Abstract
In this study, we investigate the structure of the mitochondrial F0F1-ATP synthase of the colorless alga Polytomella sp. with respect to the enzyme of its green close relative Chlamydomonas reinhardtii. It is demonstrated that several unique features of the ATP synthase in C. reinhardtii are also present in Polytomella sp. The α- and β-subunits of the ATP synthase from both algae are highly unusual in that they exhibit extensions at their N- and C-terminal ends, respectively. Several subunits of the Polytomella ATP synthase in the range of 9 to 66 kD have homologs in the green alga but do not have known equivalents as yet in mitochondrial ATP synthases of mammals, plants, or fungi. The largest of these so-called ASA (ATP Synthase-Associated) subunits, ASA1, is shown to be an extrinsic protein. Short heat treatment of isolated Polytomella mitochondria unexpectedly dissociated the otherwise highly stable ATP synthase dimer of 1,600 kD into subcomplexes of 800 and 400 kD, assigned as the ATP synthase monomer and F1-ATPase, respectively. Whereas no ASA subunits were found in the F1-ATPase, all but two were present in the monomer. ASA6 (12 kD) and ASA9 (9 kD), predicted to be membrane bound, were not detected in the monomer and are thus proposed to be involved in the formation or stabilization of the enzyme. A hypothetical configuration of the Chlamydomonad dimeric ATP synthase portraying its unique features is provided to spur further research on this topic.
F0F1-ATP synthase (EC 3.6.1.3) occurs ubiquitously on energy-transducing membranes, such as mitochondrial, thylakoid, and bacterial plasma membranes, and produces the majority of cellular ATP under aerobic conditions. The enzyme separates readily into two distinct parts: a membrane-embedded domain (F0-ATP synthase) involved in proton translocation and a water-soluble domain (F1-ATPase) that is the site of ATP synthesis or hydrolysis. The most investigated ATP synthase to date is that of Escherichia coli, consisting of eight different subunits with a stoichiometry of α3β3γδɛab2c10-12, which are all essential for its function (Foster and Fillingame, 1982; Schneider and Altendorf, 1985). Subunits-α, -β, -γ, -δ, and -ɛ constitute the F1-ATPase, whereas subunits-a, -b, and -c constitute the F0-ATP synthase. A central stalk that connects the two domains is formed by subunits-γ and -ɛ, while a second peripheral stalk that links the apex of the α3β3 hexagon to the F0 domain is constituted by subunit-b and -δ.
The mitochondrial F0F1-ATP synthase is significantly more complex than the bacterial enzyme and consists of at least 15 different proteins. In addition to the eight essential subunits found in bacteria, the mitochondrial enzyme contains several so-called supernumerary subunits (<20 kD) that are required for the structure or regulation of the enzyme. For instance, the peripheral stalk of mitochondrial ATP synthase is composed of four subunits (b, d, F6, and oligomycin sensitivity conferral protein [OSCP]; OSCP is equivalent to subunit-δ in bacteria; Collinson et al., 1994, 1996) instead of two in bacteria (b and δ). Other supernumerary subunits are involved in the dimerization or oligomerization of the ATP synthase, a process that was shown to occur in the mitochondrial membrane (Allen et al., 1989; Paumard et al., 2002; Dudkina et al., 2005; Minauro-Sanmiguel et al., 2005). In yeast (Saccharomyces cerevisiae), dimerization involves the physical association of two neighboring F0 domains via subunit 4 (subunit-b in mammals) and the associated proteins e and g (Spannagel et al., 1998; Paumard et al., 2002). These proteins as well as several other F0 subunits are found in both mammals and yeast. In plants, fewer F0 subunits have been identified so far, whereas a plant-specific subunit named FAd was found (Heazlewood et al., 2003).
Compared to other known mitochondrial ATP synthases, those of Chlamydomonad algae show several unique structural features. First, the catalytic subunits-α and -β in the green alga Chlamydomonas reinhardtii exhibit peculiar extensions at their N and C termini, respectively (Franzén and Falk, 1992; Nurani and Franzén, 1996). The α-subunit in the nonphotosynthetic alga Polytomella sp. also exhibits an extended N terminus, the sequence of which is highly similar to that of its close relative, C. reinhardtii (Atteia et al., 1997). Second, both C. reinhardtii and Polytomella sp. mitochondrial ATP synthases, solubilized with dodecyl maltoside (1%–2%), run on blue native PAGE (BN-PAGE) exclusively as a dimer of approximately 1,600 kD (Atteia et al., 2003; van Lis et al., 2003, 2005). This is in clear contrast to the mitochondrial ATP synthases of beef, yeast, or plants, which, when solubilized with 1% dodecyl maltoside, occur on BN-PAGE as two major forms corresponding to the F0F1-ATP synthase monomer (600 kD) and the F1-ATPase (400 kD; Jänsch et al., 1996; Arnold et al., 1998; Eubel et al., 2003). Third, C. reinhardtii mitochondrial ATP synthase separated by BN-PAGE contains seven proteins in the range of 10 to 61 kD that have no clear equivalents in other mitochondrial ATP synthases (Funes et al., 2002; van Lis et al., 2003) and that were given the designation ASA (ATP Synthase-Associated; Cardol et al., 2005). Moreover, a recent study on the ATP synthase of Polytomella sp. revealed the presence of nine ASA proteins, whereas typical mitochondrial ATP synthase subunits-b, -F6, and -d of the peripheral stalk and supernumerary subunits-A6L, -e, -f, and -g are missing (Vázquez-Acevedo et al., 2006). Last, electron microscopy images of the dimeric mitochondrial ATP synthase of Polytomella sp. showed the presence of a very pronounced peripheral stalk (Dudkina et al., 2005), strongly disparate from the thin stalk found in the yeast ATP synthase (Dudkina et al., 2006).
Here, we further characterize the mitochondrial ATP synthase of the colorless alga Polytomella sp. in relation to the available data on this enzyme in C. reinhardtii and follow up on the electron microscopy and biochemical data obtained for the Polytomella ATP synthase (Atteia et al., 1997, 2003; Dudkina et al., 2005, 2006; Vázquez-Acevedo et al., 2006). As the composition of the ATP synthases from the two algae appear highly similar, data on the enzyme obtained from either source are assumed to be mostly interchangeable. This work gives new insights into the structure of the mitochondrial ATP synthase of the Chlamydomonad algae and opens the way to a testable hypothesis regarding the structural features and role of the identified atypical traits.
RESULTS
Polytomella sp. and C. reinhardtii F0F1-ATP Synthase Separated by Two-Dimensional BN/SDS-PAGE
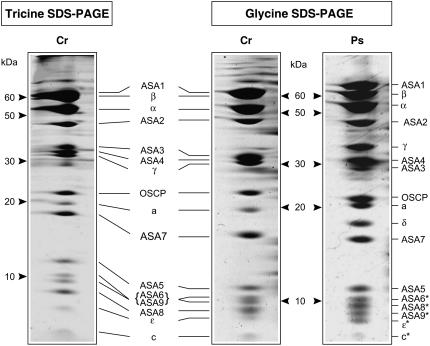
BN gel bands containing the dimeric F0F1-ATP synthase from Polytomella sp. and C. reinhardtii (1,600 kD) were applied on Gly or Tricine SDS-PAGE. The Polytomella ATP synthase is resolved into 17 distinct polypeptides on glycine SDS-PAGE, ranging from 66 kD to 7 kD (Fig. 1). Recently, on Tricine SDS-PAGE, 17 subunits, including nine ASA subunits, were found that were assigned after N-terminal sequencing (Vázquez-Acevedo et al., 2006). These N-terminal sequences corroborated those we obtained for the 12 largest proteins from the Polytomella ATP synthase (data not shown). The ASA subunit nomenclature used here is as previously reported (Cardol et al., 2005; Vázquez-Acevedo et al., 2006). The ASA designation with Ps or Cr prefix refers to the subunit in a particular alga and otherwise refers to the subunit in a general sense, having presumably a similar structure and function in both algae.
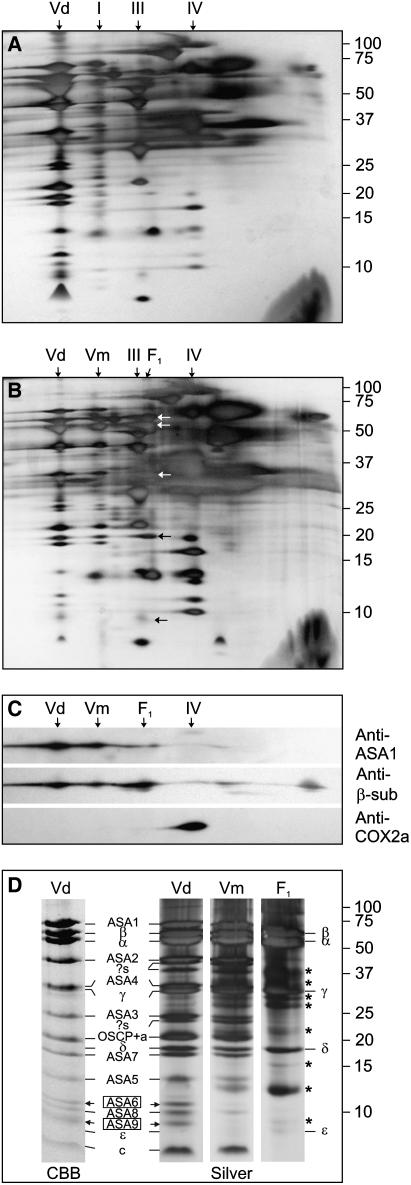
Figure 1.
Polypeptide composition of the Polytomella (Ps) F0F1-ATP synthase dimer and comparison with that of C. reinhardtii (Cr). The subunits of the ATP synthase from BN gel were resolved on 2D Gly SDS-PAGE (15% acrylamide) or on Tricine SDS-PAGE (13% acrylamide) and Coomassie Blue stained. For C. reinhardtii, Tricine SDS-PAGE was used to allow reference to earlier studies using this system (Funes et al., 2002; van Lis et al., 2003). The identities of subunits-ASA8, -ASA9, and -ɛ of C. reinhardtii on Tricine SDS-PAGE were not confirmed. The 12 largest Polytomella subunits on Gly SDS-PAGE were identified by N-terminal sequencing, whereas the rest (marked with *) were tentatively assigned based on the Tricine SDS-PAGE subunit profile (Vázquez-Acevedo et al., 2006). The C. reinhardtii subunits on Gly SDS-PAGE were identified by liquid chromatography-mass spectrometry/mass spectrometry analysis (A. Atteia, A. Adrait, S. Brugière, M. Tardiff, R. van Lis, L. Kuhn, O. Bastien, J. Garin, J. Joyard, and N. Rolland, unpublished data).
Notwithstanding some disparities, the ATP synthase subunit profiles on Gly SDS-PAGE are rather similar in the two algae. The α- and β-subunits migrate similarly in both algae (Fig. 1). Sequencing of the α- and β-subunit cDNAs of Polytomella sp. revealed the presence of atypical extensions highly similar to those of C. reinhardtii. The α-subunits in both algae contain an N-terminal extension of approximately 20 residues, whereas the β-subunits exhibit at their C termini a hydrophilic α-helical extension of about 64 residues (Fig. 2). The function of these extensions is not known. The γ-subunit of Polytomella sp. runs consistently higher on SDS-PAGE than that of C. reinhardtii (Fig. 1), although the predicted molecular masses of the γ-subunits from both algae are very similar (approximately 31 kD). Polytomella subunits-α, -β, and -γ all show highest sequence identity with their counterparts in C. reinhardtii. Subunit-δ of C. reinhardtii, which could only be detected by silver staining, migrates similarly to that of Polytomella sp. (18 kD; data not shown).
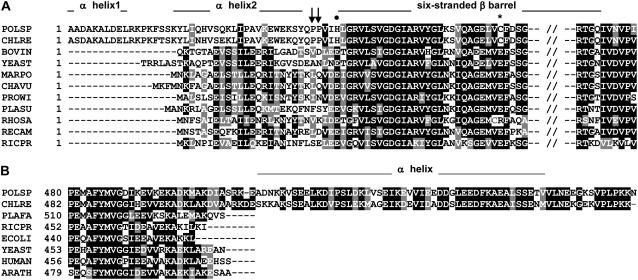
Figure 2.
Subunits-α and -β in Chlamydomonad algae exhibit extensions. A, Multiple sequence alignment of the N-terminal part of mature α-subunits. Of the two predicted N-terminal α-helices, the second α-helix is amphipathic and likely interacts with the N terminus of the OSCP (Weber et al., 2004). The arrows indicate the position of unique Pro residues in the algal proteins; •, conserved Glu residue substituted by a His residue in the algae; *, the presence of an unusual Cys residue in the algae. Sequences used are: BOVIN, Bos taurus (P19483); CHAVU, Chara vulgaris (Q7YAN8); CHLRE, C. reinhardtii (Q96550); MARPO, Marchantia polymorpha (P2685); PLASU, Platymonas subcordiformis (Q36517); POLSP, Polytomella sp. (CAI3486); PROWI, Prototheca wickerhamii (Q37628); RECAM, Reclinomonas americana (O21268); RICPR, Rickettsia prowazekii (O50288); RHOSA, Rhodomonas salina (Q9G8W0); and YEAST (P07251). B, Multiple sequence alignment of the C-terminal part of different β-subunits. Sequences used are: CHLRE, C. reinhardtii (P38482); POLSP, Polytomella sp. (CAI3487); PLAFA, Plasmodium falciparum (Q810V2); RICPR, R. prowazekii (O50290); ECOLI, E. coli (P0ABB4); YEAST (P00830); HUMAN, Homo sapiens (P06576); and ARATH, Arabidopsis (Q541W7). Note that the presence of Pro residues in predicted α-helices (as seen in the β extension) may in reality disrupt the helices.
In C. reinhardtii, ASA1 migrates together with the β-subunit at 60 kD on SDS-PAGE (van Lis et al., 2003) but runs at 66 kD in Polytomella sp. (Fig. 1). Interestingly, it was inferred from the cDNA sequence of Polytomella ASA1 (PsASA1) that the protein shares 54% sequence identity with the C. reinhardtii ASA1 (CrASA1) but has a 52-residue insertion (L186-S238, precursor protein), which accounts for the 6-kD difference observed on SDS-PAGE.
In the low molecular mass range (<15 kD), the profiles are similar and consist of subunits-ASA5, -6, -8, -9, -ɛ, and -c. C. reinhardtii ASA8 (9.9 kD) and ASA9 (12.1 kD) were newly identified on Gly SDS-PAGE using mass spectrometry (liquid chromatography-mass spectrometry/mass spectrometry) as part of an ongoing proteomics project that includes the C. reinhardtii ATP synthase separated on BN-PAGE (A. Atteia, A. Adrait, S. Brugière, M. Tardiff, R. van Lis, L. Kuhn, O. Bastien, J. Garin, J. Joyard, and N. Rolland, unpublished data). CrASA8 was identified based on the similarity of its N-terminal sequence to that of PsASA8. CrASA9 was inferred to correspond to PsASA9, because the complete sequences of all 17 C. reinhardtii ATP synthase subunits are known and the N-terminal sequences of all 17 Polytomella subunits, except PsASA9, were matched to their C. reinhardtii counterparts (Vázquez-Acevedo et al., 2006). The apparent molecular masses of PsASA9 and CrASA9 are approximately 9 and 10 kD, respectively.
Characteristics of the Chlamydomonad ASA Subunits
Using the complete amino acid sequences of all nine CrASA subunits that were retrieved from the C. reinhardtii genome sequence database (v3.0), no homologs were found after sequence similarity searches against nonredundant databases at the National Center for Biotechnology Information and others, including the genome sequences of the diatom Thalassiosira pseudonana and the green alga Ostreococcus tauri at the Joint Genome Institute (JGI). At present, the ASA subunits were found only in Chlorophycean algae (Vázquez-Acevedo et al., 2006). Conserved domain searches at the National Center for Biotechnology Information and different pattern and profile searches at the ExPASy proteomics server did not reveal any functional domains in the CrASA subunits that could hint to their role.
Based on the CrASA sequences, the set of uncharacterized ASA subunits covers a pI range of 5.66 to 9.27, a calculated molecular mass range of 60 kD (66 kD in Polytomella sp.) to 10 kD, and a grand average of hydrophobicity (GRAVY) score of −0.402 to 0.355 (Table I). ASA6 is predicted to exhibit two transmembrane (TM) segments, whereas ASA8 and ASA9 are largely hydrophilic proteins that, on the contrary, seem to contain one TM segment. Conversely, ASA2 is an overall hydrophobic protein that does not contain strongly predicted TM segments. ASA1, ASA4, and ASA6 possibly possess coiled-coil structures, which may be important for inter- or intrasubunit interactions. ASA5, ASA8, and ASA9 do not contain cleavable targeting sequences, which suggests their targeting to the ATP synthase complex directly via the intermembrane space instead of via the matrix (for review, see Herrmann and Neupert, 2003; Herrmann and Hell, 2005) or possibly via a differential translocation across the inner membrane, such as was described for the AAA protein BCS1 (Stan et al., 2003).
Table I.
Physicochemical properties of the ASA subunits of the Chlamydomonad mitochondrial ATP synthase
The presence of putative coiled-coil-forming sequences may indicate intersubunit interactions or homodimerization. N-term. (Res. No.), Residue number marking start of N terminus; MM, molecular mass calculated from sequence; GRAVY, higher scores indicate increasing hydrophobicity; TM prediction, indicates the number of predicted TM segments using SOSUI (S), TMpred (T); H, HMMTOP; Paircoil score, coiled-coil domain prediction; Ps, Polytomella sp.; Cr., C. reinhardtii.
| Protein | N-Term. (Res. No.) | Protein Identificationa | MM | pI | GRAVY | TM Prediction
|
Maximum Paircoil Score | ||
|---|---|---|---|---|---|---|---|---|---|
| S | T | H | |||||||
| kD | |||||||||
| ASA1 (Ps) | 23 | CAD90158b | 66.2 | 5.47 | −0.402 | – | – | – | 0.054 (K173-K202) |
| ASA1 (Cr) | 24 | 78831 | 60.5 | 5.66 | −0.377 | – | – | – | 0.390 (K17-K46) |
| 0.743 (G424-C453) | |||||||||
| ASA2 | 30 | 192142 | 45.6 | 9.08 | 0.346 | – | 2 | – | – |
| ASA3 | 33 | 152682 | 36.3 | 5.71 | −0.053 | – | – | – | – |
| ASA4 | 29 | 182740 | 31.2 | 6.07 | −0.184 | – | – | – | 0.383 (213A-E424) |
| ASA7 | 27 | 192157 | 19.5 | 9.27 | −0.064 | – | 1 | – | – |
| ASA5 | 1 | 184815 | 14.3 | 9.24 | −0.370 | – | 1 | – | – |
| ASA6 | 28 | 186501 | 13.3 | 9.17 | 0.355 | 2 | 6 | 2 | 0.219 (E2-L31) |
| ASA8 | 1 | 184537 | 9.9 | 9.43 | −0.309 | – | 1 | 1 | – |
| ASA9 | 1 | 191034 | 12.1 | 9.08 | −0.249 | 1 | 1 | 1 | – |
Gene model numbers of JGI Chlamy v3.0.
GenBank accession number.
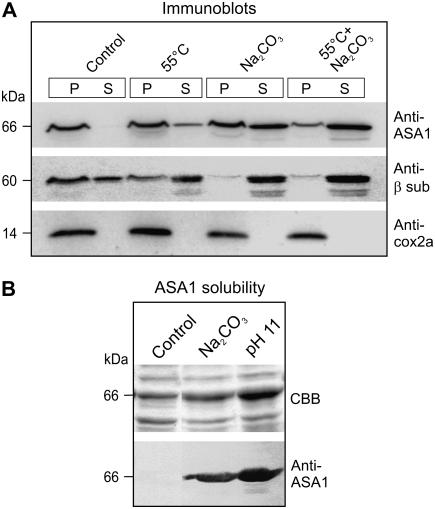
Both CrASA1 and PsASA1 are predicted to be soluble. This notion was supported by dissociation studies using mitochondrial membranes from Polytomella sp. Upon treatment of mitochondrial membranes with either heat (55°C, to dissociate the ATP synthase; see below), Na2CO3 (to release extrinsic proteins), or a combination of the two, ASA1 could be dissociated from the complex and released as a soluble protein, although heat alone results in relatively low levels of soluble ASA1 (Fig. 3A). The differential dissociation of ASA1 and the β-subunit could imply that ASA1 is anchored to the membrane independently from the F1-ATPase. Solubility studies with the overexpressed ASA1 from both algae show that the protein is scarcely soluble at physiological pH (7–8). The protein is, however, soluble at high pH (with Na2CO3 or buffer only; shown in Fig. 3B for PsASA1), which supports its extrinsic nature. A hint that ASA1 is indeed not intrinsic comes from the fact that nonionic detergents such as Triton X-100 or dodecyl maltoside do not increase the solubility of overexpressed ASA1 (data not shown).
Figure 3.
Localization of PsASA1 in the mitochondrial membrane and solubility of the overexpressed protein. A, Mitochondrial membranes from Polytomella were subjected to the indicated treatments. P and S, Pellet and soluble fractions after ultracentrifugation. Immunoblotting shows the dissociation of ASA1 from the membrane with respect to the β-subunit (extrinsic) and subunit COX2a of complex IV (intrinsic). B, Solubility studies were done by sonicating E. coli cells overexpressing PsASA1 in Tris buffer, pH 8.0 (control), in Na2CO3, pH 11.4, and in CAPS, pH 11.0, followed by centrifugation. The supernatants were analyzed by Coomassie Blue staining (CBB) and immunoblotting.
Heat Dissociation of the Strongly Dimeric ATP Synthase of Polytomella sp.
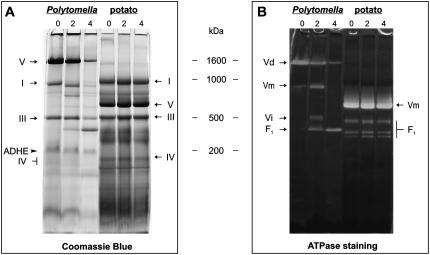
Unlike other known ATP synthases, those of Polytomella sp. and C. reinhardtii hardly dissociate into their F1 and F0 domains upon solubilization with dodecylmaltoside (Figs. 4 and 5A; Atteia et al., 2003; van Lis et al., 2003). Dissociation of the Polytomella ATP synthase dimer could, however, be observed after a short incubation of freshly isolated mitochondria at 55°C. The destabilization of the dimer was shown by BN-PAGE stained with Coomassie Blue and in-gel ATPase activity staining. The ATPase activity of untreated Polytomella mitochondria distributes into a main band of approximately 1,600 kD (Vd) and a faint dissociation product of approximately 800 kD (Vm) on BN gel (Fig. 4). After a 2-min heat treatment of Polytomella mitochondria, the intensity of Vm increased, while a new complex of approximately 400 kD (F1) appeared that showed ATPase activity. After 4 min exposure to 55°C, F1 was the major complex to exhibit ATPase activity (Fig. 4). Low amounts of ATP synthase monomer (Vm) are detected in untreated mitochondria, suggesting a slight dissociation that may be detergent induced. The effect of heat treatment on potato (Solanum tuberosum) mitochondria, used here as a control, was also followed (Fig. 4). The profile of potato protein complexes exhibiting ATPase activity shows the monomeric form of 580 kD (Vm) and three different forms of the F1 domain at 350 to 450 kD that dissociate upon solubilization (Jänsch et al., 1996). Unlike for Polytomella sp., heat treatment did not alter significantly the presence of the potato ATP synthase (sub) complexes or the other oxidative phosphorylation (OXPHOS) complexes (Fig. 4). Also, for mitochondria from C. reinhardtii, no significant heat dissociation could be observed (data not shown). The difference in heat dissociation between Polytomella sp. and these other organisms may stem from differences in subunit structure and in membrane lipid composition.
Figure 4.
BN-PAGE analysis of heat-treated mitochondria from Polytomella and potato tubers. Freshly isolated mitochondria were incubated at 55°C for 2 and 4 min and solubilized with dodecyl maltoside for BN-PAGE analysis. A, Coomassie Blue-stained BN gel. B, In-gel ATPase activity staining. The position of the OXPHOS (sub) complexes in untreated samples were taken from previous work (Jänsch et al., 1996; Atteia et al., 2003). Roman numbers I to V, OXPHOS complexes; Vi, intermediate form (uncharacterized); F1, dissociated F1 domain; ADHE, bifunctional aldehyde/alcohol dehydrogenase; 0, untreated samples; 2, 2 min; 4, 4 min.
Figure 5.
2D BN/SDS-PAGE analysis of Polytomella mitochondria incubated at 55°C. A to C, A whole lane from a BN gel was loaded on a Tricine denaturing gel (13% acrylamide). A, No heat treatment. B, A 2-min heat treatment. Arrows indicate the subunits of the F1-ATPase. C, Immunoblotting of proteins on 2D gels after 2-min heat treatment. D, Pieces from a BN gel containing the different ATP synthase forms that were present after 2 min of heat treatment were cut out and loaded on Tricine-SDS-PAGE. Dimer-specific subunits ASA6 and ASA9 are boxed. Roman numbers I to V, Vd, Vm, as in Figure 4; ?s, not identified and only detected by silver staining; *, contaminating bands resulting from overlapping protein components in the 2D protein profile. Subunit assignment on Tricine SDS-PAGE was confirmed from Vázquez-Acevedo et al. (2006).
Control and 2-min heat-treated mitochondria from Polytomella sp. were analyzed by two-dimensional (2D) BN/SDS-PAGE and silver staining using the Tricine system for its good resolution of smaller proteins (Schägger and von Jagow, 1987). The main changes induced by heat are the dissociation of the complexes that have large extramembrane entities, the ATP synthase and complex I, whereas complexes III and IV seemed relatively unaffected (Fig. 5, A and B). In the 2-min heat treatment 2D SDS-PAGE profile, the composition of Vm resembles that of Vd. Because Vm includes all the typical mitochondrial ATP synthase subunits, it likely corresponds to the monomeric ATP synthase. The polypeptide profile of F1 is typical of a F1-ATPase with its five major subunits (Fig. 5, B and D). The behavior of ASA1 with respect to the β-subunit on 2D BN/SDS-PAGE after 2-min heat treatment was followed by immunoblotting. ASA1 was mainly detected in Vd and Vm, whereas low levels of ASA1 were found past Vm but not in F1, indicating that some ASA1 may be associated in F0 subcomplexes (Fig. 5C). However, clear evidence of this was not obtained from 2D SDS-PAGE analysis, likely because protein levels were too low. Most of subunit-β was present in Vd, Vm, and F1, whereas a portion was found, unlike ASA1, in its free form at the bottom of the gel. As judged by the strong signal of the β-subunit in F1, a sizable part of the ATP synthase had dissociated beyond the monomer after 2-min heat treatment. The ASA1 signal beyond Vm is proportionally much weaker than that of the β-subunit. It is therefore likely that an important fraction of ASA1 becomes insoluble when heat dissociation progresses beyond the monomeric form.
Although most subunits of Vd are present in Vm, it was found that in the latter form, two bands around 10 kD were missing. These subunits are thus hypothesized to be important for the dimerization of the ATP synthase or for the stabilization of the dimer. By comparing our Tricine SDS-PAGE subunit profile (Fig. 5D) to that reported previously (Vázquez-Acevedo et al., 2006), one subunit was identified as ASA6 (12 kD) and the other as ASA9 (9 kD), which is assumed to match ASA9 in C. reinhardtii (see above).
DISCUSSION
The F0F1-ATP synthases of Polytomella sp. and its photosynthetic counterpart C. reinhardtii share an atypical polypeptide composition. The unusual extensions that C. reinhardtii exhibit at the N and C termini of the catalytic subunits-α and -β, respectively (Franzén and Falk, 1992; Nurani and Franzén, 1996), are shown to be also present in Polytomella sp. The fact that the α- and β-subunit extensions of both algae are highly conserved suggests functional restraints. Based on sequence homology with the IF1 inhibitor protein and the inhibitory peptide of the E. coli ɛ-subunit, a role of the β extension as reversible inhibitor of ATP hydrolysis was hypothesized (Atteia et al., 1997). Based on new sequence and secondary structure data for IF1 and subunit-ɛ, this could, however, not be further confirmed. The possibility exists that the β extension is required to interact with the ASA proteins.
In the F0F1-ATP synthase, the N termini of the α-subunit and the OSCP interact (Carbajo et al., 2005). It was proposed that four conserved residues at the N terminus of the OSCP and its equivalent in chloroplast and bacteria, subunit-δ, are important for the interaction with the α-subunit (Y10, A11, A13, R84, E. coli numbering; Hong and Pedersen, 2003). Surprisingly, these residues are not present in the OSCP of the chlorophytes, which suggests a different interaction with the α-subunit. Twin Pro at positions 41 and 42 may enable a specific reorientation to accommodate this atypical interaction, in which the α extension and/or ASA proteins may play a role.
The occurrence of the ASA proteins and the lack of typical subunits of the peripheral stalk in the mitochondrial ATP synthase, an enzyme that has been extensively studied, are puzzling. Because these atypical subunits lack significant sequence homology to known proteins, questions arise as to their function and localization within the complex and whether they are genuine subunits. In several studies on the ATP synthase of C. reinhardtii and Polytomella sp. using whole mitochondria, mitochondrial membranes, or the isolated complex, the ASA proteins were consistently found in the ATP synthase subunit profile on 2D BN/SDS-PAGE in a similar stoichiometry (Atteia et al., 2003; van Lis et al., 2003, 2005; Vázquez-Acevedo et al., 2006). In addition, in Polytomella, the respective amounts of ATP synthase and respiratory complexes vary with the pH of the growth medium, but this does not affect the presence of the ASA proteins (Atteia et al., 2003). Taken together, these data suggest that the ASA proteins are genuine subunits of the algal ATP synthases.
Different indirect clues exist as to the localization and function of at least some of the ASA subunits in the ATP synthase complex. Of the subunits that constitute the peripheral stalk in the bovine ATP synthase (OSCP, b, d, and F6), only the OSCP is present in the algae; it therefore follows that ASA subunits fulfill a structural role in forming part of the peripheral stalk. A typical mitochondrial subunit-b (approximately 20 kD) contains two TM segments at its N terminus, while the rest of the protein is hydrophilic (Arnold et al., 1998; Burger et al., 2003; Heazlewood et al., 2003). CrASA5 (13 kD) possibly contains an N-terminal TM segment with the remainder of the proteins being hydrophilic; the protein may be anchored in the membrane and protruding into the matrix, similar to subunit-b. Electron microscopy images of dimeric ATP synthase from Polytomella sp. show a very pronounced peripheral stalk or possibly two stalks (Dudkina et al., 2005). In comparison, the yeast ATP synthase possesses no pronounced peripheral stalks but a considerably more bulky F0 domain. It seems thus that the Polytomella membrane domain is too small to host the larger ASA subunits, which are more likely part of the observed pronounced peripheral stalk(s). For the Polytomella enzyme, a large globular domain is visible at the apex of the F1 domain that may be attributed to ASA1. The fact that we established ASA1 to be a soluble protein adds to the notion that the protein most likely has a considerable portion of its surface exposed to the solvent, as seems to be the case with the observed globular domain.
The dissociation of the Polytomella ATP synthase dimer (Vd) by heat treatment allowed to have insights into the composition of the monomer (Vm). Vm (800 kD) contains seven ASA subunits (1, 2, 3, 4, 7, 5, and 8), which causes its apparent molecular mass to be at least 200 kD greater than the ATP synthase monomer of yeast, beef, and Arabidopsis (Arabidopsis thaliana). Two subunits found in Vd, ASA6 and ASA9, were lacking in Vm and are therefore proposed to be involved in the dimerization. Dimer-specific PsASA9 is assumed to match CrASA9, identified by mass spectrometry (A. Atteia, A. Adrait, S. Brugière, M. Tardiff, R. van Lis, L. Kuhn, O. Bastien, J. Garin, J. Joyard, and N. Rolland, unpublished data). Both ASA6 and ASA9 are predicted to be membrane bound. In yeast, GXXXG motifs inside the single TM segments of dimer-specific subunits-g and -e are essential for ATP synthase dimerization, whereas the coiled-coil structure of subunit-e stabilizes the dimer (Arselin et al., 2003; Everard-Gigot et al., 2005). In ASA6, a possible coiled coil is predicted that could function in the dimerization. In the predicted TM segments of ASA6 and ASA9, no GXXXG motifs are present. In addition, no other specific elements were identified that could point to their role in dimer stability, which should thus be further assessed in Polytomella sp. using biochemical methods.
Heat treatment (60°C) was also used by Vázquez-Acevedo et al. (2006) to dissociate the purified Polytomella ATP synthase. Although heat seems to have a similar effect on the ATP synthase whether it is in the mitochondrial membrane or purified, discrepancies exist between the outcomes of the two approaches. First, the above-mentioned authors stated that Vd was identical to Vm (although this was not clearly shown), whereas we show Vm to be devoid of ASA6 and ASA9. The presence of dodecyl maltoside and glycerol in the preparations of purified ATP synthase can affect subunit interactions. Glycerol is known to enhance hydrophobic interactions, whereas dodecyl maltoside may favor TM segment association when used close to the critical micelle concentration of 0.2 mm (Fisher et al., 2003), possibly causing a different dissociation behavior of ASA6 and ASA9. Second, the authors observed an ASA1/3/5/8/a/c10 subcomplex of 200 kD that we have, however, never seen during several years of heat dissociation experiments; immunodetection of ASA1 at or near the expected position of the subcomplex on 2D BN/SDS-PAGE in 2-min heat-treated mitochondria is negligible (this work). The ASA1/3/5/8/a/c10 subcomplex released upon heat treatment in the presence of detergent is apparently soluble, but when membranes are heat treated, the subcomplex likely aggregates, because it cannot be solubilized from the membranes and observed on BN-PAGE.
Based on our analysis of the characteristics of the ATP synthase subunits, which are assumed to be similar in both algae, an attempt has been made to provide structural details to the working model proposed by Dudkina et al. (2005, 2006) involving the presence of two peripheral stalks (Fig. 6). Vázquez-Acevedo et al. (2006) also propose a model of the Polytomella ATP synthase that differs significantly in that it places the ASA1/3/5/8/a/c10 subcomplex entirely in the membrane and ASA subunits 2, 4, and 7 forming a single thin peripheral stalk. We propose that the ASA1/3/5/8/a/c10 subcomplex constitutes the (double) peripheral stalk mostly intact, with the other ASA subunits elsewhere in the complex (Fig. 6, legend). This may help to imagine why the subcomplex/stalk is destabilized in the absence of detergent: the large matrix-exposed and membrane-bound moieties in the stalk could easily aggregate when the interaction with F1-ATPase is disrupted by heat treatment. Complexes without large extramembrane portions such as complex III and IV seem not notably affected by heat.
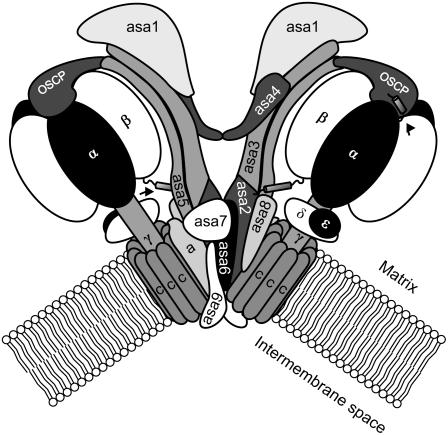
Figure 6.
Working model of dimeric mitochondrial ATP synthase in Chlamydomonad algae, based on our current knowledge of the enzyme from both C. reinhardtii and Polytomella sp. The monomers are shown rotated 180° one from another (around a vertical axis), as proposed previously for the yeast enzyme (Paumard et al., 2002). Arrows indicate the α and β extensions. The α extension is shown interacting with the OSCP and the β extension with ASA subunits of the peripheral stalk. ASA1 is placed in the matrix because it is an extrinsic protein. ASA2 is a hydrophobic protein and probably associates to the membrane. ASA3 is somewhat hydrophilic but without specific features. ASA4 is a possible homodimer-forming subunit based on its prediction for coiled-coil formation (Table I). ASA5 may be anchored in the membrane and protruding into the matrix. ASA6 and ASA9 are proposed to be dimer specific and membrane bound, whereas ASA7 and ASA8 may be associated to or traversing the membrane.
Although the ATP synthases of C. reinhardtii and Polytomella sp. are expected to be similar, the fact that the C. reinhardtii ATP synthase dimer is not readily dissociated by heat illustrates that differences exist. Also, PsASA1 and CrASA1 are highly similar in the algae, but PsASA1 contains a 52-residue insertion (this work). It is therefore desirable to obtain the complete sequences of the PsASA subunits. There are other examples of differences in the respiratory chain of the two algae, which are thought to reflect their evolutionary divergence from a photosynthetic ancestor (Round, 1980; van Lis et al., 2005).
Although questions remain as to what is the advantage of a highly stable dimer, the atypical Chlamydomonad mitochondrial ATP synthase is expected to bring new incentives to the matured field of ATP synthase research.
MATERIALS AND METHODS
Isolation of Mitochondria
Mitochondria were isolated from Polytomella sp. cells (198.80, E.G. Pringsheim) grown on acetate (van Lis et al., 2005) and from Chlamydomonas reinhardtii cells grown on Tris-acetate phosphate medium as previously described (Eriksson et al., 1995). Potato (Solanum tuberosum) tuber mitochondria were isolated as reported (von Stedingk et al., 1997), omitting the Percoll gradient step. Protein concentration was determined using the bicinchoninic acid kit (Pierce). Mitochondrial membranes from Polytomella sp. were prepared as described (Atteia et al., 2003).
BN-PAGE Analysis and ATPase Staining
Isolated mitochondria were washed in 0.25 m sorbitol, 15 mm Bis-Tris, pH 7.0, solubilized with 2% (w/v) dodecyl maltoside (4 g detergent/g protein), and supplemented with 0.5% (w/v) Coomassie Blue after ultracentrifugation for 20 min at 60,000g. Subsequently, BN-PAGE was done as described (Schägger and von Jagow, 1991; Atteia et al., 2003). For in-gel ATPase activity, BN gels were rinsed twice in HEPES-KOH 50 mm, pH 8.0, and then incubated in this buffer containing 30 mm CaCl2 and 10 mm ATP at room temperature. Calcium phosphate precipitates appear within 20 min and continue to intensify up to 1 h.
2D Gel Electrophoresis, Immunoblotting, and N-Terminal Protein Sequencing
2D analysis of BN gel lanes containing mitochondrial proteins of Polytomella sp. and C. reinhardtii was done using 15% Gly SDS-polyacrylamide gels (Laemmli, 1970) or 13% Tricine SDS-polyacrylamide gels (Schägger and von Jagow, 1987). For N-terminal protein sequencing, the gels were blotted onto polyvinylidene difluoride membrane (Millipore) and stained with Ponceau Red. The proteins were subjected to N-terminal sequencing by automated Edman degradation using a LF3000 Beckman sequencer. For immunoblotting, the proteins from Tricine 2D gels were transferred onto nitrocellulose and decorated with the antibodies indicated in Figure 5. The antibodies against the β-subunit and COX2a were previously described (van Lis et al., 2005; Atteia et al., 2006).
Dissociation Studies
Isolated mitochondria were resuspended in their own breaking buffer at a concentration of 25 mg protein/mL. Aliquots of 3 mg of mitochondria in breaking buffer, containing protease inhibitors phenylmethylsulfonyl fluoride (0.1 mm), benzamidine (0.5 mm), and hexanoic acid (1 mm), were treated at 55°C for the indicated times and placed immediately on ice. Subsequently, the samples were processed for BN-PAGE as described above. For ASA1 dissociation studies, mitochondrial membranes from Polytomella sp. were resuspended at a concentration of 1.7 mg/mL, either in PM buffer (5 mm potassium phosphate, pH 7.0, 200 mm mannitol) or in 100 mm Na2CO3, pH 11.4, with the above-mentioned protease inhibitors added. Membranes, part of which were treated at 55°C for 2 min, were left on ice for 30 min with occasional vortexing. The membranes were then centrifuged at 100,000g for 30 min, after which the supernatant was kept, whereas the membranes were collected after washing once in PM buffer. The protein samples were separated using Gly SDS-PAGE (12% acrylamide), transferred onto nitrocellulose, and analyzed by Ponceau Red staining and subsequent immunoblotting.
Screening of a Polytomella cDNA Library for ATP Synthase Subunits
Screening of a Polytomella λ-ZAPII cDNA library (Atteia et al., 2005) was carried out by plaque hybridization following standard protocols, using as probes the cDNAs coding for the corresponding subunits of C. reinhardtii. The isolated cDNAs were sequenced and deposited in the DDBJ/EMBL/GenBank databases.
Overexpression, Antibody Production, and Solubility Assays of ASA1
Using the cDNA of ASA1 as template, a PCR product containing the coding sequence for the entire mature protein from C. reinhardtii was obtained with the forward primer 5′-GGAATTCCATATGTATGTGACCGCCCTGAAGG-′3 and reverse primer 5′-ATGCTGCTCGAGCGCGCCGCGGCC-3′, containing, respectively, the NdeI and XhoI restriction sites (underlined) for subsequent cloning. The PCR product was cloned into the expression vector pET24a (Novagen). A PCR product containing the coding sequence for the entire mature protein from Polytomella sp. was obtained with forward primer 5′-GATCCATGGGCTACCTTGCCCCCCTCCGC-′3 and reverse primer 5′-CTAAGATCTGTTACCGTTGACGAGATCGGG-3′, containing, respectively, the NcoI and BglII restriction sites (underlined) for subsequent cloning. The PCR product was first cloned into the vector pQE60 (Qiagen), from which it was excised using restriction enzymes NcoI and HindIII (partial restriction, as HindIII cuts once in the PsASA1 cDNA). This fragment now contained a C-terminal His tag, which was then cloned into expression vector pACYCDuet (Novagen). Overexpression of CrASA1 and PsASA1 was done using the Escherichia coli strain BL21 (DE3; Stratagene) at 37°C for 4 h, induced by 1 mm isopropyl β-d-1-thiogalactopyranoside. For antibody production, CrASA1 was purified using nickel-nitrilotriacetic acid agarose resin (Sigma) according to standard protocols but in the presence of 2 m urea to maintain complete solubility of the protein. Antibodies against CrASA1 were produced in rabbit at Charles River Laboratories. For solubility studies, E. coli cell pellets overexpressing ASA1 were resuspended in lysis buffer containing 300 mm NaCl, 1 mg/mL lysozyme, and 1 mm phenylmethylsulfonyl fluoride in addition to either 20 mm Tris, pH 8.0, 100 mm Na2CO3, pH 11.4, or 50 mm 3-(cyclohexylamino)propanesulfonic acid (CAPS), pH 11.0, incubated for 30 min on ice and lysed using sonication. Additives such as nonionic detergents were used with Tris buffer at pH 8.0. After centrifugation, the supernatants were run on Gly SDS-PAGE (10% acrylamide) and either stained with Coomassie Blue or transferred onto nitrocellulose and immunoblotted using the anti-ASA1 antibody.
Sequence Analysis
Molecular mass, pI, GRAVY scores, and amino acid composition were determined using the ProtParam program. For the prediction of secondary structures, the SOPMA program was used (Combet et al., 2000), and the Paircoil program was used for the prediction of coiled coil regions (Berger et al., 1995). For TM segment prediction, the SOSUI (Hirokawa et al., 1998), TMPred (Hofmann and Stoffel, 1993), and HMMTOP (Tusnady and Simon, 2001) programs were applied. Multiple sequence alignments were performed with ClustalW (Thompson et al., 1997) and refined manually. These software programs are all available from the ExPASy proteomics Web site. For the analysis of α-helices, use was made of the helical wheel projection and amphipathy moment plot options of the program Winpep v.3.01 (Hennig, 1999). Representations of multiple alignments were done using the BOXSHADE program. The genome sequences of C. reinhardtii (v 3.0), Thalassiosira pseudonana, and Ostreococcus tauri are available at the Joint Genome Institute.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers CAI3486 (α-subunit), CAI3487 (β-subunit), CAF3602 (γ-subunit), and CAD90158 (ASA1).
Acknowledgments
We thank Drs. S.I. Beale (Brown University) and D. Drapier (Institut de Biologie Physico-Chimique) for constructive comments on the manuscript. The gene sequence data were produced by the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) and are provided for use in this publication/correspondence only.
This work was supported by Consejo Nacional de Ciencia y Tecnología (grant no. 41328–Q to G.M.-H.) and by the Centre National de la Recherche Scientifique-Département des Sciences de la Vie (A.A.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Robert van Lis (rvanlis@yahoo.fr).
References
- Allen RD, Schroeder CC, Fok AK (1989) An investigation of mitochondrial inner membranes by rapid-freeze deep-etch techniques. J Cell Biol 108 2233–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schägger H (1998) Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J 17 7170–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arselin G, Giraud MF, Dautant A, Vaillier J, Brethes D, Coulary-Salin B, Schaeffer J, Velours J (2003) The GxxxG motif of the transmembrane domain of subunit e is involved in the dimerization/oligomerization of the yeast ATP synthase complex in the mitochondrial membrane. Eur J Biochem 270 1875–1884 [DOI] [PubMed] [Google Scholar]
- Atteia A, Dreyfus G, González-Halphen D (1997) Characterization of the α and β subunits of the F0F1-ATP synthase from the alga Polytomella spp., a close relative of Chlamydomonas reinhardtii. Biochim Biophys Acta 1320 275–284 [DOI] [PubMed] [Google Scholar]
- Atteia A, van Lis R, Beale SI (2005) Enzymes of the heme biosynthetic pathway in the nonphotosynthetic alga Polytomella sp. Eukaryot Cell 4 2087–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atteia A, van Lis R, Gelius-Dietrich G, Adrait A, Garin J, Joyard J, Rolland N, Martin W (2006) Pyruvate formate-lyase and a novel route of eukaryotic ATP synthesis in Chlamydomonas mitochondria. J Biol Chem 281 9909–9918 [DOI] [PubMed] [Google Scholar]
- Atteia A, van Lis R, Mendoza-Hernández G, Henze K, Martin M, Riveros-Rosas H, González-Halphen D (2003) Bifunctional aldehyde/alcohol dehydrogenase (ADHE) in chlorophyte algal mitochondria. Plant Mol Biol 53 175–188 [DOI] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS (1995) Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA 92 8259–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Lang BF, Braun HP, Marx S (2003) The enigmatic mitochondrial ORF ymf39 codes for ATP synthase chain b. Nucleic Acids Res 31 2353–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbajo RJ, Kellas FA, Runswick MJ, Montgomery MG, Walker JE, Neuhaus D (2005) Structure of the F1-binding domain of the stator of bovine F1F0-ATP synthase and how it binds an alpha-subunit. J Mol Biol 351 824–838 [DOI] [PubMed] [Google Scholar]
- Cardol P, Gonzalez-Halphen D, Reyes-Prieto A, Baurain D, Matagne RF, Remacle C (2005) The mitochondrial oxidative phosphorylation proteome of Chlamydomonas reinhardtii deduced from the genome sequencing project. Plant Physiol 137 447–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson IR, Skehel JM, Fearnley IM, Runswick MJ, Walker JE (1996) The F1F0-ATP synthase complex from bovine heart mitochondria: the molar ratio of the subunits in the stalk region linking the F1 and F0 domains. Biochemistry 35 12640–12646 [DOI] [PubMed] [Google Scholar]
- Collinson IR, van Raaij MJ, Runswick MJ, Fearnley IM, Skehel JM, Orriss GL, Miroux B, Walker JE (1994) ATP synthase from bovine heart mitochondria: in vitro assembly of a stalk complex in the presence of F1-ATP synthase and in its absence. J Mol Biol 242 408–421 [DOI] [PubMed] [Google Scholar]
- Combet C, Blanchet C, Geourjon C, Deléage G (2000) NPS@: network protein sequence analysis. Trends Biochem Sci 25 147–150 [DOI] [PubMed] [Google Scholar]
- Dudkina NV, Heinemeyer J, Keegstra W, Boekema EJ, Braun HP (2005) Structure of dimeric ATP synthase from mitochondria: an angular association of monomers induces the strong curvature of the inner membrane. FEBS Lett 579 5769–5772 [DOI] [PubMed] [Google Scholar]
- Dudkina NV, Sunderhaus S, Braun HP, Boekema EJ (2006) Characterization of dimeric ATP synthase and cristae membrane ultrastructure from Saccharomyces and Polytomella mitochondria. FEBS Lett 580 3427–3432 [DOI] [PubMed] [Google Scholar]
- Eriksson M, Gadeström P, Samuelsson G (1995) Isolation, purification, and characterization of mitochondria from Chlamydomonas reinhardtii. Plant Physiol 107 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H, Jänsch L, Braun HP (2003) New insights into the respiratory chain of plant mitochondria: supercomplexes and a unique composition of complex II. Plant Physiol 133 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard-Gigot V, Dunn CD, Dolan BM, Brunner S, Jensen RE, Stuart RA (2005) Functional analysis of subunit e of the F1Fo-ATP synthase of the yeast Saccharomyces cerevisiae: importance of the N-terminal membrane anchor region. Eukaryot Cell 4 346–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher LE, Engelman DM, Sturgis JN (2003) Effect of detergents on the association of the glycophorin a transmembrane helix. Biophys J 85 3097–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster DL, Fillingame RH (1982) Stoichiometry of subunits in the H+-ATP synthase complex of Escherichia coli. J Biol Chem 257 2009–2015 [PubMed] [Google Scholar]
- Franzén L-G, Falk G (1992) Nucleotide sequence of cDNA clones encoding the beta subunit of mitochondrial ATP synthase from the green alga Chlamydomonas reinhardtii: the precursor protein encoded by the cDNA contains both an N-terminal presequence and a C-terminal extension. Plant Mol Biol 19 771–780 [DOI] [PubMed] [Google Scholar]
- Funes S, Davidson E, Claros MG, van Lis R, Perez-Martinez X, Vázquez-Acevedo M, King MP, González-Halphen D (2002) The typically mitochondrial DNA-encoded ATP6 subunit of the F1F0-ATP synthase is encoded by a nuclear gene in Chlamydomonas reinhardtii. J Biol Chem 277 6051–6058 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Whelan J, Millar AH (2003) The products of the mitochondrial orf25 and orfB genes are FO components in the plant F1F0 ATP synthase. FEBS Lett 540 201–205 [DOI] [PubMed] [Google Scholar]
- Hennig L (1999) WinGene/WinPep: user-friendly software for the analysis of aminoacid sequences. Biotechniques 26 1170–1172 [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Hell K (2005) Chopped, trapped or tacked: protein translocation into the IMS of mitochondria. Trends Biochem Sci 30 205–211 [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Neupert W (2003) Protein insertion into the inner membrane of mitochondria. IUBMB Life 55 219–225 [DOI] [PubMed] [Google Scholar]
- Hirokawa T, Boon-Chieng S, Mitaku S (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14 378–379 [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W (1993) TMbase: a database of membrane spanning proteins segments. Biol Chem Hoppe Seyler 374 166 [Google Scholar]
- Hong S, Pedersen PL (2003) ATP synthases: insights into their motor functions from sequence and structural analyses. J Bioenerg Biomembr 35 95–120 [DOI] [PubMed] [Google Scholar]
- Jänsch L, Kruft V, Schmitz UK, Braun HP (1996) New insights into the composition, molecular mass and stoichiometry of the protein complexes of plant mitochondria. Plant J 9 357–368 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Minauro-Sanmiguel F, Wilkens S, Garcia JJ (2005) Structure of dimeric mitochondrial ATP synthase: novel F0 bridging features and the structural basis of mitochondrial cristae biogenesis. Proc Natl Acad Sci USA 102 12356–12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurani G, Franzén L-G (1996) Isolation and characterization of the mitochondrial ATP synthase from Chlamydomonas reinhardtii: cDNA sequence and deduced protein sequence of the alpha subunit. Plant Mol Biol 31 1105–1116 [DOI] [PubMed] [Google Scholar]
- Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brethes D, di Rago JP, Velours J (2002) The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J 21 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round FE (1980) The evolution of pigmented and unpigmented unicells: a reconsideration of the Protista. Biosystems 12 61–69 [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166 368–379 [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199 223–231 [DOI] [PubMed] [Google Scholar]
- Schneider E, Altendorf K (1985) All three subunits are required for the reconstitution of an active proton channel (F0) of Escherichia coli ATP synthase (F1F0). EMBO J 4 515–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spannagel C, Vaillier J, Arselin G, Graves PV, Grandier-Vazeille X, Velours J (1998) Evidence of a subunit 4 (subunit b) dimer in favor of the proximity of ATP synthase complexes in yeast inner mitochondrial membrane. Biochim Biophys Acta 1414 260–264 [DOI] [PubMed] [Google Scholar]
- Stan T, Brix J, Schneider-Mergener J, Pfanner N, Neupert W, Rapaport D (2003) Mitochondrial protein import: recognition of internal import signals of BCS1 by the TOM complex. Mol Cell Biol 23 2239–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady GE, Simon I (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17 849–850 [DOI] [PubMed] [Google Scholar]
- van Lis R, Atteia A, Mendoza-Hernández G, González-Halphen D (2003) Identification of novel mitochondrial protein components of Chlamydomonas reinhardtii: a proteomic approach. Plant Physiol 132 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lis R, González-Halphen D, Atteia A (2005) Divergence of the mitochondrial electron transport chains from the green alga Chlamydomonas reinhardtii and its colorless close relative Polytomella sp. Biochim Biophys Acta 1708 23–34 [DOI] [PubMed] [Google Scholar]
- Vázquez-Acevedo M, Cardol P, Cano-Estrada A, Lapaille M, Remacle C, González-Halphen D (2006) The mitochondrial ATP synthase of chlorophycean algae contains eight subunits of unknown origin involved in the formation of an atypical stator-stalk and in the dimerization of the complex. J Bioenerg Biomembr 38 271–282 [DOI] [PubMed] [Google Scholar]
- von Stedingk EM, Pavlov PF, Grinkevich VA, Glaser E (1997) Mitochondrial protein import: modification of sulfhydryl groups of the inner mitochondrial membrane import machinery in Solanum tuberosum inhibits protein import. Plant Mol Biol 35 809–820 [DOI] [PubMed] [Google Scholar]
- Weber J, Muharemagic A, Wilke-Mounts S, Senior AE (2004) Analysis of sequence determinants of F1F0-ATP synthase in the N-terminal region of alpha subunit for binding of delta subunit. J Biol Chem 279 25673–25679 [DOI] [PubMed] [Google Scholar]