flowers of the psd1 psd2-1 psd3-1 triple mutant was also not changed as compared to wild type (Fig. 7B). Fatty acid composition of PE and PS in plants is characterized by a high proportion of linoleic acid (18:2) and α-linolenic acid (18:3). However, PS isolated from the ER or the plasma membrane of plants was shown to be enriched in very long chain fatty acids, i.e. docosanoic acid (22:0) and tetracosanoic acid (24:0; Bohn et al., 2001; Vincent et al., 2001). Fatty acid composition of PE and PS isolated from whole leaves or flowers in wild type and psd1 psd2-1 psd3-1 was very similar (Table I), suggesting that total deficiency in PS decarboxylase activity did not affect incorporation of lipid precursors from different pools into PE or PS.
Figure 7.
Lipid composition of the psd1 psd2-1 psd3-1 triple mutant. Total lipids were isolated from whole tissues or organelles, separated by TLC, and lipids quantified by GC analysis of fatty acid methyl esters. A, Lipid composition in leaves of wild-type Col-0 and psd1 psd2-1 psd3-1. B, Lipid composition in whole flowers of wild-type Col-0 and psd1 psd2-1 psd3-1. C, Mitochondria were isolated from dark-grown seedlings of wild-type Col-0 and psd1 psd2-1 psd3-1 and used for the determination of phospholipid composition.
Table I.
Fatty acid composition of PE and PS in the psd1 psd2 psd3 triple mutant
Lipids were isolated from leaves or flowers, separated by two-dimensional TLC from wild type or psd1 psd2-1 psd3-1, and quantified by GC of fatty acid methyl esters. Data are given in mol% and represent mean ± sd of three measurements. Very long chain fatty acids (22:0 and 24:0) were only detected in PS but were absent from PE and all other membrane lipids analyzed.
| Fatty Acid | Leaves
|
Flowers
|
||
|---|---|---|---|---|
| Wild-Type Col-0 | psd1 psd2 psd3 | Wild-Type Col-0 | psd1 psd2 psd3 | |
| PE | ||||
| 16:0 | 21.3 ± 2.6 | 21.8 ± 0.5 | 21.3 ± 0.8 | 19.7 ± 1.0 |
| 16:1 | 0.4 ± 0.2 | 0.1 ± 0.1 | 0.5 ± 0.4 | 0.4 ± 0.2 |
| 16:2 | 0.2 ± 0.1 | 0.2 ± 0.0 | 0.3 ± 0.2 | 0.5 ± 0.1 |
| 16:3 | 1.0 ± 0.4 | 0.8 ± 0.5 | 0.1 ± 0.2 | 0.3 ± 0.0 |
| 18:0 | 0.9 ± 0.8 | 1.4 ± 0.1 | 1.6 ± 0.5 | 1.4 ± 0.1 |
| 18:1 | 2.6 ± 0.5 | 2.5 ± 0.1 | 2.6 ± 1.1 | 2.5 ± 0.3 |
| 18:2 | 38.5 ± 1.0 | 38.5 ± 1.1 | 36.1 ± 0.9 | 35.0 ± 0.7 |
| 18:3 | 33.8 ± 3.3 | 33.0 ± 0.9 | 35.8 ± 2.9 | 38.7 ± 2.5 |
| PS | ||||
| 16:0 | 13.9 ± 1.0 | 15.0 ± 2.1 | 16.1 ± 5.2 | 13.6 ± 1.2 |
| 16:1 | 1.4 ± 0.6 | 1.2 ± 0.4 | 0.6 ± 0.4 | 0.8 ± 0.1 |
| 16:2 | 0.9 ± 0.4 | 1.4 ± 1.1 | 0.9 ± 0.2 | 0.7 ± 0.1 |
| 16:3 | 0.6 ± 0.1 | 0.8 ± 0.1 | 0.6 ± 0.8 | 0.2 ± 0.2 |
| 18:0 | 4.5 ± 0.8 | 4.0 ± 0.1 | 4.5 ± 1.1 | 5.3 ± 0.7 |
| 18:1 | 2.7 ± 0.7 | 2.4 ± 0.3 | 2.3 ± 0.5 | 1.8 ± 0.2 |
| 18:2 | 27.6 ± 1.0 | 26.9 ± 1.4 | 27.8 ± 3.1 | 25.0 ± 0.2 |
| 18:3 | 32.4 ± 2.2 | 35.3 ± 2.5 | 38.5 ± 5.1 | 41.6 ± 3.7 |
| 22:0 | 3.8 ± 0.5 | 3.5 ± 0.4 | 2.2 ± 0.2 | 3.1 ± 1.0 |
| 24:0 | 7.2 ± 1.3 | 6.6 ± 1.1 | 1.3 ± 0.1 | 2.0 ± 0.4 |
In yeast, mitochondria represent an essential site for PE synthesis via the PS decarboxylase pathway. To elucidate the role of PS decarboxylase in plants for mitochondrial PE synthesis, mitochondria were isolated from dark-grown seedlings of the Arabidopsis psd1 and psd1 psd2-1 psd3-1 triple mutant and employed for lipid analysis. The amount of PE was slightly decreased in psd1 (data not shown), and it was even further reduced in psd1 psd2-1 psd3-1 (Fig. 7C). Concomitantly, the amount of PS was increased in mitochondria of psd1 psd2-1 psd3-1.
DISCUSSION
Two classes of PS decarboxylases can be distinguished in different organisms based on sequence similarity (Fig. 1). Eukaryotic organisms contain PS decarboxylases that localize to the endomembrane system and carry a long N-terminal extension. A second class of PS decarboxylases includes prokaryotic and mitochondrial enzymes. The close relationship between mitochondrial and bacterial PS decarboxylases can be explained by the common origin according to the endosymbiont hypothesis. Here, we show that in addition to PSD1, which localizes to the mitochondria (Rontein et al., 2003b), Arabidopsis contains two further PS decarboxylases (PSD2 and PSD3) of eukaryotic type that localize to the tonoplast and ER, respectively (Fig. 3). All three enzymes encode functionally active PS decarboxylases (Fig. 2). Analysis of single and multiple T-DNA insertion mutants for all three genes revealed that in leaves, about two-thirds of total PS decarboxylase activity is associated with PSD3, while PSD2 shows only very low activity. PSD1 is the major PS decarboxylase enzyme of mitochondria, and it also contributes to about one-third of total leaf PSD activity (Fig. 6).
PE represents one of the major phospholipids in all organisms. It is abundant in the plasma membrane, ER, tonoplast, nuclear membrane, and mitochondria but is absent from plastids. Three pathways have been described to contribute to PE synthesis: (1) the transfer of phosphoethanolamine from CDP-ethanolamine onto diacylglycerol by aminoalcoholphospho-transferase (Kennedy pathway); (2) decarboxylation of PS by PS decarboxylase; and (3) base exchange reaction of PS with ethanolamine. In yeast, mitochondria are essential for PE production for the entire cell because PS synthesized at the ER is transported to the mitochondria for subsequent decarboxylation by PSD. PE produced in yeast mitochondria is redistributed to the entire cell. No change of PE content was observed in leaves or flowers of the Arabidopsis psd1 psd2 psd3 triple mutant (Fig. 7), indicating that in contrast to other organisms, PS decarboxylase is not a major pathway for PE synthesis in plants. These results are in line with data recently obtained by Mizoi et al. (2006), who showed that the Kennedy pathway predominantly contributes to PE synthesis in Arabidopsis. However, a significant reduction in mitochondrial PE content was observed in the triple mutant psd1 psd2 psd3 (Fig. 7). Therefore, the PS decarboxylase pathway is essential for normal PE accumulation in plant mitochondria. Arabidopsis contains two genes encoding aminoalcoholphospho-transferase that are predicted to localize to the ER and mitochondria, respectively (Goode and Dewey, 1999; Beisson et al., 2003). Furthermore, the fact that aminoalcoholphospho-transferase activity was detected in mitochondria strongly suggests that these organelles are capable of synthesizing PE via the Kennedy pathway (Sparace and Moore, 1981). The reduced PE content had no visible effect on the ultra-structure of mitochondria in leaves or flowers of the triple mutant, as observed by electron microscopy (data not shown). A considerable reduction of PE synthesis in mitochondria was observed in the Arabidopsis psd1 psd2 psd3 triple mutant, but mitochondrial PE content in the psd1 mutant was only slightly affected (Fig. 6). In yeast, PE content was strongly reduced in the yeast psd1 psd2 double mutant, while phospholipid metabolism in the psd1 single mutant was less severely affected. Thus, a fraction of PE in mitochondria originates from PS decarboxylation in mitochondrial membranes, but PS decarboxylation in ER/tonoplast membranes also contributes to mitochondrial PE production. Therefore, similar to yeast, two pathways for PS decarboxylation and transport of phospholipid to the mitochondria occur in Arabidopsis. Presumably, mitochondrial-associated membrane domains of the ER are an important site for PS decarboxylation and phospholipid transfer from the ER to the mitochondria (Schumacher et al., 2002). Furthermore, this scenario might explain why isolated mitochondria of the Arabidopsis psd1 mutant and microsomal membranes of the psd2 psd3 double mutant still contained residual PSD activity. Therefore, similar to yeast and animal cells, a specific transport system must exist in plants involved in phospholipid transfer from the ER to the mitochondria (Heikinheimo and Somerharju, 1998; Wu and Voelker, 2001). However, the factors regulating ER to mitochondria lipid trafficking in plants are unknown and will be the focus of future studies.
MATERIALS AND METHODS
Origin of PS Decarboxylase cDNAs
The PSD1 (At4g16700) full-length open reading frame was amplified from first-strand cDNA derived from Arabidopsis (Arabidopsis thaliana) leaf mRNA (Superscript cDNA synthesis kit; Invitrogen) by PCR using the primers PD288 and PD246 (Table II). The PCR product was ligated into the SphI, HindIII sites of pQE30 (Qiagen), resulting in the vector pQE30-PSD1. The coding region of PSD2 (At5g57190) was amplified from single-strand cDNA by PCR with the primers PD97 and PD98, thereby introducing SphI and KpnI sites at the 5′ end and 3′ end, respectively. The PSD2 PCR fragment was subcloned into pGEMTeasy (Promega), resulting in the construct pGEMTeasy-PSD2 and sequenced on both strands. The EST (GenBank accession no. AV527283) of PSD3 (At4g25970) was obtained from the Kazusa DNA Research Institute. Sequencing revealed that this clone contained the authentic PSD3 cDNA in the EcoRI and XhoI sites of pBluescriptIISK−. The full-length coding sequences for At-PSD2 (At5g57190) and At-PSD3 (At4g25970) were deposited in GenBank with the accession numbers EF203902 and EF203901, respectively.
Table II.
Oligonucleotides used for PCR amplification
Sequences of primers are presented in 5′ to 3′ orientation.
| Oligonucleotide | Sequence |
|---|---|
| PD97 | CAT GCA TGC GTT TAT CTT CCT GGT GCG TTA GG |
| PD98 | GTC GGT ACC TCA GTG GCA AAG ATT TTC AGA TC |
| QP1ECF | CAC GAG CTC AAA ATA GGA CTC CGG |
| QP1R | CAC TCT GCA GGC TCA AGT ACA C |
| LBb1 | GCG TGG ACC GCT TGC TGC AAC T |
| PD90 | ATG CTC GCT CAA CTG CTC ATG AAG TAT AG |
| PD91 | ATG CTC GCT CAA CTG CTC ATG AAG TAT AG |
| PD88 | CTG CGA CAT TCT CCA AAG GAA ATG ATC AAC |
| PD89 | GTT CAA CTC AAC AGA GAA TGA GAG AGC CA |
| JL202 | CAT TTT ATA ATA ACG CTG CGG ACA TCT AC |
| PD185 | GAA ATC TAA CCC ATA TGA AGC CTG CCT AT |
| Lb3Fa | TAG CAT CTG AAT TTC ATA ACC AAT CTC GAT ACA C |
| PD288 | AAA GCA TGC ATG AAA CCT CGT TTT CC TCA AAA |
| PD246 | GGT AAG CTT CAT TCC TCT TTC CAT CTT CCC AA |
| PD243 | GCC GAA TTC ATG AAA CCT CGT TTT CCT CAA AA |
| PD244 | AAT GGA TCC CGT TCC TCT TTC CAT CTT CCC AA |
| ZP2F | CAC GAA TTC AAC ATG CGT TTA TCT TCC |
| ZP2R | CTC ATG ATC AAC GAT CAA TGT AGA CCT AG |
| ZP1F | CTC AGG TAC CGA GAA TCA TGG G |
| ZP1R | CTC ATG ATC AAC GGG CTC AAG TAC AC |
| MCS-1 | CCA TGG ATC TAG ATC TGC AGG TAC CAA TTG AAG CTT |
| MCS-2 | AAG CTT CAA TTG GTA CCT GCA GAT CTA GAT CCA TGG |
| PD344 | GCCACTGAACTGATTACCAGAGA |
| PD345 | GAGTTATTATGCACGAAACAAGAGG |
| PD348 | CACACTCCACTTGGTCTTGCGT |
| PD349 | TGGTCTTTCCGGTGAGAGTCTTCA |
| PD243 | GCCGAATTCATGAAACCTCGTTTTCCTCAAAA |
| PD244 | AATGGATCCCGTTCCTCTTTCCATCTTCCCAA |
PS Decarboxylase Assay
PS decarboxylase activity was measured by incubating protein extracts with radioactive PS (sn-1,2-dioleoyl-3-phosphoryl-l-[3-14C]Ser, Amersham). Each reaction (total volume of 200 μL) contained 1 mg protein in 50 mm Tris-HCl, pH 7.0, 0.1% Triton X-100, 0.05 μCi (130 ng) radioactive PS, and 1.3 μg nonradioactive PS. Reactions were incubated at 28°C for 10 min. Lipids were extracted after addition of 100 μL 1 m KCl, 0.2 m H3PO4, and 200 μL chloroform:methanol (2:1) and separated by TLC on silica plates according to Hawrot and Kennedy (1975) in chloroform:methanol:water (65:25:4). The conversion of PS to PE was analyzed by autoradiography using the Phosphoimager system (BAS1500; Fuji).
Lipid Measurements
Lipids were extracted from leaves and flowers with chloroform-methanol and separated via two-dimensional TLC with chloroform:methanol:water (65:25:4) and chloroform:acetone:methanol:acetic acid:water (50:20:10:10:5) according to Benning et al. (1995). Lipids isolated from mitochondria were separated by one-dimensional TLC as described in Dörmann et al. (1995). Lipids isolated from TLC were methylated with HCl/methanol and methyl esters quantified by GC using pentadecanoic acid (15:0) as internal standard (Browse et al., 1986).
Yeast Complementation
The yeast (Saccharomyces cerevisiae) strains were RYY51 (ura3 his3 trp1 leu2 lys2 psd1-1∷TRP1 psd2-1∷HIS3) and the corresponding wild type, SEY6210 (ura3 his3 trp1 leu2 lys2; Trotter and Voelker, 1995). The vector for plant PSD constructs was pVT103-U, which contains the URA-3 gene for selection and the ADH1 promoter to drive gene expression (Vernet et al., 1987). The full open reading frames for Arabidopsis PSD2 and PSD3 were amplified from the respective cDNAs by PCR using Pfu DNA polymerase and subcloned into pVT103-U for expression in yeast.
Expression of PSD cDNAs in Escherichia coli
The full-length PSD3 cDNA was released from pBluescriptIISK− (see above) with XbaI and KpnI and ligated into pKK-MCS. This vector is a derivative of pKK233-2 (GenBank accession no. U02439; Amann and Brosius, 1985) into which a new polylinker containing NcoI, XbaI, BglII, PstI, KpnI, MfeI, and HindIII restriction sites (generated by annealing of the oligonulceotides MCS-1 and MCS-2; Table II) was inserted into the NcoI and HindIII sites. The C-terminal part of PSD3 was amplified by PCR with the primers QP1ECF and QP1R and ligated into the SacI and PstI sites of pQE100 (Qiagen). Subsequently, the cDNA was released with BamHI and BglII and ligated into the BglII site of pKK-MCS. Constructs were transformed into the E. coli strain EH150 carrying a temperature-sensitive mutation of the psd gene (Hawrot and Kennedy, 1976). Protein expression in E. coli was induced with 0.5 mm isopropylthio-β-galactoside and cells harvested by centrifugation (6,000g, 5 min). After resuspending in 100 mm KH2PO4, pH 7.0, cells were lysed by ultrasound. Total protein extracts (35 μg protein) were used per assay. In contrast to assays done with plant protein (see above), PSD activity of transformed E. coli EH150 cells was measured at 42°C.
Isolation of Microsomes and Mitochondria
Arabidopsis leaves were homogenized in 50 mm Tris-HCl, pH 7.0, at 4°C by grinding in a mortar with sand. Cell debris was separated by centrifugation (1,200g, 1 min). After centrifugation of the supernatant at 100,000g for 1 h, a microsomal membrane fraction was obtained by resuspending the pellet in 50 mm Tris-HCl, pH 7.0.
A simplified method of Klein et al. (1998) was used for isolation of mitochondria. Briefly, Arabidopsis seeds were germinated and the plants grown in 300 mL of 2× Murashige and Skoog (1962) medium, 3% (w/v) Suc, 20 mm isobutyric acid for 3 weeks in darkness. Plant material was harvested and homogenized with an ultra-turrax in extraction buffer (450 mm Suc, 1.5 mm EGTA, 15 mm MOPS-KOH, pH 7.4, 10 mm dithiothreitol, and 6 g/L polyvinylpyrrolidone). After filtration through Miracloth (Calbiochem), cell debris was removed by centrifugation (2,000g, 5 min). A crude mitochondrial pellet was obtained by centrifugation (16,000g, 10 min), resuspended in washing buffer (300 mm Suc, 1 mm EGTA, 10 mm MOPS-KOH, pH 5.2), and loaded onto a Percoll step gradient (18%, 23%, and 40%). After centrifugation at 12,000g for 45 min, mitochondria were harvested at the 40%/23% Percoll interphase. Mitochondria were washed in resuspension buffer (400 mm mannitol, 1 mm EGTA, 10 mm Tricine-KOH, pH 7.2) and centrifuged again (14,000g, 10 min). Protein concentration in fractions used for enzyme assays was determined according to Bradford (1976).
Arabidopsis Growth Conditions and Isolation of T-DNA Insertion Mutants
Arabidopsis plants were grown on soil at 120 μmol m−2 s−1, 16 h light/day, 60% humidity, and 20°C. A mutant carrying a T-DNA insertion in the PSD1 gene (Salk_064716) was obtained from the SALK collection (Arabidopsis Biological Resource Center, The Ohio State University, Columbus, OH; Alonso et al., 2003; ecotype Columbia [Col-0]). PCR analysis with PSD1 gene-specific primers (PD189 and PD176) and an insert-specific primer (LBb1) was used to isolate homozygous mutant plants.
The α mutant population of the University of Wisconsin (ecotype Wassilewskija; Madison, WI; Krysan et al., 1999) was screened for plants carrying T-DNA insertions in the genes PSD2 and PSD3. Screening was done by PCR of pooled genomic DNA using the primers PD90 and PD91 for PSD2 and the primers PD88 and PD89 for PSD3, each in combination with the T-DNA-specific primer JL202 (Table II). The mutants were identified as plant PD90#11 in pool CSJ4362 and plant PD89#20 in pool CSJ2259 for psd2 and psd3, respectively.
Additional mutant alleles carrying T-DNA insertions in the genes PSD2 and PSD3 were obtained from Syngenta (SAIL 93b_G04.b.1a and SAIL 851_E09.b.1a, respectively, ecotype Col-0; Sessions et al., 2002). Identification of homozygous mutant plants was done by PCR analysis with gene-specific primers and an insertion-specific primer (Lb3Fa).
The position of the insertion in the genomic region was confirmed by sequencing of the PCR fragments. Homozygous mutant lines were obtained by segregation and PCR analysis. PCR amplification of homozygous plants using the gene-specific and insert-specific primers resulted in the synthesis of the fragment containing the genomic DNA/T-DNA junction, but due to the large size of the insertion, no band was obtained when using the two gene-specific primers. Furthermore, Southern-blot hybridization using genomic probes confirmed that mutant plants carried homozygous insertions.
Northern Blot and Semiquantitative PCR
Northern blotting was done with total RNA isolated from different Arabidopsis organs (Sambrook et al., 1989). The blots were hybridized with different PSD cDNA probes (see above). RNA for semiquantitative real-time PCR was isolated using the Plant RNA Purification Reagent kit (Invitrogen/Gibco BRL). Residual DNA was removed with RNAse-free DNAse (Roche Diagnostics GmbH), and mRNA isolated with the Oligotex mRNA Mini kit (Qiagen). Isolated mRNA was used as template for the synthesis of cDNAs with the Superscript III First-Strand Synthesis system for RT-PCR kit (Invitrogen/Gibco BRL). Semiquantitative real-time PCR was done as described for PCR (see above) using the primer pairs PD344/PD345 and PD348/PD349 for PSD2 and ubiquitin, respectively. The expected PDS2 amplicon was approximately 120 bp of size.
Confocal Microscopy of GFP Fusion Proteins
The PSD1 cDNA was amplified from clone pQE30-PSD1 with the primers PD243 and PD244 (Table II) and ligated into the EcoRI and BamHI sites of pEZR-K-LN (Gert-Jan de Boer, Carnegie Institution, Stanford, California) as an N-terminal, translational fusion with GFP. The PCR fragment of PSD2 obtained by PCR amplification of pGEMTeasy-PSD2 with the primers ZP2F and ZP2R was digested with EcoRI and BclI and ligated into the EcoRI and BamHI sites of pEZR-K-LN, resulting in the plasmid pEZR-PSD2. The PSD3 cDNA was amplified by PCR using the primers ZP1F and ZP1R from clone AV527283, introducing new KpnI and BclI restriction sites. This fragment was ligated into the KpnI, BamHI sites of pEZR-K-LN, resulting in the plasmid pEZR-PSD3.
Plasmid DNA was coated onto 1.0-μm gold micro-carriers (Bio-Rad). Transfer into Arabidopsis leaves was achieved by bombardment in a Biolistic PSD-1000/He particle delivery system (Bio-Rad) using 1,100 psi rupture discs according to the manufacturer's protocol. Leaves were incubated on Murashige and Skoog (1962) medium in the dark for 2 d and fluorescence signals analyzed under a Leica DM-IRBE confocal microscope with excitation at 495 nm and emission at 519 nm. Fusion constructs with GFP or DsRed as controls were obtained for localization to the ER (KDEL-GFP; Scott et al., 1999) and to the mitochondria (pre-101-GFP) and tonoplast (pTPK1-DsRed2; Katrin Czempinski and Bernd Müller-Röber, University of Potsdam, Germany).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EF203901 and EF203902.
Acknowledgments
We thank Professor Elmar Hartmann (Free University of Berlin) for his generous support of this project in its initial phase. The constructs pre-101-GFP and pTPK1-DsRed2 were kindly provided by Katrin Czempinski and Bernd Müller-Röber, University of Potsdam, Germany.
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. SFB 429, B6) and by the Max Planck Society.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Peter Dörmann (doermann@mpimp-golm.mpg.de).
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Amann E, Brosius J (1985) ATG vectors for regulated high-level expression of cloned genes in Escherichia coli. Gene 40 183–190 [DOI] [PubMed] [Google Scholar]
- Beisson F, Koo AJK, Ruuska S, Schwender J, Pollard M, Thelen JJ, Paddock T, Salas JJ, Savage L, Milcamps A, et al (2003) Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol 132 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C, Huang Y-H, Gage DA (1995) Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch Biochem Biophys 317 103–111 [DOI] [PubMed] [Google Scholar]
- Birner R, Bürgermeister M, Schneiter R, Daum G (2001) Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol Biol Cell 12 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn M, Heinz E, Lüthje S (2001) Lipid composition and fluidity of plasma membranes isolated from corn (Zea mays L) roots. Arch Biochem Biophys 387 35–40 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Browse J, McCourt PJ, Somerville CR (1986) Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem 152 141–145 [DOI] [PubMed] [Google Scholar]
- Clancey CJ, Chang S-C, Dowhan W (1993) Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant. J Biol Chem 268 24580–24590 [PubMed] [Google Scholar]
- Delhaize E, Hebb DM, Richards KD, Lin J-M, Ryan PR, Gardner RC (1999) Cloning and expression of a wheat (Triticum aestivum L) phosphatidylserine synthase cDNA overexpression in plants alters the composition of phospholipids. J Biol Chem 274 7082–7088 [DOI] [PubMed] [Google Scholar]
- Dennis EA, Kennedy EP (1972) Intracellular sites of lipid synthesis and the biogenesis of mitochondria. J Lipid Res 13 263–267 [PubMed] [Google Scholar]
- Dörmann P, Hoffmann-Benning S, Balbo I, Benning C (1995) Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7 1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K, Umeda M (2000) An essential role for a membrane lipid in cytokinesis: regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine. J Cell Biol 149 1215–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode JH, Dewey RE (1999) Characterization of aminoalcoholphosphotransferases from Arabidopsis thaliana and soybean. Plant Physiol Biochem 37 445–457 [Google Scholar]
- Hawrot E, Kennedy EP (1975) Biogenesis of membrane lipids: mutants of Escherichia coli with temperature-sensitive phosphatidylserine decarboxylase. Proc Natl Acad Sci USA 72 1112–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrot E, Kennedy EP (1976) Conditional lethal phosphatidylserine decarboxylase mutants of Escherichia coli. Mapping of the structural gene for phosphatidylserine decarboxylase. Mol Gen Genet 148 271–279 [DOI] [PubMed] [Google Scholar]
- Hawrot E, Kennedy EP (1978) Phospholipid composition and membrane function in phosphatidylserine decarboxylase mutants of Escherichia coli. J Biol Chem 253 8213–8220 [PubMed] [Google Scholar]
- Heikinheimo L, Somerharju P (1998) Preferential decarboxylation of hydrophilic phosphatidylserine species in cultured cells. Implications on the mechanism of transport to mitochondria and cellular aminophospholipid species compositions. J Biol Chem 273 3327–3335 [DOI] [PubMed] [Google Scholar]
- Klein M, Binder S, Brennicke A (1998) Purification of mitochondria from Arabidopsis. Methods Mol Biol 82 49–53 [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge O, Nishijima M, Akamatsu Y (1991) A cloned gene encoding phosphatidylserine decarboxylase complements the phosphatidylserine biosynthetic defect of a Chinese hamster ovary cell mutant. J Biol Chem 266 6370–6376 [PubMed] [Google Scholar]
- Li Q-X, Dowhan W (1988) Structural characterization of Escherichia coli phosphatidylserine decarboxylase. J Biol Chem 263 11516–11522 [PubMed] [Google Scholar]
- Marshall MO, Kates M (1973) Biosynthesis of phosphatidyl ethanolamine and phosphatidyl choline in spinach leaves. FEBS Lett 31 199–202 [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Okakda M, Horikoshi Y, Matsuzaki H, Kishi T, Itaya M, Shibuya I (1998) Cloning, sequencing, and disruption of the Bacillus subtilis psd gene coding for phosphatidylserine decarboxylase. J Bacteriol 180 100–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoi J, Nakamura M, Nishida I (2006) Defects in CTP:phosphorylethanolamine cytidylyltransferase affect embryonic and postembryonic development in Arabidopsis. Plant Cell 18 3370–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore TS Jr (1975) Phosphatidylserine synthesis in castor bean endosperm. Plant Physiol 56 177–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Pavlidis P, Ramaswami M, Tanouye MA (1994) The Drosophila easily shocked gene: A mutation in a phospholipid synthetic pathway causes seizure, neuronal failure, and paralysis. Cell 79 23–33 [DOI] [PubMed] [Google Scholar]
- Rontein D, Nishida I, Tashiro G, Yoshioka K, Wu W-I, Voelker DR, Basset G, Hanson AD (2001) Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme. J Biol Chem 276 35523–35529 [DOI] [PubMed] [Google Scholar]
- Rontein D, Rhodes D, Hanson AD (2003. a) Evidence from engineering that decarboxylation of free serine is the major source of ethanolamine moieties in plants. Plant Cell Physiol 44 1185–1191 [DOI] [PubMed] [Google Scholar]
- Rontein D, Wu W-I, Voelker DR, Hanson AD (2003. b) Mitochondrial phosphatidylserine decarboxylase from higher plants. Functional complementation in yeast, localization in plants, and overexpression in Arabidopsis. Plant Physiol 132 1678–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schumacher MM, Choi J-Y, Voelker DR (2002) Phosphatidylserine transport to the mitochondria is regulated by ubiquitination. J Biol Chem 277 51033–51042 [DOI] [PubMed] [Google Scholar]
- Scott AC, Wyatt S, Tsou P-L, Robertson N, Allen NS (1999) A model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques 26 1125–1132 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparace SA, Moore TS Jr (1981) Phospholipid metabolism in plant mitochondria. II. Submitochondrial sites of synthesis of phosphatidylcholine and phosphatidylethanolamine. Plant Physiol 67 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen R, Nanowski TS, Beigneux A, Kulinski A, Young SG, Vance JE (2005) Disruption of the phosphatidylserine decarboxylase gene in mice causes embryonic lethality and mitochondrial defects. J Biol Chem 280 40032–40040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter PJ, Pedretti J, Voelker DR (1993) Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J Biol Chem 268 21416–21424 [PubMed] [Google Scholar]
- Trotter PJ, Pedretti J, Voelker DR (1995) Phosphatidylserine decarboxylase 2 of Saccharomyces cerevisiae. Cloning and mapping of the gene, heterologous expression, and creation of the null allele. J Biol Chem 270 6071–6080 [DOI] [PubMed] [Google Scholar]
- Trotter PJ, Voelker DR (1995) Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J Biol Chem 270 6062–6070 [DOI] [PubMed] [Google Scholar]
- Vance JE, Steenbergen R (2005) Metabolism and functions of phosphatidylserine. Prog Lipid Res 44 207–234 [DOI] [PubMed] [Google Scholar]
- van Golde LMG, Raben J, Batenburg JJ, Fleischer B, Zambrano F, Fleischer S (1974) Biosynthesis of lipids in Golgi complex and other subcellular fractions from rat liver. Biochim Biophys Acta 360 179–192 [DOI] [PubMed] [Google Scholar]
- Vernet T, Dignar D, Thomas DY (1987) A family of yeast expression vectors containing the phage f1 intergenic region. Gene 52 225–233 [DOI] [PubMed] [Google Scholar]
- Vincent P, Maneta-Peyret L, Cassagne C, Moreau P (2001) Phosphatidylserine delivery to endoplasmic reticulum-derived vesicles of plant cells depends on two biosynthetic pathways. FEBS Lett 498 32–36 [DOI] [PubMed] [Google Scholar]
- Voelker DR (1997) Phosphatidylserine decarboxylase. Biochim Biophys Acta 1348 236–244 [DOI] [PubMed] [Google Scholar]
- Webb MS, Green BR (1991) Biochemical and biophysical properties of thylakoid acyl lipids. Biochim Biophys Acta 1060 133–158 [Google Scholar]
- Wu W-I, Voelker DR (2001) Characterization of phosphatidylserine transport to the locus of phosphatidylserine decarboxylase 2 in permeabilized yeast. J Biol Chem 276 7114–7121 [DOI] [PubMed] [Google Scholar]