Abstract
A lectin different from the previously described mannose-binding agglutinins has been isolated from the liverwort Marchantia polymorpha. Biochemical characterization of the purified lectin combined with the data from earlier transcriptome analyses demonstrated that the novel M. polymorpha agglutinin is not related to any of the known plant lectin families, but closely resembles the Agaricus bisporus-type lectins, which hitherto have been found exclusively in fungi. Immunolocalization studies confirmed that lectin is exclusively associated with plant cells, ruling out the possibility of a fungal origin. Extensive screening of publicly accessible databases confirmed that, apart from fungi, the occurrence of A. bisporus-type lectins is confined to M. polymorpha and the moss Tortula ruralis. Expression of a typical fungal protein in a liverwort and a moss raises the question of the origin of the corresponding genes. Regardless of the evolutionary origin, the presence of a functional A. bisporus lectin ortholog in M. polymorpha provides evidence for the expression of an additional carbohydrate-binding domain in Viridiplantae.
Modern plant biology research provided a fairly detailed overview of the occurrence and identity of carbohydrate-binding proteins in plants. Apart from a few exceptions, all of the approximately 500 currently known plant lectins can be classified into seven families of structurally and evolutionarily related proteins (for reviews, see Van Damme et al., 1998, 2004). Three lectin families, namely, the amaranthins, the jacalin-related lectins, and the Cucurbitaceae phloem lectins, are apparently confined to plants. Until recently, legume lectins were also considered typical plant proteins because no similar lectins were found in other organisms. However, structural analyses (Velloso et al., 2003) clearly demonstrated that the so-called endoplasmic reticulum-Golgi intermediate compartment proteins, which are absent from plants, but found in animals and fungi (Dodd and Drickamer, 2001), share a common ancestor and, accordingly, should be considered homologs of the legume lectins. In contrast, homologs of the monocot Man-binding lectins have been isolated from a fish (Fugu rubripes; Tsutsui et al., 2003) and from the slimemold Dictyostelium discoideum (Jung et al., 1996). Hevein domains have also been identified in several proteins from metazoa (e.g. Caenorhabditis elegans) and fungi, whereas domains corresponding to the sugar-binding chain of type-2 ribosome-inactivating proteins are widespread among both prokaryotes and eukaryotes. These observations raise some important questions about the evolutionary origin of plant lectins. Apart from the ricin-B domain and, to a lesser extent the hevein domain, there is little evidence for classical vertical inheritance from a common prokaryotic ancestor. In addition, evolution of the lectin families within the Viridiplantae is still poorly understood because, apart from a few exceptions, all plant lectins were identified in flowering plants. For the time being, the documented occurrence of lectins (with a known sequence) outside flowering plants is limited to jacalin-related lectins in a cycad (Cycas revoluta; Yagi et al., 2002), a fern Phlebodium aureum (Tateno et al., 2003), and a monocot Man-binding lectin in the gymnosperm Taxus media (Kai et al., 2004) and the liverwort Marchantia polymorpha (Peumans et al., 2002).
Here, we present evidence that the liverwort M. polymorpha and the moss Tortula ruralis express proteins that can be considered orthologs of the Agaricus bisporus lectin family. Biochemical characterization of the purified lectin confirmed that some of the orthologs expressed by M. polymorpha are fully active agglutinins with a carbohydrate-binding specificity similar to that of the A. bisporus agglutinin (ABA). Immunolocalization studies clearly demonstrated that lectin is exclusively associated with M. polymorpha cells, ruling out the possibility that the protein is produced by a contaminating fungus. These findings not only provide firm evidence for the expression in plants of a lectin type that hitherto has been found exclusively in fungi, but also add a novel family to the list of previously identified plant lectin families. No other plant sequences could be identified that encode similar proteins, indicating that within the Viridiplantae the occurrence of the A. bisporus-type lectin is confined to lower plants. The impact of these novel findings on the unraveling of the evolution of lectins in general and plant lectins in particular is discussed.
RESULTS AND DISCUSSION
ABA Orthologs Are Found in the Liverwort M. polymorpha and the Moss T. ruralis
Screening publicly accessible databases for the occurrence in lower plants of homologs/orthologs of lectins found in flowering plants and fungi led to the identification of ESTs from the liverwort M. polymorpha and the moss T. ruralis that encode proteins with marked sequence similarity (>40% and >70% sequence identity and similarity, respectively) to ABA. To corroborate the possible occurrence of other nonfungal ABA-related lectins, protein and DNA databases were extensively searched using the sequences of all known fungal and M. polymorpha and T. ruralis ABA orthologs as a query. No additional nonfungal ABA-related proteins could be retrieved.
Immature Female Sex Organs of the Liverwort M. polymorpha Express Multiple ABA Orthologs
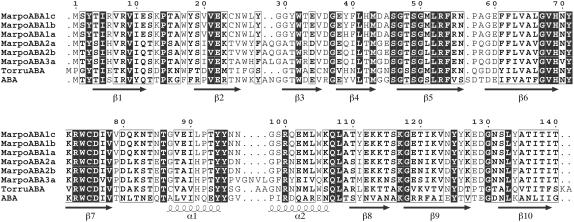
A total number of 17 ESTs encoding proteins similar to ABA were identified in the M. polymorpha EST database (containing a total number of 1,415 EST entries). Due to high overall sequence identity/similarity, the M. polymorpha proteins are considered ABA orthologs. All 17 ESTs were retrieved from a library prepared from immature female sex organs (Nagai et al., 1999). None was found in the library prepared from immature male sex organs (Nishiyama et al., 2000). In the library from immature female sex organs, the ESTs encoding ABA orthologs represent 1.75% of the library total (970). The occurrence of ESTs encoding proteins similar to ABA was already noticed by Nagai et al. (1999) in their original report of the analysis of ESTs from M. polymorpha immature female sexual organs. According to this report, these ESTs were grouped in four contigs (nos. 3, 17, 51, and 55). However, a more detailed analysis of the nucleotide sequences revealed that these 17 ESTs should be assembled into five different contigs (see Supplemental Figs. S1–S3), which, according to the length of the encoded polypeptide, can be subdivided into two subgroups. The first subgroup comprises three contigs (referred to as MarpoABA1a, MarpoABA1b, and MarpoABA1c, respectively; see Supplemental Figs. S1 and S2) encoding an ABA ortholog of 140 amino acid residues (Fig. 1). The second subgroup comprises two contigs (referred to as MarpoABA2a and MarpoABA2b, respectively), encoding an ABA ortholog of 142 amino acid residues (see Supplemental Fig. S3).
Figure 1.
Structural alignment of the amino acid sequences of M. polymorpha ABA orthologs (MarpoABA), the Tortula ABA ortholog (TorruABA), and ABA. Strands of β-sheet (β1–β10) and stretches of α-helix (α1, α2) occurring along the polypeptide chain of ABA have been indicated.
Analyses of the complete deduced amino acid sequences allowed calculating the major physicochemical parameters of the corresponding proteins (see Supplemental Table S1) and, in addition, indicated that all M. polymorpha ABA orthologs are synthesized without a signal peptide.
Rehydrating Gametophytes of the Desiccation-Tolerant Bryophyte T. ruralis Express a Single ABA Ortholog
Two ESTs encoding a putative ortholog of ABA were identified in the T. ruralis EST database (containing a total number of 9,306 EST entries). Both ESTs have an identical nucleotide sequence and encode a protein of 144 residues (further referred to as TorruABA). The deduced sequence of TorruABA shares approximately 50% and 80% sequence identity and similarity, respectively, with M. polymorpha ABA orthologs and lacks a signal peptide. TorruABA aligns best with the MarpoABA1 group (see Supplemental Fig. S4). The major physicochemical parameters of TorruABA could be calculated from the complete deduced amino acid sequence (see Supplemental Table S1).
PCR Amplification and Sequence Analysis of Genomic Sequences Encoding M. polymorpha ABA Orthologs
To confirm the presence of genomic sequences corresponding to the contigs assembled from the deposited ESTs, DNA was extracted from young female sexual organs of M. polymorpha and genomic sequences encoding the ABA orthologs amplified by PCR. Analysis of the PCR fragments confirmed the presence in the genome of sequences corresponding to the open reading frames (ORFs) of MarpoABA1a, MarpoABA1b, MarpoABA1c, and MarpoABA2a and MarpoABA2b. In addition, a PCR fragment with a novel sequence representing a third subgroup (referred to as MarpoABA3a; see Supplemental Fig. S5) encoding a lectin polypeptide of 146 amino acid residues was obtained (Fig. 1). Although it cannot be precluded that some genomic sequences were not amplified or detected, the results of PCR amplification experiments confirm that, as could already be inferred from the EST data, the expression of ABA orthologs in M. polymorpha is controlled by a gene family.
PCR amplification experiments were also set up with DNA isolated from an axenic culture of M. polymorpha. Analysis of the amplified fragments confirmed that the DNA of the axenically grown M. polymorpha cells contains the same lectin sequences as the DNA from the field-grown sample, which strongly indicated that lectin is synthesized by plant cells and not derived from a possible fungal contaminant.
A Previously Characterized M. polymorpha Lectin Corresponds to MarpoABA2
The first lectin to be characterized from a lower plant was isolated from the liverwort M. polymorpha. Using a combination of classical protein purification techniques, Adam and Becker (1993) purified a lectin from M. polymorpha gametophytes and characterized the protein in some detail. The lectin behaved as a monomeric protein with a Mr of 16,135 (as determined by mass spectrometry [MS]) and is not glycosylated. Agglutination assays further indicated that the lectin agglutinates erythrocytes of different mammalia and that this agglutination activity cannot be inhibited by any simple sugar but only by complex carbohydrate structures. Both the Mr of the polypeptide and the specificity of the lectin are reminiscent of the ABA-type fungal lectin. Moreover, the molecular mass as determined by MS is almost identical to the values calculated from the deduced sequences of M. polymorpha ABA (Supplemental Table S1). It is very likely, therefore, that the M. polymorpha lectin described by Adam and Becker (1993) corresponds to an ABA ortholog.
To corroborate the identity, lectin was reisolated and characterized. The purified protein yielded a single polypeptide band of approximately 15 kD upon SDS-PAGE (data not shown) and eluted with an apparent Mr of approximately 30 kD upon gel filtration on a Superose-12 column, indicating that the native lectin is a dimeric protein.
MS of the purified lectin yielded a main peak of 16,102.0 ± 0.55 D and three minor peaks with Mrs of 16,118.5 (±1.22) D, 16,133.0 (±1.65) D, and 16,350.0 (±0.63) D, respectively (see Supplemental Fig. S6). Because the protein was N-terminally blocked (as was already reported by Adam and Becker, 1993), partial sequencing was achieved by MS. The purified lectin sample was digested with trypsin and the fragments were analyzed by MS and tandem MS (MS/MS). Comparison of the peptide mass fingerprint with the calculated masses of the tryptic peptides of the different M. polymorpha ABA orthologs indicated that the major component of purified lectin corresponds to MarpoABA2b. MarpoABA2a, which is identical to MarpoABA2b except that residue 136 is Tyr instead of Phe (and hence has a mass that is 16 D higher), most probably corresponds to the second peak (16,118.5 D) in the electrospray ionization (ESI) spectrum (which differs by 16 D from the first peak). The peak at 16,133.0 D in the ESI spectrum might represent an oxidized form. No match could be found for the 16,350.0-D component. Possibly, this peak is due to the presence of a yet unidentified molecular species.
Analysis of the tryptic peptides by MS/MS yielded no indication of the presence of the (expressed) MarpoABA1 group. This obvious absence from affinity-purified lectin preparation is in good agreement with the results of modeling/docking experiments (see below), which indicated that the major carbohydrate-binding site of MarpoABA1 isoforms is (in contrast to that of the MarpoABA2 isoforms) not functional so that they are not capable of binding the immobilized ligands.
The data presented in this section demonstrate that lectin isolated from M. polymorpha corresponds to the MarpoABA2 orthologs. Although the M. polymorpha ABA ortholog cannot be considered a novel type of lectin, its identification provides firm evidence for the expression of a typical fungal lectin in a (lower) plant.
Carbohydrate-Binding Specificity of the M. polymorpha ABA Ortholog
Hapten inhibition assays indicated that the agglutination activity of the M. polymorpha ABA ortholog is not sensitive to any common simple mono- or oligosaccharide, but is completely inhibited by low concentrations (in the μg/mL range) of animal glycoproteins like asialomucin and asialofetuin. These observations confirm the results of the preliminary specificity studies reported by Adam and Becker (1993), but do not allow comparison of the specificity of the M. polymorpha lectin to that of its fungal counterparts. Therefore, the specificity of the M. polymorpha ABA orthologs was corroborated in more detail using the high-performance glycan array systems developed by the Consortium for Functional Glycomics (see Supplemental Fig. S7). In screening with the printed array (PA) platform, lectin strongly interacted with nine glycans (Table I). All nine glycans contain β-d-Gal or α-Gal/N-acetyl galactosamine (GalNAc). Unsubstituted β-d-Gal and α-GalNAc were nearly equally reactive. Addition of β1-3-linked Gal to α-GalNAc (giving rise to Galβ1-3GalNAcα or Thomson Friedenreich or T-antigen) slightly increased the interaction with lectin. Substitution of T at O3 with sulfate, β1-3-linked GlcNAc, or α2-3-linked N-acetyl neuraminic acid (Neu5Ac) had no negative effect, indicating that lectin tolerates substitution. The same holds true for substitution of the α-GalNAc at O6 with sulfate or α2-6-linked Neu5Ac. In contrast, substitution of α-GalNAc at O6 with β1-6-linked NeuAc reduced binding by approximately one-half. The results of the ELISA array (EA) were less clear cut. Of the six glycans common to both the PA and the EA, only two, namely, T-antigen and Sulfo-T, showed binding on the EA. These obvious differences in reactivity most likely rely on differences in both the construction of the two glycan arrays and the binding assay itself. First, the PA has a higher density (though unknown amount) of glycans in each spot than the EA. Second, the lectin concentration used in the screening was 10 times higher in the PA assay than in the EA assay (300 μg mL−1 and 30 μg mL−1, respectively). As a result, the effect of a possible multivalency of the lectin-glycan interactions is much higher in the PA assay than in the EA assay. The fact that a number of glycan epitopes that do not bind in the EA assay exhibit a clear reaction in the PA assay strongly indicates that the presence of high-density polyvalent glycotopes strongly enhances the affinity of the lectin. Both the identity of the reactive glycans and the apparent preferential binding to multivalent epitopes indicate that the M. polymorpha lectin closely resembles ABA for what concerns its specificity (Wu et al., 2003).
Table I.
Overview of the results of the binding assays of MarpoABA to the Consortium glycan arrays
| Glycans | PA
|
EA
|
||
|---|---|---|---|---|
| Glycan No.a | RFUb | Glycan No.a | S/Nb | |
| Neu5Acα2-3Galβ1-3[6OSO3]GalNAcα- | 163 | 40,809 | – | – |
| Neu5Acα2-3Galβ1-3GalNAcα- | 166 | 39,096 | – | – |
| [3OSO3]Galβ1-3GalNAcα- | 11 | 38,911 | 161 | 1.40 |
| GlcNAcβ1-3Galβ1-3GalNAcα- | 122 | 37,850 | – | – |
| Neu5Acα2-3Galβ1-3(Neu5Acα2-6)GalNAcα- | 165 | 37,378 | 157 | 0.92 |
| Galβ1-3GalNAcα- | 56 | 33,681 | 69 | 2.18 |
| β-d-Gal- | 46 | 32,572 | 5 | 0.65 |
| α-GalNAc- | 80 | 30,639 | 8 | 0.63 |
| Neu5Acβ1-6GalNAcα- | 198 | 19,026 | 152 | 0.69 |
| 6-Su-GalNAcα- | –c | – | 63 | 2.53 |
| 6-Su-Neu5Acα2-3(Galβ1-3)GalNAcα- | – | – | 222 | 1.71 |
Glycan number refers to the number of the glycan in the respective arrays.
Binding activity in PA and EA is expressed as RFU (relative fluorescence units) and S/N (signal-to-noise ratio), respectively.
Not present on the array.
Immunolocalization Confirms That the M. polymorpha ABA Orthologs Are Synthesized by Plant Cells and Do Not Originate from a Contaminating Fungus
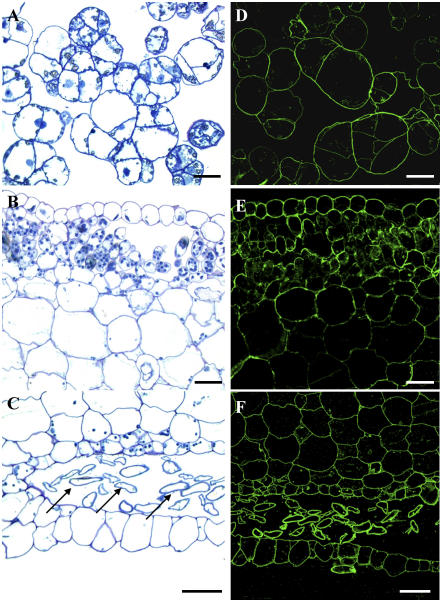
The presence of ABA orthologs in a liverwort raised the question of whether the lectin and the corresponding ESTs/genes are possibly derived from a contaminating fungus. To address this critical question, lectin was localized in axenically grown callus and field-grown thallus tissue by confocal microscopy using highly specific antibodies. Figure 2 clearly demonstrates that in both callus and thallus tissue section immunolabeling is exclusively associated with plant cells. Closer examination of micrographs of thallus sections indicates stronger labeling in the upper and lower epidermal cell layers, the rhizoids, and the small parenchyma cells located below the upper epidermal cell layer as compared to the large parenchyma cells. No signal was detected in the control samples (data not shown). These findings unambiguously demonstrate that ABA orthologs are synthesized by plant cells and do not originate from a fungal contaminant. Moreover, there is no evidence whatsoever for the presence of fungal hyphae in sections from field-grown thalli.
Figure 2.
Localization of MarpoABA in thalli and callus of M. polymorpha. A to C, Transverse sections of callus cells (A) and of upper (B) and lower (C) cell layers of the liverwort thallus after staining with toluidine blue. Note the rich content of parenchyma cells below the epidermal layer. Arrows in C indicate transverse sections of rhizoids. D to F, Confocal images of immunolabeled sections from the corresponding callus (D) and thallus (E and F) cells showing immunolabeling essentially restricted to the cell contours. Scale bars = 20 μm.
The fluorescence patterns shown in Figure 2 give no decisive answer that the M. polymorpha ABA ortholog is like the fungal ABA ortholog from Arthrobotrys oligospora (Rosén et al., 1997) confined to the cytoplasmic/nuclear compartment. At first sight, lectin seems to be associated with the cell wall of M. polymorpha cells. However, further studies are required to corroborate the exact location of lectin in plant cells.
Molecular Modeling of the Structure of the M. polymorpha and T. ruralis ABA Orthologs
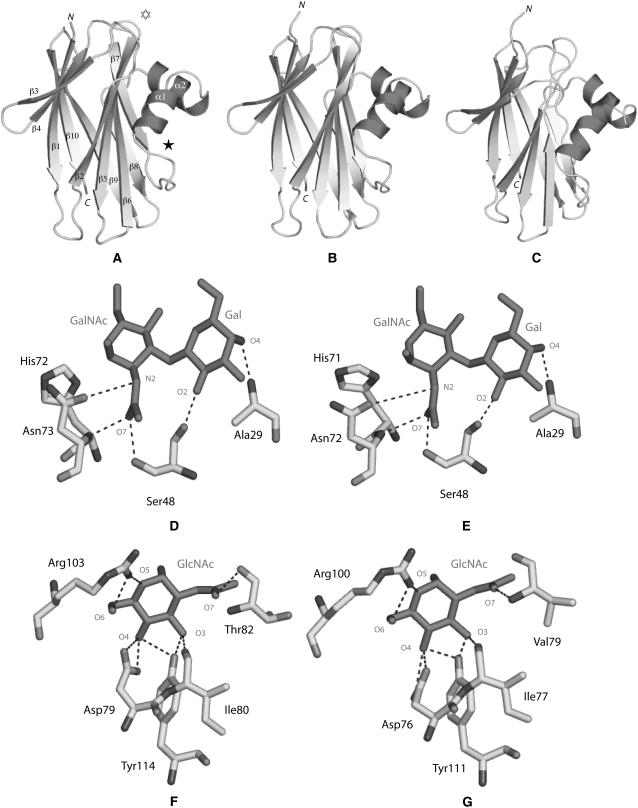
To check whether the binding sites of M. polymorpha and T. ruralis ABA orthologs can be functional, their overall fold and three-dimensional structure was determined by molecular modeling using the atomic coordinates of ABA as a model. The modeled MarpoABA1a and TorruABA exhibit an overall three-dimensional fold similar to that found in ABA (Carrizo et al., 2005; Fig. 3, A–C).
Figure 3.
Molecular modeling of ABA orthologs from Marchantia and Tortula. A, Ribbon diagram of the ABA protomer showing the β-sandwich organization made of two bundles of β-sheets ([β1, β3, β4, β9, β10] and [β2, β5, β6, β7, β8]) interconnected by a helix-loop-helix consisting of two α-helices, α1 and α2. N and C correspond to the N- and C-terminal ends of the polypeptide chain, respectively. The location of the T-antigen-binding site (white star) and GlcNAc-binding site (black star) are indicated by stars. B, Ribbon diagram of the modeled MarpoABA2a. C, Ribbon diagram of the modeled TorruABA. D, Network of hydrogen bonds (dark dashed lines) connecting the T-antigen disaccharide (Galβ1,3GalNAc; black sticks) to the amino acid residues forming the carbohydrate-binding site of ABA. E, Docking of the T-antigen disaccharide (Galβ1,3GalNAc; black sticks) in the carbohydrate-binding site of the modeled MarpoABA2a. F, Network of hydrogen bonds (dark dashed lines) connecting GlcNAc (black sticks) to the amino acid residues forming the carbohydrate-binding site of ABA. G, Docking of GlcNAc (black sticks) in the carbohydrate-binding site of the modeled MarpoABA1a.
In ABA, the amino acid residues Ala-29, Ser-48, and Asn-73, which form the main carbohydrate-binding site, create a network of five hydrogen bonds with both the T-antigen (Galβ1,3GalNAc1α-O-Ser) and the T-antigen disaccharide (Galβ1,3GalNAc; Fig. 3D; Carrizo et al., 2005). An additional hydrogen bond mediated by a water molecule is formed between His-72 and the GalNAc moiety of both the T-antigen and the T-antigen disaccharide. Another water-mediated hydrogen bond occurs between Arg-107 and the GalNAc moiety of the T-antigen disaccharide. All these amino acid residues are strictly conserved in MarpoABA2a and MarpoABA2b. Accordingly, one can reasonably expect that MarpoABA2a and MarpoABA2b interact in a similar way with both the T-antigen and the T-antigen disaccharide as ABA itself. Docking experiments confirm that these interactions can take place (Fig. 3E). In addition, the results of glycan array-binding assays provide direct experimental evidence that a mixture of MarpoABA2a and MarpoABA2b interacts with the T-antigen disaccharide (Table I).
In contrast to MarpoABA2a and MarpoABA2b, MarpoABA1a, MarpoABA1b, and MarpoABA1c, as well as TorruABA, lack the Ala-29 residue and hence interact more weakly with the T-antigen. Due to the replacement of Asn-73 by Val, MarpoABA3a is most probably unable to bind the T-antigen (Supplemental Table S2). These predictions are in good agreement with the absence of MarpoABA1a, MarpoABA1b, MarpoABA1c, and MarpoABA3 isoforms from the affinity-purified lectin preparation.
Besides its major T-antigen-binding site, ABA contains a second (minor) site that interacts with GlcNAc (Carrizo et al., 2005; Nakamura-Tsuruta et al., 2006). This GlcNAc-binding site comprises the five amino acid residues Asp-79, Ile-80, Thr-82, Arg-103, and Tyr-114, which establish a network of eight hydrogen bonds with O3, O4, O5, O6, and O7 of GlcNAc, respectively (Fig. 3F). MarpoABA1a, MarpoABA1b, and MarpoABA1c possess identical or similar residues at homologous positions and, according to docking experiments, interact with GlcNAc in a similar way as ABA (Fig. 3G). Replacement of Thr-82 by a Val residue in MarpoABA1a, MarpoABA1b, and MarpoABA1c probably has little effect on the binding of GlcNAc because both Thr and Val residues interact with O7 of GlcNAc through their N atom, which is similarly positioned at the end of the conserved β7 sheet in all the lectins of the ABA family (Fig. 1). Replacement of the key residue Tyr-114 (of ABA) by an Ile-113 (MarpoABA2a, MarpoABA2b), a Met-113 (MarpoABA3a), or a Leu-113 (TorruABA) residue eliminates two stabilizing hydrogen bonds with O5 and O6 of GlcNAc, respectively, and hence severely hampers or even prevents the interaction of these lectins with GlcNAc. In summary, results of the modeling/docking experiments (Supplemental Table S2) indicate that MarpoABA2a and MarpoABA2b possess a fully functional T-antigen and a weakly active GlcNAc-binding site, whereas MarpoABA1a, MarpoABA1b, and MarpoABA1c contain fully active GlcNAc-binding sites, but T-antigen binding with reduced activity and MarpoABA3a possesses neither a functional T-antigen- nor a GlcNAc-binding site. In TorruABA, both binding sites most probably have reduced activity. Although not conclusive, results of the modeling/docking experiments have some predictive value with respect to the agglutinating activity of the different lectins. Therefore, it should be taken into consideration that the agglutinating activity of ABA is mainly (or even exclusively) determined by the T-antigen-binding site (because no agglutination occurs in the presence of free T-antigen). Only MarpoABA2a and MarpoABA2b possess lectin activity comparable to that of ABA. MarpoABA1a, MarpoABA1b, MarpoABA1c, and TorruABA possibly display (strongly) reduced lectin activity, whereas MarpoABA3a is devoid of lectin activity.
Why Does M. polymorpha Express Lectins with Specificity toward the T-Antigen Disaccharide and GlcNAc?
Identification of ABA orthologs in M. polymorpha raises the question of why this liverwort expresses cytoplasmic/nuclear lectins with specificity toward the T-antigen disaccharide. At present, the biological role of the fungal ABA orthologs is not fully understood. Studies with A. oligospora indicated that, in this fungus, the ABA ortholog behaves as a cytoplasmic storage protein (Rosén et al., 1997), whereas a role as an insecticidal protein was attributed to the ABA ortholog from Xerocomus chrysenteron (Trigueros et al., 2003). Taking into consideration that all studied fungal ABA orthologs have similar specificity, the defensive role proposed for the X. chrysenteron agglutinin might also apply to other members of this lectin family. In this respect, ABA-type fungal lectins can be regarded as functional homologs of potent insecticidal T- and/or Tn-antigen-binding plant lectins (like jacalin and the Glechoma hederacea lectin; Czapla and Lang, 1990; Wang et al., 2003; Singh et al., 2006). However, neither of these two types of typical plant lectins has been found in M. polymorpha. Although purely speculative, one can imagine that the liverwort used a typical fungal protein as an alternative for the insecticidal lectins expressed in flowering plants. This reasoning does not apply to the defense against sucking insects because, as has been demonstrated previously, M. polymorpha expresses fully active orthologs of the Galanthus nivalis agglutinin (Peumans et al., 2002), which is a potent defense protein against sucking insects (like aphids and white flies). Possibly, simultaneous expression of both an ABA- and a G. nivalis agglutinin-type lectin offers M. polymorpha protection against a broad range of chewing and sucking insects.
CONCLUSION
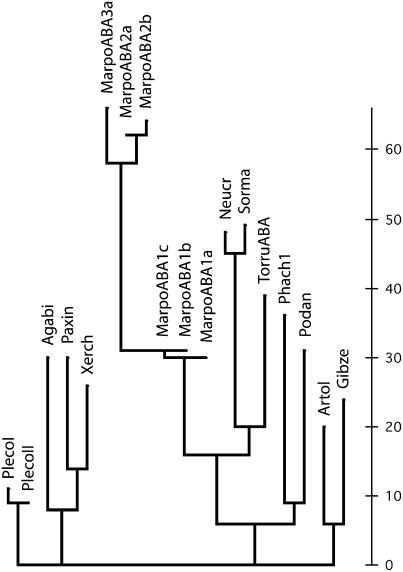
M. polymorpha expresses functional orthologs of a lectin that hitherto was exclusively found in fungi. Construction of a phylogenetic tree from the available sequences not only confirms that the M. polymorpha/T. ruralis ABA orthologs belong to the same protein family as the fungal ABA orthologs, but also indicates that some fungal proteins are more closely related to the M. polymorpha/T. ruralis proteins than to other fungal ABA orthologs (Fig. 4; see Supplemental Fig. S8). Identification of homologous/orthologous lectins in both plants and fungi is not novel because representatives of the ricin-B family, for example, have already been isolated from flowering plants and fungi (as well as from numerous animals and prokaryotes). However, the finding that ABA-related proteins are apparently confined to fungi and some lower plants is unexpected, but highly relevant for what concerns the molecular evolution of lectins in general and plant lectins in particular. The apparent absence of homologous/orthologous genes from the genomes of all (sequenced) bacterial and animal genomes is difficult to explain in terms of classical vertical inheritance from a common (prokaryotic) ancestor. In addition, the question arises as to why no orthologs of the M. polymorpha and T. ruralis lectin genes are present in the genomes of embryophyta. Regardless of the answer to this question, identification of ABA orthologs in M. polymorpha and T. ruralis confirms the previously made observations (based on transcriptome analysis) that some lower plants express proteins that are absent from higher plants. Such observations were not only made for M. polymorpha and T. ruralis, but also for the moss Physcomitrella patens. In the latter case, only about 50% of the expressed protein genes could be matched to an Arabidopsis (Arabidopsis thaliana) homolog (Rensing et al., 2002).
Figure 4.
Phylogenetic tree of ABA orthologs from fungi and plants. MarpoABA and TorruABA refer to the expressed plant proteins from M. polymorpha and T. ruralis, respectively. Basidiomycota lectins are Agabi (ABA; AAA85813); paxin (Paxillus involutus expressed proteins; AAT91249); PlecoI (Pleurotus cornucopiae lectin PCL-F1; BAB63922); PlecoII (Pleurotus cornucopiae lectin PCL-F2; BAB63923); Phach1 (Phanerochaete chrysosporium hypothetical protein encoded by complement AADS01000313.1:6707–7153); and Xerch (Xerocomus chrysenteron lectin XCL1; AL73235). Ascomycota lectins are Artol (Arthrobotrys oligospora lectin; CAA65781); Gibze (Gibberella zeae hypothetical expressed protein EAA76455); Neucr (Neurospora crassa hypothetical expressed protein EAA30976); Podan (Podospora anserina hypothetical protein CAD60779); and Sorma (Sordaria macrospora hypothetical protein CAH03681). Figures refer to GenBank accession numbers. The scale bar indicates the number of amino acid changes.
MATERIALS AND METHODS
Retrieval of Sequences
Sequences encoding ABA-related proteins were retrieved by BLAST searches using the amino acid sequence of ABA as a query. In a later stage, the deduced sequences of the ABA orthologs found in Marchantia polymorpha and Tortula ruralis were used as a query. All retrieved EST sequences were analyzed individually. In the absence of complete ESTs, contigs were reconstructed from ESTs showing overlaps of at least 200 identical nucleotides. Searches were completed on December 15, 2005. The following databases were screened for the presence of EST and/or genomic sequences encoding plant orthologs of ABA: National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), cosmoss.org (http://www.cosmoss.org), Phytome (http://www.phytome.org/search.php), The Institute for Genomic Research (http://tigrblast.tigr.org/tgi), and Solanaceae Genomics network (http://www.sgn.cornell.edu).
Plant Material
M. polymorpha thalli were collected locally in the beginning of December. Immediately after collection, thalli were exhaustively rinsed with tap water to remove soil particles and organic debris trapped between the rhizoids. The washed thalli were further processed manually to remove dead tissue at the proximal end. An axenic culture of M. polymorpha was obtained from Okayama University and maintained as described before (Ishihara et al., 2003).
Isolation of the M. polymorpha ABA Ortholog
Freshly harvested and processed thalli (500 g) were transferred into 5 L of a solution of 20 mm unbuffered 1,3-diaminopropane containing 0.1% thiourea and homogenized with a blender. The homogenate was filtered through cheesecloth, centrifuged (3,000g for 15 min), and the supernatant filtered through filter paper (Whatman 3MM). The cleared filtrate was diluted with an equal volume of distilled water and loaded on a column of Q Fast Flow (Amersham Biosciences; 5 cm × 5 cm; approximately 100-mL bed volume) equilibrated with 20 mm unbuffered 1,3-diaminopropane. After passing the extract, the column was washed with 500 mL of 20 mm unbuffered 1,3-diaminopropane and the bound proteins were eluted with 250 mL of 0.5 m NaCl in 0.1 m Tris-HCl (pH 7.8). This partially purified protein fraction was adjusted to pH 7.5 with 1 n HCl and loaded on a column of asialomucin-Sepharose 4B (2.6 cm × 5 cm; 25-mL bed volume) equilibrated with phosphate-buffered saline (PBS). After loading the protein fraction, the column was washed with PBS until the A280 fell below 0.01 and the lectin eluted with 100 mL 20 mm unbuffered 1,3-diaminopropane. Because this lectin fraction was still strongly colored (due to the presence of some brown materials that were aspecifically bound to the affinity column), it was rechromatographed on the same affinity matrix to improve the purity. The pH of the first eluate was adjusted to 7.5 with 1 n acetic acid and solid NaCl added to a final concentration of 0.2 m. The brown precipitate that formed upon lowering the pH was removed by centrifugation (3,000g for 15 min in 50-mL Falcon tubes) and the supernatant loaded on a small column (1.6 cm × 5 cm; approximately 10-mL bed volume) of asialomucin-Sepharose 4B. After washing with PBS until the A280 fell below 0.01, the bound lectin was desorbed with 20 mL 20 mm unbuffered 1,3-diaminopropane. To concentrate the affinity-purified lectin, the eluate was loaded on a small column (1 cm × 2 cm; 1.5-mL bed volume) of Q Fast Flow equilibrated with 20 mm unbuffered 1,3-diaminopropane and the lectin eluted with 3 mL of 0.4 m NaCl in 0.1 m Tris-HCl, pH 7.8. This concentration step also improved the purity of the lectin because most of the impurities present in the lectin fraction after the second affinity chromatography did not elute with the lectin. Because the concentrated lectin solution was still slightly colored, a gel filtration chromatography step was added to the purification scheme. The fraction eluted from the Q Fast Flow column was chromatographed on a column (2.6 cm × 70 cm; approximately 350-mL bed volume) of Sephacryl 100 using 20 mm unbuffered 1,3-diaminopropane as a running buffer. Under these conditions, the lectin eluted in a symmetrical peak well before the colored compounds. The fractions containing the lectin were pooled, dialyzed against water, and lyophilized. Approximately 2 mg of pure lectin were obtained starting from 500 g of thalli.
Analytical Techniques
The purified lectin was analyzed by SDS-PAGE in 15% (w/v) acrylamide gels as described by Laemmli (1970). Analytical gel filtration was performed on a Superose-12 column (Amersham Biosciences) using PBS (1.5 mm KH2PO4, 10 mm Na2HPO4, 3 mm KCl, 140 mm NaCl, pH 7.4) containing 0.2 m Gal as running buffer. Banana thaumatin-like protein (20 kD) and β-glucanase (30 kD) were used as Mr markers. Total neutral sugar was determined by the phenol-H2SO4 method with d-Glc as standard (Dubois et al., 1956).
For N-terminal amino acid sequencing, purified proteins were separated by SDS-PAGE and electroblotted on a polyvinylidene difluoride membrane. Polypeptides were excised from the blots and sequenced on a model 477A protein sequencer interfaced with a model 120A online analyzer (Applied Biosystems).
MS Analysis of Intact Lectin
The Mr of the intact lectin was determined using ESI-MS on a Micromass quantitative time-of-flight I MS (Waters) equipped with an automated nanoelectrospray source (Advion). A solution of 5 pmol μL−1 was prepared in 50% acetonitrile/0.1% formic acid in water of which 2 μL were loaded on the ESI chip and sprayed using a capillary voltage of 1,100 V. Spectra were collected for 3 min with 1-s scans covering a m/z range from 500 to 2,000 D/E. The spectrum was processed using the Masslynx software delivered with the instrument.
MS Identification of the Lectin
Lectin polypeptides were excised from the gel and treated for tryptic digestion according to a protocol described previously (Devreese et al., 2002). After digestion, the supernatant was recovered and the gel slices reextracted twice with 50 μL of 60% acetonitrile/0.1% formic acid. The combined fractions were dried in a Speedvac concentrator (Thermo Savant).
The peptide mixture was applied on a 4700 Proteomics analyzer, a matrix-assisted laser-desorption ionization-time-of-flight mass spectrometer (Applied Biosystems), equipped with a Nd:YAG laser at a rate of 200 Hz. One microliter of the digest mixture (after reconstitution in 12 μL 0.1% formic acid) was applied, mixed with 1 μL 50 mm α-cyano-4-hydroxycinnamic acid in 50% acetonitrile and 0.1% trifluoroacetic acid on a matrix-assisted laser-desorption ionization target plate. Prior to analysis, the mass spectrometer was externally calibrated with a mixture of angiotensin I, Glu-fibrinopeptide B, adenocorticotropic hormone (1–17), and adenocorticotropic hormone (18–39). For MS/MS experiments, the instrument was externally calibrated with fragments of Glu-fibrinopeptide. The MS results were compared to the theoretical masses of the peptides encoded by the lectin genes.
PCR Amplification of Genomic DNA Fragments
DNA was extracted from thalli, female sexual organs of M. polymorpha, and axenically cultured callus cells using the protocol described by Stewart and Via (1993). Alternatively, DNA was extracted using the FastDNA spin kit in an automatic homogenizer (FastPrep Instrument; MP Biomedicals and Qbiogene) following the manufacturer's recommendations. Genomic sequences encoding ABA orthologs were amplified by PCR using primers (1) derived from the 5′- and 3′-untranslated sequence; or (2) derived from the 5′ and 3′ ORF of ABA orthologs of M. polymorpha. The reaction mixture for amplification of genomic DNA sequences contained 10 mm Tris-HCl, pH 8.3, 50 mm KCl, 1.5 mm MgCl2, 100 mg L−1 gelatin, 0.4 mm of each dNTP, 2.5 units of Taq polymerase (Invitrogen), 500 ng of genomic DNA, and 20 μL of the appropriate primer mixtures (20 μm) in a 100-μL reaction volume. After denaturation of the DNA for 5 min at 95°C, amplification was performed for 30 cycles through a regime of 15-s template denaturation at 92°C followed by 30-s primer annealing at 50°C and 1-min primer extension at 72°C. PCR fragments were purified using the Qiaquick PCR purification kit (Qiagen) and cloned in pCR2.1-TOPO cloning vector using the TOPO-TA cloning kit from Invitrogen. Plasmids were isolated from purified single colonies on a miniprep scale using the alkaline lysis method (Mierendorf and Pfeffer, 1987) and sequenced by the dideoxy method (Sanger et al., 1977).
Preparation of Monospecific Antibodies
Polyclonal antibodies were raised against the M. polymorpha ABA orthologs in a female New Zealand White rabbit. The animal was injected subcutaneously with 1 mg of the purified lectin dissolved in PBS and emulsified in 1 mL of Freund's complete adjuvant. Four booster injections with 0.2 mg lectin in 1 mL of PBS were given with 10-d intervals. Ten days after the final injection, blood was collected from an ear marginal vein and the crude serum was prepared by standard techniques. Western-blot analysis of crude extracts from M. polymorpha ABA thallus tissue demonstrated that the crude antiserum (200-fold diluted) reacted exclusively with a single polypeptide of the same size as the purified lectin (see Supplemental Fig. S9), indicating that the antibodies are monospecific. Accordingly, the antiserum was suitable for immunolocalization experiments without the need for further purification.
Immunolocalization of ABA Orthologs
Samples of axenically cultured callus cells and small pieces of thallus were fixed with aldehyde (2% of formaldehyde and 0.25% of glutaraldehyde) in 0.05 m cacodylate buffer (pH 7.2) for 24 h at 4°C, washed with cacodylate buffer, and dehydrated in ethanol series (20%, 40%, 60%, 75%, 80%, 95%, and 100% of ethanol). After dehydration, tissue samples were stepwise infiltrated with mixtures of LR White resin in ethanol (1:2, 1:1, and 2:1, respectively) and embedded in pure LR White resin. Polymerization was finally performed overnight at 70°C. Transverse semithin (2 μm in thickness) sections were prepared using a Reichert ultraCutE microtome. Semithin sections were mounted on glass slides for bright-field and laser-scanning confocal microscopy examinations. Some sections were stained with toluidine blue for bright-field microscopy examination.
Sections were blocked for 2 h at room temperature in PBSTA (0.14 m NaCl, 2.7 mm KCl, 7.8 mm Na2HPO4, 1.5 mm KH2PO4, 1% Tween 20, 1% bovine serum albumin [BSA]; pH 7.2) and incubated overnight at 4°C with 10-fold diluted (in PBSTA) anti-Marpola antibodies. After washing with the same buffer, sections were incubated for 2 h at room temperature with 1,000-fold diluted (in PBSTA) anti-rabbit IgG coupled to Alexa 488 (Molecular Probes), washed, and dried. Possible autofluorescence and unspecific binding of the secondary labeled antibody were checked in a control experiment in which the primary anti-Marpola antibody was omitted.
For bright-field microscopy, images of toluidine blue-stained sections were taken using a CCD camera (color Coolview; Photonic Science). For immunolabeling, confocal images were acquired with a SP2 confocal laser-scanning system equipped with an upright microscope (Leica) and a 40× (PL APO, N.A. 1.25) oil immersion objective. The 488-nm ray line of an argon laser was used to detect the labeling. Light emitted from the Alexa 488 probe was collected in the range between 500 and 540 nm. The laser line intensity and the photomultiplier tube setting were kept constant for both the control and the labeled samples.
Hemagglutination and Hapten Inhibition Assays
Agglutination assays were carried out in small glass tubes in a final volume of 50 μL containing 40 μL of a 1% suspension of red blood cells and 10 μL of extracts or lectin solutions. To determine the specific agglutination activity, lectin was serially diluted with 2-fold increments. Agglutination was assessed visually after 1 h at room temperature. Erythrocytes were treated with trypsin as described previously (Van Damme et al., 1987).
The carbohydrate-binding specificity of the lectin was determined by hapten inhibition of the agglutination of trypsin-treated rabbit erythrocytes. To 10 μL of a solution of MarpoABA (10 μg mL−1 in PBS) 10-μL aliquots of solutions of sugars (0.5 m in PBS) or glycoproteins (5 mg mL−1 in PBS) were added. After preincubation for 1 h at 25°C, 30 μL of a 1% suspension of trypsin-treated rabbit erythrocytes were added and the agglutination evaluated after 1 h. To determine the inhibitory potency of the most active monosaccharides and glycoproteins, assays were repeated with serially diluted stock solutions of sugars and glycoproteins. The concentration required for 50% inhibition of the agglutination of trypsin-treated rabbit erythrocytes was determined visually.
Glycan Array Screening
A purified preparation of lectin was labeled at a concentration of 2 mg mL−1 with an Alexa Fluor 488 protein labeling kit (Molecular Probes) according to manufacturer's protocol. The labeled lectin was applied to a PD-10 column (Sephadex G-25; Amersham Biosciences) and separated from free label. Protein concentration was determined by Lowry determination and Alexa 488-labeling efficiency determined at 485-nm excitation/535-nm emission. The MarpoABA-Alexa 488 was applied to the Consortium streptavidin/biotin array and to the PA for binding specificity determination (http://www.functionalglycomics.org/static/consortium/resources/resourcecoreh.shtml). Labeled MarpoABA was screened on both arrays in binding buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm CaCl2, 2 mm MgCl2, 0.05% Tween 20, 1% BSA).
Lectin was screened on the streptavidin/biotin array (EA V3) as previously described (Bochner et al., 2005). Briefly, biotinylated glycosides (Korchagina and Bovin, 1992a) were bound to streptavidin-coated microtiter plates in four replicates. Precoated plates were washed three times with 100 μL of wash buffer (binding buffer minus BSA) prior to incubation. A stock solution of MarpoABA-Alexa 488 (30 μg mL−1) was added to each well and incubated at room temperature for 1 h. The plates were washed and read in 25 μL of wash buffer on a Victor-2 1420 multilabel counter (Perkin-Elmer Life Sciences) at 485-nm excitation/535-nm emission.
Lectin was screened on the Consortium PA (V1) as previously described (Blixt et al., 2005). Briefly, an aliquot of the same preparation of MarpoABA-Alexa 488 (300 μg mL−1) was applied in a volume of 50-μL binding buffer onto the preprinted slide, cover slipped, and incubated protected from light for 1 h. The slide was washed successively in wash buffer, wash buffer minus Tween 20, and deionized water, and then dried under a stream of nitrogen. The slide was read on a ScanArray express microarray scanner and the image was analyzed using ScanArray express software (Perkin-Elmer Life Sciences).
Molecular Modeling of ABA Orthologs from Marchantia and Tortula
Hydrophobic cluster analysis, molecular modeling, and docking experiments were carried out using standard techniques. Three-dimensional models of proteins were built using the atomic coordinates of ABA (Carrizo et al., 2005). Full details are described in the Supplemental Data.
Phylogenetic Analysis
Multiple amino acid sequence alignments based on ClustalX (Thompson et al., 1997) were carried out using SeqPup (D.G. Gilbert, Indiana University) and MacClade (Maddison and Maddison, 1992) was used to build a parsimony phylogenetic tree.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EF076035 to EF076040.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of the nucleotide sequences of the five contigs assembled from the available EST sequences of Marchantia.
Supplemental Figure S2. Alignment of the nucleotide sequences of the ORFs of the MarpoABA1 group (A) and their deduced amino acid sequences (B).
Supplemental Figure S3. Alignment of the nucleotide sequences of the ORFs of the MarpoABA2 group (A) and their deduced amino acid sequences (B).
Supplemental Figure S4. Alignment of the deduced amino acid sequences of the T. ruralis ABA ortholog with MarpoABA1a.
Supplemental Figure S5. Alignment of the nucleotide sequences of the ORFs of the five contigs assembled from the available EST sequences and an additional genomic sequence amplified by PCR.
Supplemental Figure S6. MS of M. polymorpha ABA orthologs.
Supplemental Figure S7. Analysis of carbohydrate-binding specificity of M. polymorpha ABA orthologs using ELISA glycan array and printed glycan array.
Supplemental Figure S8. Sequences of ABA orthologs from fungi and plants.
Supplemental Figure S9. Western-blot analysis of an ABA ortholog in crude extracts of M. polymorpha thalli.
Supplemental Table S1. Overview of the main physicochemical parameters of the M. polymorpha and T. ruralis ABA orthologs.
Supplemental Table S2. Overview of the activity of the T-antigen dissacharide and GlcNAc-binding site of M. polymorpha and T. ruralis ABA orthologs.
Note Added in Proof
In a recently (September 6, 2006) released set of M. polymorpha EST sequences derived from a M. polymorpha thallus E cDNA library, four clones were identified that are identical to MarpoABA1a (M. polymorpha thallus cDNA clones lwa2d05, lwa16o18, lwa32n17, and lwa25g22). This finding confirms the expression of the corresponding protein in thallus tissue.
Supplementary Material
Acknowledgments
We thank Prof. Monaco (University of Verona, Italy) for making available the x-ray coordinates of apoABA, ABA in complex with the T-antigen disaccharide, and ABA in complex with the T-antigen disaccharide and GlcNAc prior to release at the Protein Data Bank. We also want to thank Dr. Katsuyuki Yamato (Kyoto University, Japan) for providing the cDNA clone encoding MarpoABA2a (GenBank accession no. C95977) and Angela Lee of Core H of the Consortium for Functional Glycomics for expert technical assistance.
This work was supported by the Fund for Scientific Research-Flanders (project no. G.0201.04) and the Research Council of Ghent University. The glycan array analysis was conducted by the Protein-Glycan Interaction Core H of the Consortium for Functional Glycomics funded by the National Institute of General Medical Sciences (grant no. GM62116).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instruction for Authors (www.plantphysiol.org) is: Els J.M. Van Damme (elsjm.vandamme@ugent.be).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adam KP, Becker H (1993) A lectin from the liverwort Marchantia polymorpha L. Experientia 49 1098–1100 [DOI] [PubMed] [Google Scholar]
- Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, et al (2005) Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci USA 101 17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BS, Alvarez RA, Mehta P, Bovin NV, Blixt O, White JR, Schnaar RL (2005) Glycan array screening reveals a candidate ligand for Siglec-8. J Biol Chem 280 4307–4312 [DOI] [PubMed] [Google Scholar]
- Carrizo ME, Capaldi S, Perduca M, Irazoqui FJ, Nores GA, Monaco HL (2005) The antineoplastic lectin of the common edible mushroom (Agaricus bisporus) has two binding sites, each specific for a different configuration at a single epimeric hydroxyl. J Biol Chem 280 10614–10623 [DOI] [PubMed] [Google Scholar]
- Czapla TH, Lang BA (1990) Effect of plant lectins on the larval development of European corn borer (Lepidoptera pyralidae) and southern corn rootworm (Coleoptera chrysomelidae). J Econ Entomol 83 2480–2485 [Google Scholar]
- Devreese B, Vanrobaeys F, Smet J, Van Beeumen J, Van Coster R (2002) Mass spectrometric identification of mitochondrial oxidative phosphorylation-subunits separated by two dimensional blue-native polyacrylamide gel electrophoresis. Electrophoresis 23 2525–2533 [DOI] [PubMed] [Google Scholar]
- Dodd RB, Drickamer K (2001) Lectin-like proteins in model organisms: implications for evolution of carbohydrate-binding activity. Glycobiology 11 71R–79R [DOI] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F (1956) Colorimetric method for determination of sugar and related substances. Anal Chem 28 350–356 [Google Scholar]
- Ishihara K, Hamada H, Hirata T, Nakajima N (2003) Biotransformation using plant cultured cells. J Mol Catal B Enzym 23 145–170 [Google Scholar]
- Jung E, Fucini P, Stewart M, Noegel AA, Schleicher M (1996) Linking microfilaments to intracellular membranes: the actin-binding and vesicle-associated protein comitin exhibits a mannose-specific lectin activity. EMBO J 15 1238–1246 [PMC free article] [PubMed] [Google Scholar]
- Kai G, Zhao L, Zheng J, Zhang L, Miao Z, Sun X, Tang K (2004) Isolation and characterization of a new mannose-binding lectin gene from Taxus media. J Biosci 29 399–407 [DOI] [PubMed] [Google Scholar]
- Korchagina E, Bovina NV (1992) Synthesis of spaced trisaccharides with blood group A and B specificity, their fragments, and structural analogs. Bioorg Khim 18 283–298 [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR (1992) MacClade: Analysis of Phylogeny and Character Evolution, Version 3.0. Sinauer Associates, Sunderland, MA
- Mierendorf RC, Pfeffer D (1987) Direct sequencing of denatured plasmid DNA. Methods Enzymol 152 556–562 [DOI] [PubMed] [Google Scholar]
- Nagai J, Yamato KT, Sakaida M, Yoda H, Fukuzawa H, Ohyama K (1999) Expressed sequence tags from immature female sexual organ of a liverwort, Marchantia polymorpha. DNA Res 6 1–11 [DOI] [PubMed] [Google Scholar]
- Nakamura-Tsuruta S, Kominami J, Kuno A, Hirabayashi J (2006) Evidence that Agaricus bisporus agglutinin (ABA) has dual sugar-binding specificity. Biochem Biophys Res Commun 347 215–220 [DOI] [PubMed] [Google Scholar]
- Nishiyama R, Yamato KT, Miura K, Sakaida M, Okada S, Kono K, Takahama M, Sone T, Takenaka M, Fukuzawa H, et al (2000) Comparison of expressed sequence tags from male and female sexual organs of Marchantia polymorpha. DNA Res 7 165–174 [DOI] [PubMed] [Google Scholar]
- Peumans WJ, Barre A, Bras J, Rougé P, Proost P, Van Damme EJM (2002) The liverwort contains a lectin that is structurally and evolutionary related to the monocot mannose-binding lectins. Plant Physiol 129 1054–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Rombauts S, Van de Peer Y, Reski R (2002) Moss transcriptome and beyond. Trends Plant Sci 7 535–538 [DOI] [PubMed] [Google Scholar]
- Rosén S, Sjollema K, Veenhuis M, Tunlid A (1997) A cytoplasmic lectin produced by the fungus Arthrobotrys oligospora functions as a storage protein during saprophytic and parasitic growth. Microbiology 143 2593–2604 [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T, Wu JH, Peumans WJ, Rougé P, Van Damme EJM, Alvarez RA, Blixt O, Wu AM (2006) Carbohydrate specificity of an insecticidal lectin isolated from the leaves of Glechoma hederacea (ground ivy) towards mammalian glycoconjugates. Biochem J 393 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CN Jr, Via LE (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14 748–750 [PubMed] [Google Scholar]
- Tateno H, Winter HC, Petryniak J, Goldstein IJ (2003) Purification, characterization, molecular cloning, and expression of novel members of jacalin-related lectins from rhizomes of the true fern Phlebodium aureum (L) J. Smith (Polypodiaceae). J Biol Chem 278 10891–10899 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acids Res 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigueros V, Lougarre A, Ali-Ahmed D, Rahbé Y, Guillot J, Chavant L, Fournier D, Paquereau L (2003) Xerocomus chrysenteron lectin: identification of a new pesticidal protein. Biochim Biophys Acta 1621 292–298 [DOI] [PubMed] [Google Scholar]
- Tsutsui S, Tasumi S, Suetake H, Suzuki Y (2003) Lectins homologous to those of monocotyledonous plants in the skin mucus and intestine of pufferfish, Fugu rubripes. J Biol Chem 278 20882–20889 [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Allen AK, Peumans WJ (1987) Isolation and characterization of a lectin with exclusive specificity towards mannose from snowdrop (Galanthus nivalis) bulbs. FEBS Lett 215 140–144 [Google Scholar]
- Van Damme EJM, Barre A, Rougé P, Peumans WJ (2004) Cytoplasmic/nuclear plant lectins: a new story. Trends Plant Sci 9 484–489 [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Peumans WJ, Barre A, Rougé P (1998) Plant lectins: a composite of several distinct families of structurally and evolutionary related proteins with diverse biological roles. CRC Crit Rev Plant Sci 17 575–692 [Google Scholar]
- Velloso LM, Svensson K, Pettersson RF, Lindqvist Y (2003) The crystal structure of the carbohydrate-recognition domain of the glycoprotein sorting receptor p58/ERGIC-53 reveals an unpredicted metal-binding site and conformational changes associated with calcium ion binding. J Mol Biol 334 845–851 [DOI] [PubMed] [Google Scholar]
- Wang W, Hause B, Peumans WJ, Smagghe G, Mackie A, Fraser R, Van Damme EJM (2003) The Tn antigen-specific lectin from ground ivy is an insecticidal protein with an unusual physiology. Plant Physiol 132 1322–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AM, Wu JH, Herp A, Liu JH (2003) Effect of polyvalencies of glycotopes on the binding of a lectin from the edible mushroom, Agaricus bisporus. Biochem J 371 311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi F, Iwaya T, Haraguchi T, Goldstein IJ (2002) The lectin from leaves of Japanese cycad, Cycas revoluta Thunb. (gymnosperm) is a member of the jacalin-related family. Eur J Biochem 269 4335–4341 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.