Abstract
A 22-kb DNA locus of Legionella pneumophila is described that contains 18 genes, 16 of which are required for macrophage killing (icm genes). In this paper two previously described icm loci were linked by the discovery of five genes located between the two loci. Four of the newly described genes are required for macrophage killing (icmMLKE) and one is dispensable. The 16 icm genes appeared to be organized as six individual genes (icmR, icmQ, icmG, icmC, icmD, and icmF), and four operons (icmTS, icmPO, icmMLKE, and icmJB). Four icm genes (icmP, icmO, icmL, and icmE) show significant sequence similarity to plasmid genes involved in conjugation, whereas the other icm genes were found not to bear any sequence similarity to database entries. We found that L. pneumophila can mediate plasmid DNA transfer at a frequency of 10−3 to 10−4 per donor. Strains containing null mutations in two icm genes (icmT and icmR) showed a severe reduction in conjugation frequency and macrophage killing. Strains containing an insertion in four other icm genes (icmF, icmE, icmC, and dotA) were shown to have a less severe defect in conjugation. Mutations in the other 11 icm genes had no effect on conjugation frequency. We currently do not know whether conjugation itself plays a role in macrophage killing. It is possible either that small plasmids can take advantage of an existing secretion system to be mobilized or that DNA transfer is required for human macrophage killing by L. pneumophila.
Legionella pneumophila, the causative agent of Legionnaires’ disease, is a facultative intracellular pathogen with a broad host range. The bacteria are able to infect, multiply within, and kill human macrophages, as well as free-living amoebae (1, 2). When inside host cells, L. pneumophila are found within a specialized phagosome that does not fuse with lysosomes (3). The bacteria multiply within the specialized phagosome, until the cell eventually lyses, releasing bacteria that can start new rounds of infection.
Several years ago, a collection of 55 L. pneumophila mutants defective for macrophage killing were isolated from a large (n = 4,500) pool of Tn903dIIlacZ insertions, and classified into 16 DNA hybridization groups (4). One of these groups (group 1), which contains 10 of the insertion mutants, was previously characterized as the icmA–dotA region (5, 6). More recently, five additional DNA hybridization groups were characterized as two separate regions (ref. 7; M.P. and H.A.S., unpublished results). One of the regions (6.5 kb), was shown to contain nine insertion mutations located on a single DNA hybridization group (group 3), that contains six icm genes (7). The second region (11 kb) was shown to contain 18 insertion mutations located on four contiguous DNA hybridization groups (groups 2, 6, 4, and 17), which contain an additional six icm genes (M.P. and H.A.S., unpublished results).
The aim of this study was to complete the characterization of the additional DNA hybridization groups. We found that the two regions described above are connected to one another, and two additional DNA hybridization groups (groups 7 and 9) were found to be located between them. In summary, a 22-kb DNA region was found to contain 18 genes, 16 of which were shown to be required for macrophage killing by L. pneumophila. All together, these genes account for all the insertion mutants with severe defects in macrophage killing and intracellular multiplication.
MATERIALS AND METHODS
Bacterial Strains, Media, and Tissue Culture Manipulations.
Bacterial strains used in this work are described in Table 1. Bacterial media and antibiotic concentrations were used as described before (7). Bacterial matings were done as previously described (4), but similar numbers of recipient and donor were used. The human leukemia cell line HL-60 (9) was used during this study. Cytotoxicity assays and intracellular growth assays were done as previously described (7).
Table 1.
L. pneumophila strains
| Strain | Genotype and features | Ref. or source |
|---|---|---|
| 25D | Icm− avirulent mutant | 8 |
| GS3001 | JR32 icmS3001::Kan | 7 |
| GS3002 | JR32 icmP3002::Kan | 7 |
| GS3003 | JR32 icmO3003::Kan | 7 |
| GS3004 | JR32 icmO–lphA3004::Kan | This study |
| GS3005 | JR32 lphA3005::Kan | This study |
| GS3006 | JR32 lphA3006::Kan | This study |
| GS3007 | JR32 lphA3007::Kan | This study |
| GS3008 | JR32 icmM3008::Kan | This study |
| GS3009 | JR32 icmL3009::Kan | This study |
| GS3010 | JR32 icmK3010::Kan | This study |
| JR32 | Salt-sensitive isolate of AM511 | 4 |
| JR32R | Rifampicin derivative of JR32 | This study |
| LELA1718 | JR32 icmF1718::Tn903dIIlacZ | 4 |
| LELA3118 | JR32 dotA3118::Tn903dIIlacZ | 4 |
| LELA4432 | JR32 icmE4432::Tn903dIIlacZ | 4 |
| LELA3244 | JR32 icmD3244::Tn903dIIlacZ | 4 |
| LELA3278 | JR32 icmR3278::Tn903dIIlacZ | 4 |
| LELA3393 | JR32 icmB3393::Tn903dIIlacZ | 4 |
| LELA3463 | JR32 icmQ3463::Tn903dIIlacZ | 4 |
| LELA4004 | JR32 icmX4004::Tn903dIIlacZ | 4 |
| LELA4086 | JR32 icmT4086::Tn903dIIlacZ | 4 |
| MW635 | JR32 icmG635::Kan | ∗ |
| MW645 | JR32 icmC645::Kan | ∗ |
| MW656 | JR32 icmJ656::Kan | ∗ |
M.P. and H.A.S., unpublished work.
Screening of L. pneumophila Cosmid Library.
The L. pneumophila pLAFR1 library (10) was screened by using two probes. A 0.3-kb BssHII fragment from pGS-Lp-32 (7) was used as a probe for group 3. pAB-24, which contains the EcoRI fragment from LELA4432 (4), was used as a probe of group 9. Several positive cosmids were identified, and four of them (pGS-cos-1, pGS-cos-20, pGS-cos-51, and pGS-cos-60) were analyzed and used for generating different subclones.
Plasmid Construction.
Three subclones were constructed from pGS-cos-20, in the L. pneumophila vector pMMB207αb (7). A 3174-bp PstI fragment (bases 5764–8938 in the sequence) was cloned to generate pGS-Lc-40, which contains the lphA and icmMLK genes. A 4740-bp SmaI–BsaBI fragment (bases 8432–13172 in the sequence) was cloned to generate pGS-Lc-42, which contains the icmE gene. A 8751-bp PflMI–BsaBI fragment (bases 4421–13172 in the sequence) was cloned to generate pGS-Lc-47, which contains part of icmO, lphA, icmMLKE, and icmG. In all these subclones the Ptac promoter is in the opposite orientation to the direction of transcription of the icm genes. In-frame deletions in icmM, icmL, and icmK were constructed by PCR, as described before (7). The plasmids pGS-Lp-40-D2, pGS-Lp-40-D3, and pGS-Lp-40-D4 contain in-frame deletions in icmM (codons 9–87), icmL (codons 27–207), and icmK (codons 18–219), respectively. The in-frame deletion in icmE was constructed by using two Eco47III sites located in the icmE coding region (codons 316–537) to generate pGS-Lc-47-D5.
Construction of Strains by Allelic Exchange.
Allelic exchange was performed as previously described (7). The strains GS3007, GS3008, GS3009, and GS3010 contain the kanamycin-resistance cassette in lphA (after codon 241), icmM (after codon 82), icmL (after codon 98), and icmK (after codon 242), respectively. The strain GS3005 contains the kanamycin-resistance cassette between icmO and lphA (base 6027 in the sequence). In the strains GS3004 and GS3006 part of lphA and part of the region between icmO and lphA was deleted, and the kanamycin-resistance cassette was inserted instead. In GS3004 the region between nucleotides 5844 and 6531 was deleted and in GS3006 the region between nucleotides 6027 and 6427 was deleted.
Sequence Analysis and Accession Number.
The sequence determined was analyzed as described before (7). Sequence data (22 kb) of the complete icmTSRQPO, lphA, icmMLKEGCDJB, tphA, and icmF locus have been assigned GenBank data accession no. Y15044.
RESULTS
Seven DNA Hybridization Groups That Cover 22 kb Are Contiguous.
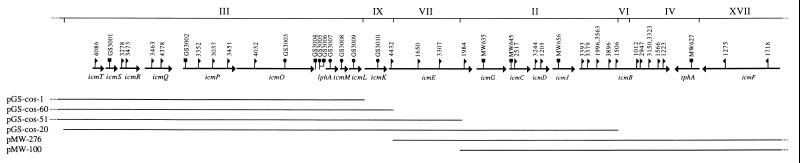
To determine whether additional DNA hybridization groups are linked to the two regions described before (ref. 7; M.P. and H.A.S., unpublished results), and to characterize additional DNA hybridization groups, an L. pneumophila cosmid library (10) was screened with probes from hybridization groups 3 and 9. Several positive cosmids were identified and analyzed with restriction enzymes and by Southern hybridization with several probes (data not shown). We found that the two regions are connected to one another, and two additional DNA hybridization groups (groups 7 and 9) were found to be located between them. The whole region (22 kb) was found to contain seven contiguous DNA hybridization groups, in the following order: 3, 9, 7, 2, 6, 4, and 17 (Fig. 1). This region corresponds to the insertion sites of 30 of the 55 mutants that were originally isolated (4). Three of these cosmids (pGS-cos-1, pGS-cos-51, and pGS-cos-60) (Fig. 1), contain a 16-kb EcoRI fragment upstream of group 3. This EcoRI fragment does not correspond to any of the 16 DNA hybridization groups originally isolated (4). To test whether any of the known DNA hybridization groups are located downstream of group 17, the cosmids pMW-275 and pMW-276 (M.P. and H.A.S., unpublished results) were analyzed (Fig. 1). In both of these cosmids a 1.4-kb EcoRI fragment was found to be located downstream from group 17; this EcoRI fragment does not correspond to any of the 16 DNA hybridization groups originally isolated (4). Nevertheless it is possible that genes involved in macrophage killing are located to the left and right of the 22-kb region described.
Figure 1.
Linkage map of the icmTSRQPO-lphA-icmMLKEGCDJB-tphA-icmF locus, and cosmids used for map construction. Coding regions are indicated by bold arrows. The DNA hybridization groups are indicated by roman numerals. The sites of the Tn903dIIlacZ insertions (LELA strains) are indicated by flags (showing the direction of the lacZ gene fusion), and the sites of the kanamycin-resistance cassettes are indicated by circles for insertion and squares for deletion substitutions.
According to the DNA sequence, this region was found to contain 18 open reading frames (ORFs) on a 22-kb DNA region (Fig. 1). Only 10 of these ORFs were found to contain transposon insertions from the original collection. In two previous reports, three of the ORFs that do not contain transposon insertions were already shown to be required for macrophage killing (icmS, icmG, and icmJ), and one ORF (tphA) was shown to be dispensable (ref. 7; M.P. and H.A.S., unpublished results). As will be described below, we tested whether the other four ORFs that do not contain transposon insertions are required for macrophage killing. Introduction of an insertion cassette into three of these ORFs (icmM, icmL, and icmK) resulted in strains that cannot kill HL-60-derived macrophages, and the fourth ORF (lphA) was found to be dispensable for macrophage killing. In summary, the 22-kb DNA region contains 18 genes, 16 of which are required for macrophage killing.
Protein Similarities and Properties.
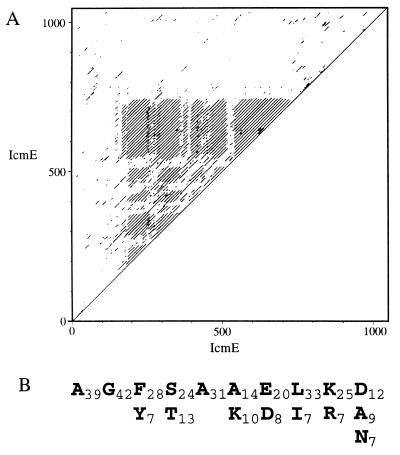
Sequence similarity searches against the EMBL, GenBank, and Swiss-Prot databases were done with these genes and the corresponding proteins. As was described previously, icmP and icmO were found to have significant sequence similarity (Table 2) with the trbA and trbC genes from Salmonella plasmid R64. The trbA and trbC genes were shown to be indispensable for R64 DNA transfer (11). Two of the genes described here (icmL and icmE) were also found to have significant sequence similarity with plasmid-encoded genes. icmL was found to have significant sequence similarity (Table 2) with a partial ORF (of unknown function) from Escherichia coli plasmid R446 (12). The C-terminal part of IcmE (amino acids 825-1048) was found to have sequence similarity (Table 2) with the TrbI protein from the plasmid RK2 and other TrbI proteins. TrbI proteins are known to be involved in plasmid DNA transfer (13). IcmE was found to contain an interesting feature in the part without sequence similarity to TrbI. This protein contains multiple repeats of 10 amino acids, between amino acids 192 and 724, as presented in Fig. 2.
Table 2.
Protein sequence similarity
| Protein | Homolog | Plasmid | Identity, % | Similarity,* % | Overlap of amino acids | Function |
|---|---|---|---|---|---|---|
| IcmP | TrbA | R64 (IncI1) | 23 | 35 | 376 (whole protein) | DNA transfer |
| IcmO | TrbC | R64 (IncI1) | 24 | 36 | 783 (whole protein) | DNA transfer |
| IcmL | Orf3 | R446 (IncM) | 32 | 41 | 129 (N-terminal) | ? |
| IcmE | TrbI | RK2 (IncP) | 16 | 27 | 223 (C-terminal) | DNA transfer |
| LphA | YIAD | 26 | 43 | 190 (whole protein) | Lipoprotein | |
| TphA | ProP | 30 | 44 | 408 (whole protein) | Transporter |
Similar amino acids are I, L, V, and M; H, K, and R; D and E; T and S; and Q and N.
Figure 2.
(A) Dot plot analysis of the IcmE protein, showing the multiple repeat present in the middle of the protein between amino acids 192 and 724. (B) Amino acid content of the 10-amino acid multiple repeat (42 repeats); the number of times that an amino acid appears in the repeat is shown. Only amino acids that appear 7 or more times are presented.
Two genes (lphA and tphA) that were found not to be required for macrophage killing were also found to have significant sequence similarity to proteins in the GenBank database. LphA (for lipoprotein homolog) was found to have significant sequence similarity (Table 2) to an E. coli lipoprotein (a protein with unknown function named YIAD). TphA (for transport protein homolog) was found to have significant sequence similarity (Table 2) to a family of H+/amino acid symport proteins (e.g., E. coli ProP).
Only four genes (of 16) that were shown to be required for macrophage killing were found to have significant sequence similarity in the sequence databases. All four are plasmid-encoded genes whose function is connected with plasmid DNA transfer. The other 12 genes are novel genes. Several properties of all the proteins from this region are presented in Table 3.
Table 3.
Protein properties
| Protein | Size, aa | pI* | Predicted location† | Motifs‡ |
|---|---|---|---|---|
| IcmT | 87 | 12.0 | IM-1 | |
| IcmS | 115 | 4.4 | Cyt | |
| IcmR | 122 | 4.7 | Cyt | |
| IcmQ | 192 | 9.6 | Cyt | |
| IcmP | 376 | 8.3 | IM-2 | RGD |
| IcmO | 783 | 5.3 | IM-2 | A/G |
| LphA | 189 | 9.9 | OM-31 | Lipoprotein |
| IcmM | 94 | 6.4 | IM-1 | |
| IcmL | 212 | 9.4 | IM-1 | |
| IcmK | 360 | 5.9 | Perp-26 | |
| IcmE | 1,048 | 5.6 | IM-1 | |
| IcmG | 269 | 7.8 | IM-1 | |
| IcmC | 194 | 9.1 | IM-4 | |
| IcmD | 132 | 9.8 | IM-1 | |
| IcmJ | 208 | 7.5 | IM-1 | |
| IcmB | 1,009 | 5.4 | IM-1 | A/G |
| TphA | 418 | 9.3 | IM-12 | Transporter |
| IcmF | 973 | 7.0 | IM-2 | A/G |
Calculated with compute pI/MW program (14).
Location was determined by the psort program (15). IM-n, inner membrane and number of transmembrane domains; Cyt, cytoplasm; OM and Perp, outer membrane and periplasm, respectively, followed by location of the N-terminal signal sequence cleavage site.
Motif search was done with the prosite program (16). A/G, ATP- or GTP-binding site; RGD, Arg-Gly-Asp sequence.
The Region Between icmO and lphA and the lphA Gene.
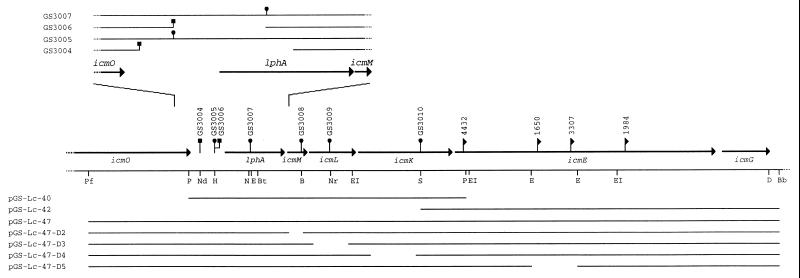
In a previous report (7) and in unpublished work of M.P. and H.A.S., the region between icmT and icmO, as well as the region between icmG and icmF, was characterized (Fig. 1). In the next two paragraphs the characterization of the genes located between icmO and icmG (Fig. 3) is described.
Figure 3.
Detailed restriction map of the icmO-lphA-icmMLKEG locus. Coding regions are indicated by bold arrows. The site of the Tn903dIIlacZ insertions (LELA strains) are indicated by flags, and the sites of the kanamycin insertions (GS strains) are indicated by circles for insertions and squares for deletion substitutions. The restriction enzymes are as follows: B, BamHI; Bb, BsaBI; Bt, BstEII; D, DraIII; EI, EcoRI; E, Eco47III; N, NheI; Nd, NdeI; Nr, NruI; P, PstI; Pf, PflMI; S, SmaI (only relevant sites are marked). In the lower part, plasmids used for complementation studies are shown. In the upper part, chromosomal changes in the icmO-lphA region are presented.
A relatively long region (407 bp) of DNA that apparently does not encode a protein was found between icmO and lphA (Fig. 3). As mentioned above, icmP and icmO were found to bear sequence similarity to trbA and trbC from Salmonella plasmid R64. On this plasmid, the oriT operon is located down-stream of the trbABC operon (after the nikAB operon) (11). The possibility that the region between icmO and lphA, as well as the lphA gene, might have a role in macrophage killing was tested. A kanamycin-resistance cassette was introduced into the region between icmO and lphA, and into the lphA coding region (Fig. 3 Upper). Both these strains (GS3005 and GS3007) were found to be indistinguishable from wild-type for macrophage killing and intracellular multiplication. To further analyze this region, two deletion substitutions were constructed. In these deletions nearly all of the intergenic region between icmO and lphA and part of lphA were deleted (Fig. 3 Upper). These strains (GS3004 and GS3006) were also found to be indistinguishable from wild type for macrophage killing and intracellular multiplication. Therefore we conclude that the lphA gene product is dispensable for macrophage killing and intracellular multiplication.
The icmMLKE Operon.
Four genes (icmMLKE) were found to be located between lphA and icmG, but only icmE was found to contain Tn903dIIlacZ insertions (LELA4432, LELA1650, LELA3307, and LELA1984) from the original collection (Fig. 1). To test whether icmMLK are also required for macrophage killing, a kanamycin-resistance cassette was introduced into them by allelic exchange (Fig. 3). The three strains generated (GS3008, GS3009, and GS3010) were found to be completely defective for macrophage killing (Table 4). No complementation was observed for these strains with pGS-Lc-40 (Fig. 3). Macrophage killing was observed only at high multiplicities of infection (>10) when pGS-Lc-42 (Fig. 3) was introduced to insertion mutants in icmE (Table 4). Because of this result, the plasmid pGS-Lc-47, which contains part of icmO, lphA, icmMLKE, and icmG (Fig. 3), was constructed. This plasmid exhibits an increased ability to complement all the insertion mutations in icmMLKE (Table. 4). [icmG was shown to be complemented with a plasmid that contain only icmG (M.P. and H.A.S., unpublished results), indicating that the lack of complementation of the icmE insertion mutants with pGS-Lc-42 is not due to polarity of the mutation on icmG. In addition, an icmG mutant was shown to have only a partial reduction in the ability to kill human macrophages (M.P. and H.A.S., unpublished results), an observation that gives additional support to the idea that icmE insertions are not acting by polarity on icmG.]
Table 4.
Complementation of icm mutants
| Strain (Gene) | Plasmid* | Killing† |
|---|---|---|
| JR32 | — | +++ |
| 25D | — | − |
| GS3008 (icmM) | pMMB207αb | − |
| pGS-Lc-40 (icmM+L+K+) | − | |
| pGS-Lc-47 (icmM+L+K+E+) | ++ | |
| pGS-Lc-47-D2 (icmL+K+E+) | − | |
| pGS-Lc-47-D5 (icmM+L+K+) | + | |
| GS3009 (icmL) | pMMB207αb | − |
| pGS-Lc-40 (icmM+L+K+) | − | |
| pGS-Lc-47 (icmM+L+K+E+) | ++ | |
| pGS-Lc-47-D2 (icmL+K+E+) | − | |
| pGS-Lc-47-D3 (icmM+K+E+) | − | |
| pGS-Lc-47-D5 (icmM+L+K+) | + | |
| GS3010 (icmK) | pMMB207αb | − |
| pGS-Lc-40 (icmM+L+K+) | − | |
| pGS-Lc-47 (icmM+L+K+E+) | ++ | |
| pGS-Lc-47-D3 (icmM+K+E+) | − | |
| pGS-Lc-47-D4 (icmM+L+E+) | − | |
| pGS-Lc-47-D5 (icmM+L+K+) | + | |
| LELA4432 (icmE) | pMMB207αb | − |
| pGS-Lc-42 (icmE+) | +/− | |
| pGS-Lc-47 (icmM+L+K+E+) | ++ | |
| pGS-Lc-47-D4 (icmM+L+E+) | + | |
| pGS-Lc-47-D5 (icmM+L+K+) | + |
The plasmid pGS-Lc-47-D5 contains an in-frame deletion in icmE; the plasmid pGS-Lc-40 does not contain the icmE gene.
As determined by macrophage cytotoxicity assay.
To test whether the icmMLK genes are required for macrophage killing or whether insertions in these genes act by polarity on icmE, nonpolar in-frame deletions were constructed in each of them. The plasmids pGS-Lc-47-D2, pGS-Lc-47-D3, and pGS-Lc-47-D4 containing in-frame deletions in icmM, icmL, and icmK, respectively, were tested for their ability to complement GS3008, GS3009, and GS3010, respectively (Table 4). All these plasmids failed to complement the mutants. To test whether the plasmids that contain the in-frame deletions express the proteins located down-stream from the in-frame deletion, the same plasmids were used for complementation of strains that contain an insertion in a down-stream gene. When we tried to complement GS3009, which contains an insertion in icmL, with pGS-Lc-47-D2, which contains an in-frame deletion in icmM, no complementation was observed. The same result was obtained when we tried to complement GS3010, which contains an insertion in icmK, with pGS-Lc-47-D3, which contains an in-frame deletion in icmL. Partial complementation was observed when LELA4432, which contains an insertion in icmE, was complemented with pGS-Lc-47-D4, which contains an in-frame deletion in icmK (Table 4). These results do not allow us to distinguish between the possibility that icmM and icmL are themselves required for macrophage killing, or that the insertions located in them result in strains that are attenuated for macrophage killing only because of polarity on icmK and icmE. All other attempts to clarify this point were unsuccessful.
A 222-codon in-frame deletion was also constructed in icmE (codons 316–537) located in the multiple repeats region (codons 192–724). This plasmid, pGS-Lc-47-D5, partially complemented icmE insertion mutants, indicating that the protein product produced from pGS-Lc-47-D5 was still partially functional. Plasmid pGS-Lc-47-D5 also partially complemented GS3008, GS3009, and GS3010, which contain insertions in icmM, icmL, and icmK, respectively (Table 4).
Several icm Genes Are Required for Plasmid Conjugation.
The sequence similarity found between four icm genes and plasmid transfer genes (Table 2) led us to test whether two L. pneumophila strains are able to conjugate plasmids between one another. As we described, the IncQ plasmid pMMB207αb can be conjugated from L. pneumophila JR32 wild-type strain to a rifampicin-resistant JR32 recipient strain (JR32R), at a frequency of 10−3 to 10−4 per donor (7). Table 5 contains a complete analysis of conjugation frequency of insertion mutants in all the icm genes found in this region. These genes can be divided into three groups according to their affect on conjugation frequency: (i) no effect on conjugation frequency; (ii) moderate reduction (10- to 100-fold) in conjugation frequency—icmF, icmE, and icmC; and (iii) severe reduction (>1000-fold) in conjugation frequency—icmT and icmR. The conjugation frequency was restored close to wild-type level when complementing plasmids were introduced to the icmT, icmR, and icmF mutants and partial complementation was observed with icmE and icmC mutants. We also tested the involvement of two genes (icmX and dotA) from the icmA-dotA locus (5, 6); a dotA mutant was found to have a reduction of about 100-fold in conjugation frequency, whereas an insertion in icmX had no effect on conjugation frequency.
Table 5.
Conjugation frequency of icm mutants
DISCUSSION
L. pneumophila is a broad host range facultative intracellular pathogen that overcomes many natural host defense mechanisms, enabling it to cause disease in humans. Like Mycobacterium tuberculosis (17), Chlamydia psittaci (18), and Toxoplasma gondii (19), L. pneumophila infects human cells, survives, and multiplies within a specialized vacuole that does not fuse with secondary lysosomes (3). In nature, L. pneumophila and related species are found within protozoa, sometimes as symbionts. It is not known whether Legionellaceae are able to be free living, but there are several line of evidence suggesting that in natural environments Legionella grow exclusively within other organisms (2).
We described here a 22-kb DNA locus (Fig. 1) containing 18 genes, 16 of which were shown to be required for macrophage killing (ref. 7; M.P. and H.A.S., unpublished results; and this study). Protein and DNA sequence similarity searches found that 12 icm genes bear no significant sequence similarity to known prokaryotic or eukaryotic proteins. The same result was also found for the proteins that were described in the icmA-dotA locus. In contrast, IcmP, IcmO, IcmL, and IcmE show significant degree of sequence similarity (Table 2) with plasmid-encoded proteins that are involved in plasmid DNA transfer. One protein (IcmW) from the icmA-dotA locus was also found to have low sequence similarity (32% identity and 39% similarity, over 82 amino acids) to VirD3 protein from the Agrobacterium rhizogenes plasmid pRia4b and to the conjugal protein TrbB (34% identity and 46% similarity, over 59 amino acids) from A. tumefaciens plasmid pTiA6NC.
Because of the sequence similarities described above, the involvement of icm genes in L. pneumophila conjugation was characterized. We found that two icm genes (icmT and icmR) are completely required for plasmid conjugation, and three icm genes (icmF, icmE, and icmC) are partially required for plasmid conjugation. Plasmids harboring these genes were able to complement strains containing an insertion in these genes for both human macrophage killing and bacterial conjugation.
Sequence similarity between conjugation-related genes and genes involved in pathogen–host cell interactions has been described in plants and animals (20). The conjugation system of the IncN plasmid pKM101 was adapted by the plant pathogen A. tumefaciens to transfer DNA into plants and by the human pathogen Bordetella pertussis for the export of the pertussis toxin. In these three systems a high sequence similarity and similar gene organization were found (21). In the case of A. tumefaciens it was shown that mutations in several vir genes (genes involved T-DNA transfer into plants) can reduce the conjugation frequency of a heterologous RSF1010 plasmid between two strains of A. tumefaciens (22, 23), even though an additional system for plasmid conjugation (tra and trb genes) was shown to be located on the plasmid that contains the vir genes (24). The three systems described above are involved in secretion of macromolecules. The fact that several L. pneumophila icm genes were found be required for bacterial conjugation may indicate that secretion of a macromolecule (of an unknown nature) plays an important role in the ability of L. pneumophila to multiply within and kill human macrophages. It is important to note that the two genes (icmT and icmR) in which insertions most dramatically reduce conjugation frequency bear no significant sequence similarity to any known genes. Whereas icmP, icmO, icmL, and icmM which do exhibit significant sequence similarity to conjugation-related genes, are dispensable for plasmid conjugation but not for macrophage killing. We currently do not know whether conjugation itself plays a role in macrophage killing. It is possible that small plasmids take advantage of an existing secretion system to be mobilized, or that DNA transfer is required for human macrophage killing by L. pneumophila.
Another possibility is that icm gene products are not involved directly in plasmid DNA transfer but that the reduction in conjugation frequency is due to interaction of icm gene products with a different plasmid conjugation system. We found this possibility to be very unlikely for two reasons: the strains tested contain insertions close to the 5′ end of the icm genes, so that a negative dominant effect of an aberrant icm gene product is doubtful. The possibility of interaction at the regulatory level is also highly unlikely, as the effect on conjugation frequency was observed with six different genes. Therefore we think that the effect of icm gene on plasmid conjugation is a direct effect, in the sense that small plasmids are mobilized by an existing secretion system, built by the icm gene products.
It is known that proteins involved in secretion, as well as bacterial pathogenicity factors, can be encoded by mobile genetic elements. Shigella-like toxins of E. coli, cholera toxin of Vibrio cholerae, and others, are phage encoded (25, 26). Several virulence factors of Gram-negative as well as Gram-positive bacteria are plasmid encoded (27, 28). In addition, virulence factors (such as the type III secretion system) of several Gram-negative bacteria (such as uropathogenic E. coli, Yersinia pesitis, and Salmonella typhimurium) were shown to be encoded from chromosomal pathogenicity islands (29). Pathogenicity islands are thought to be acquired by horizontal gene transfer in which a large fragment of DNA is mobilized and integrated into the chromosome. In this case the G+C content of the integrated region was shown to differ from the chromosomal one. The G+C content of L. pneumophila icm locus was found to be 39%, which is the same as the L. pneumophila chromosome. The G+C content of trbA and trbC genes (which are the only two genes is which sequence similarity was found over the whole coding sequence) from the Salmonella plasmid R64, was found to be 49%. The similarity of icm genes G+C content to the overall genome G+C content may indicate that the icm locus is not a typical pathogenicity island.
It seems likely that the intracellular life style of L. pneumophila (in protozoa and animals) minimizes the occasions for horizontal gene transfer between it and other bacteria. The sequence similarities that were found in the icm region may represent sporadic events of horizontal gene transfer that occurred long ago and ended in the acquisition of several genes, but the majority of the icm genes do not show a relation to genes of other organisms. The data presented here may indicate that L. pneumophila has evolved a previously unrecognized system for survival and replication inside a wide variety of protozoan and mammalian host cells.
Acknowledgments
We are grateful to Ms. Carmen Rodriguez for excellent technical assistance. This work was supported by a grant from the National Institutes of Health (AI23549). G.S. is supported by a long-term fellowship from the European Molecular Biology Organization.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. Y15044).
References
- 1.Horwitz M A, Silverstein S C. J Clin Invest. 1980;60:441–450. doi: 10.1172/JCI109874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fields B S. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 3.Horwitz M A. J Exp Med. 1983;158:2108–2126. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sadosky A B, Wiater L A, Shuman H A. Infect Immun. 1993;61:5361–5373. doi: 10.1128/iai.61.12.5361-5373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brand B C, Sadosky A B, Shuman H A. Mol Microbiol. 1994;14:797–808. doi: 10.1111/j.1365-2958.1994.tb01316.x. [DOI] [PubMed] [Google Scholar]
- 6.Berger K H, Merriam J J, Isberg R R. Mol Microbiol. 1994;14:809–822. doi: 10.1111/j.1365-2958.1994.tb01317.x. [DOI] [PubMed] [Google Scholar]
- 7.Segal G, Shuman H A. Infect Immun. 1997;56:5057–5066. doi: 10.1128/iai.65.12.5057-5066.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horwitz M A. J Exp Med. 1987;166:1310–1328. doi: 10.1084/jem.166.5.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins S J, Ruscetti F W, Gallagher R E, Gallo R C. Proc Natl Acad Sci USA. 1978;75:2458–2462. doi: 10.1073/pnas.75.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szeto L, Shuman H A. Infect Immun. 1990;58:2585–2592. doi: 10.1128/iai.58.8.2585-2592.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furuya N, Komano T. J Bacteriol. 1996;178:1491–1497. doi: 10.1128/jb.178.6.1491-1497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tietze E, Tschape H. J Basic Microbiol. 1994;34:105–116. doi: 10.1002/jobm.3620340206. [DOI] [PubMed] [Google Scholar]
- 13.Lessl M, Balzer D, Pansegrau W, Lanka E. J Biol Chem. 1992;267:20471–20480. [PubMed] [Google Scholar]
- 14.Bjellqvist B, Hughes G J, Pasquali C, Paquet N, Ravier F, Sanchez J, Frutiger S, Hochstrasser D F. Electrophoresis. 1993;14:1023–1031. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- 15.Nakai K, Kanehisa M. Proteins Struct Funct Genet. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 16.Bairoch A. Nucleic Acids Res. 1992;20:2013–2018. doi: 10.1093/nar/20.suppl.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armstrong J A, D’Arcy Hart P. J Exp Med. 1971;134:713–740. doi: 10.1084/jem.134.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friis R R. J Bacteriol. 1972;180:706–721. doi: 10.1128/jb.110.2.706-721.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones T C, Hirsch J G. J Exp Med. 1972;136:1173–1184. doi: 10.1084/jem.136.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winans S C, Burns D L, Christie P J. Trends Microbiol. 1996;4:64–68. doi: 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pohlman R F, Genetti H D, Winans S C. Mol Microbiol. 1994;14:655–668. doi: 10.1111/j.1365-2958.1994.tb01304.x. [DOI] [PubMed] [Google Scholar]
- 22.Beijersbergen A, Dulk-Ras A D, Schilperoort R A, Hooykaas P J J. Science. 1992;256:1324–1327. doi: 10.1126/science.256.5061.1324. [DOI] [PubMed] [Google Scholar]
- 23.Gelvin S B, Habeck L L. J Bacteriol. 1990;172:1600–1608. doi: 10.1128/jb.172.3.1600-1608.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alt-Morbe J, Stryker J L, Fuqua C, Li P L, Farrand S K, Winans S C. J Bacteriol. 1996;178:4248–4257. doi: 10.1128/jb.178.14.4248-4257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waldor M K, Mekalanos J J. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 26.Cheetham B F, Katz M E. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 27.Lambert de Rouvroit C, Sluiters C, Cornelis G R. Mol Microbiol. 1992;6:395–409. [PubMed] [Google Scholar]
- 28.Venkatesan M M, Buysse J M, Oaks E V. J Bacteriol. 1992;174:1990–2001. doi: 10.1128/jb.174.6.1990-2001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]