Abstract
Organogenesis in plants is controlled by meristems. Axillary meristems, which give rise to branches and flowers, play a critical role in plant architecture and reproduction. Maize (Zea mays) and rice (Oryza sativa) have additional types of axillary meristems in the inflorescence compared to Arabidopsis (Arabidopsis thaliana) and thus provide an excellent model system to study axillary meristem initiation. Previously, we characterized the barren inflorescence2 (bif2) mutant in maize and showed that bif2 plays a key role in axillary meristem and lateral primordia initiation in the inflorescence. In this article, we cloned bif2 by transposon tagging. Isolation of bif2-like genes from seven other grasses, along with phylogenetic analysis, showed that bif2 is a co-ortholog of PINOID (PID), which regulates auxin transport in Arabidopsis. Expression analysis showed that bif2 is expressed in all axillary meristems and lateral primordia during inflorescence and vegetative development in maize and rice. Further phenotypic analysis of bif2 mutants in maize illustrates additional roles of bif2 during vegetative development. We propose that bif2/PID sequence and expression are conserved between grasses and Arabidopsis, attesting to the important role they play in development. We provide further support that bif2, and by analogy PID, is required for initiation of both axillary meristems and lateral primordia.
Organogenesis in plants is controlled by meristems (Steeves and Sussex, 1989). In the shoot, organ primordia are produced in the peripheral zone of the meristem, whereas the central zone supports further growth. Axillary meristems, which arise in the axils of leaves, play an important role in organogenesis by producing branches and flowers. During vegetative development, axillary meristems produce several leaf primordia and then arrest as dormant buds (Esau, 1967; Steeves and Sussex, 1989). During inflorescence development, axillary meristems develop into flowers or branches and the subtending leaf, called a bract, is often so reduced as to be cryptic or invisible (Bonnett, 1948; Long and Barton, 2000). Genetic and hormonal factors that regulate the initiation, arrest, and subsequent outgrowth of axillary meristems have been identified and, in particular, auxin transport has been shown to play a critical role (Grbic, 2005; McSteen and Leyser, 2005; Paponov et al., 2005; Reinhardt, 2005; Schmitz and Theres, 2005; Beveridge, 2006).
Treatment of plants with auxin transport inhibitors shows that auxin transport is required for leaf and floral meristem initiation (Okada et al., 1991; Reinhardt et al., 2000; Scanlon, 2003). In Arabidopsis (Arabidopsis thaliana), pinformed1 (pin1), pinoid (pid), yucca (yuc), and monopteros (mp) mutants have similar phenotypes to auxin transport-inhibited plants forming a pin-shaped inflorescence with very few flowers (Okada et al., 1991; Bennett et al., 1995; Przemeck et al., 1996; Cheng et al., 2006). PIN1 encodes a transmembrane protein that has been shown to transport auxin (Galweiler et al., 1998; Petrasek et al., 2006). PID encodes a Ser/Thr kinase that is proposed to positively regulate auxin transport, possibly through direct effects on PIN1 localization or trafficking (Christensen et al., 2000; Benjamins et al., 2001; Friml et al., 2004; Lee and Cho, 2006). The YUCCA gene family of flavin monooxygenases regulates the rate-limiting step in auxin biosynthesis (Zhao et al., 2001; Cheng et al., 2006). MP is a member of the auxin response factor transcription factor family that activates transcription when auxin is present (Hardtke and Berleth, 1998; Tiwari et al., 2003). Auxin is thus seen to play a key role in floral meristem initiation in Arabidopsis.
Both maize (Zea mays) and rice (Oryza sativa) inflorescences are more branched than Arabidopsis due to the presence of additional axillary meristem types (Bonnett, 1948; Cheng et al., 1983; Clifford, 1987; Irish, 1997; McSteen et al., 2000; Shimamoto and Kyozuka, 2002). Maize is monecious with two types of inflorescence, male (tassel) and female (ear). The tassel forms at the apex of the plant and consists of several long lateral branches and a central main spike (Fig. 1A). On the spike and the lateral branches, short branches, called spikelet pairs, form. Two florets are produced in each spikelet. The ear forms from an axillary meristem in the axil of the leaf about five nodes below the tassel. Rice inflorescences have similarities with maize inflorescences (Shimamoto and Kyozuka, 2002; Bommert et al., 2005b). Long lateral branches form, but the main apex aborts early in development. The branches bear spikelets singly, instead of in pairs, and terminate in a spikelet. Each spikelet bears one fertile floret, although there is evidence of two additional sterile florets (Stapf, 1917; Malcomber et al., 2006). Branching in the rice inflorescence is controlled by three types of axillary meristem—branch, spikelet, and floral meristem—whereas branching in the maize inflorescence is controlled by four types of axillary meristem—branch, spikelet pair, spikelet, and floral meristem. Therefore, maize and rice are excellent model systems for studying axillary meristem initiation during inflorescence development.
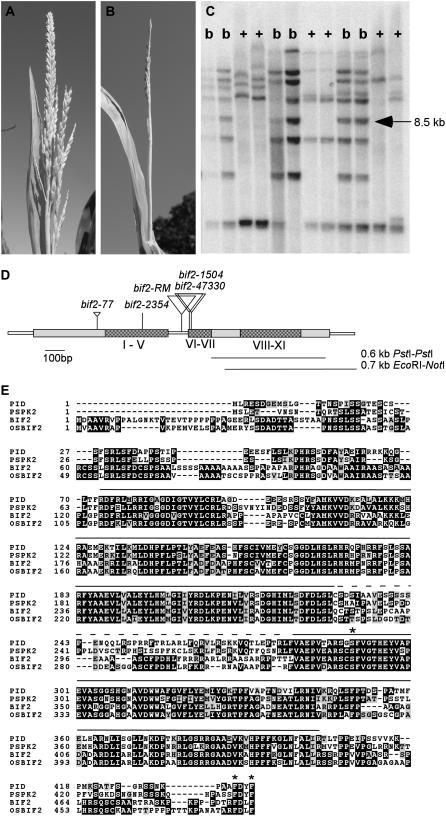
Figure 1.
Cloning of bif2 by transposon tagging, A, Normal tassel with several long lateral branches at the base of the main spike. Spikelet pairs cover the branches and main spike. B, bif2 tassel with no long lateral branches. A few single spikelets are visible on the main spike. C, DNA gel-blot analysis of EcoRI-digested genomic DNA probed with Mu1. An 8.5-kb band (arrowed) is present in bif2 mutants (b) and absent in normal cousins (+). D, Schematic of the bif2 gene showing exons as large rectangles and UTRs as narrow rectangles. The 11 subdomains of the kinase catalytic domain are shaded. The positions of transposon insertions are indicated with triangles. Probes used in Southern and northern hybridization (0.7-kb EcoRI-NotI restriction fragment) and RNA in situ hybridization (0.6-kb PstI restriction fragment) are indicated as lines beneath the map. E, Multiple sequence alignment of PID (Arabidopsis), PsPK2 (pea), bif2 (maize), and Osbif2 (rice). The conserved kinase subdomains are indicated with a line over the alignment. The insertion domain required for localization of PID is indicated with a dotted line over the alignment. The amino acids shown to be important for activation of PID by PDK1 are indicated with asterisks.
In maize and rice, mutants that fail to initiate branches, spikelets, and florets during inflorescence development have been used to identify genes that regulate axillary meristem initiation (Coe et al., 1988; Doebley et al., 1995; Komatsu et al., 2001, 2003a; McSteen and Hake, 2001; Ritter et al., 2002; Li et al., 2003; Gallavotti et al., 2004). barren inflorescence2 (bif2) mutants make fewer ear shoots, branches, spikelets, florets, and floral organs, indicating that bif2 is required for axillary meristem and lateral organ initiation in the maize inflorescence (Fig. 1B; McSteen and Hake, 2001). barren stalk1 (ba1) in maize and its rice ortholog lax panicle1 (lax1) are required for initiation of axillary meristems during both vegetative and inflorescence development (Komatsu et al., 2001, 2003a; Ritter et al., 2002; Gallavotti et al., 2004). ba1/lax1 are members of a grass-specific basic helix-loop-helix gene family, suggesting that some of the genes required for axillary meristem initiation are not conserved with Arabidopsis (Komatsu et al., 2003a; Gallavotti et al., 2004; Malcomber et al., 2006). The maize and rice co-orthologs of PIN1 have been cloned, but no loss-of-function phenotypes in the inflorescence have been reported (Xu et al., 2005; Carraro et al., 2006). The term co-ortholog (Sonnhammer and Koonin, 2002) is used rather than ortholog because multiple rounds of genome duplication have occurred in both monocot and eudicot lineages (Sonnhammer and Koonin, 2002; Bowers et al., 2003; Paterson et al., 2004; Yu et al., 2005).
In this article, we describe the cloning of bif2 from maize and seven other grasses and show by phylogenetic analysis that bif2 is a co-ortholog of PID from Arabidopsis. Expression studies show that bif2 is expressed in all axillary meristems and lateral primordia in both maize and rice. Previous phenotypic analyses showed that bif2 is required for inflorescence development. Here we report that bif2 also plays a role during vegetative development.
RESULTS
Cloning bif2 by Transposon Tagging
To identify a transposon insertion in bif2, transposon-induced alleles were subjected to DNA gel-blot analyses using a number of different restriction enzymes and probes from the most common Mutator (Mu) elements (Chandler and Hardeman, 1992; Lisch, 2002). DNA gel-blot analysis of the bif2-47330 allele identified an 8.5-kb Mu1 hybridizing fragment present in mutants and absent from wild-type cousins (Fig. 1C). This fragment was cloned by constructing a subgenomic DNA λ library in this size range and screening with Mu1. After purification and excision, the 8.5-kb fragment was restriction mapped and subcloned. A 0.7-kb EcoRI-NotI unique restriction fragment flanking the Mu1 insertion hybridized to the same 8.5-kb fragment on the DNA gel blot and the expected 7.1-kb fragment lacking Mu1 (1.4 kb) in normal cousins, indicating that the correct fragment had been cloned (data not shown). The 8.5-kb restriction fragment was sequenced and found to contain the full-length bif2 gene, including 4 kb of promoter sequence (Fig. 1D). Four cDNA clones were identified from a cDNA library made from immature tassels using the same 0.7-kb EcoRI-NotI restriction fragment as a probe. Full-length cDNA was obtained using reverse transcription (RT)-PCR on B73 tassel RNA with gene-specific primers. The predicted bif2 gene encodes a Ser/Thr kinase similar to PID of Arabidopsis (Christensen et al., 2000).
To characterize the molecular defect in bif2 alleles, PCR was performed on mutant alleles using gene-specific primers and a Mu degenerate primer. Mu1 elements were identified near the beginning of the second exon in bif2-47330, 20 bp downstream of this insertion in bif2-1504 and in the intron of bif2-RM (Fig. 1D). Using gene-specific primers, bif2-77 was determined to have a 168-bp insertion at amino acid position 108 that produces a 9-bp host site duplication. This insertion is not similar to sequences in the current databases, indicating that it may be a novel transposon. The bif2-77 insertion introduces four in-frame stop codons into BIF2 likely causing truncation of the protein before the kinase domain and therefore should be nonfunctional. bif2-2354 contains a single-nucleotide change compared to its Mo17 progenitor, causing substitution of the conserved Pro 193 with Leu. Sequence changes in five independent alleles confirm that we had cloned the bif2 locus. All alleles have similar severity of phenotype and are predicted to be null alleles.
bif2 Encodes a Ser/Thr Protein Kinase Co-Orthologous to PID of Arabidopsis
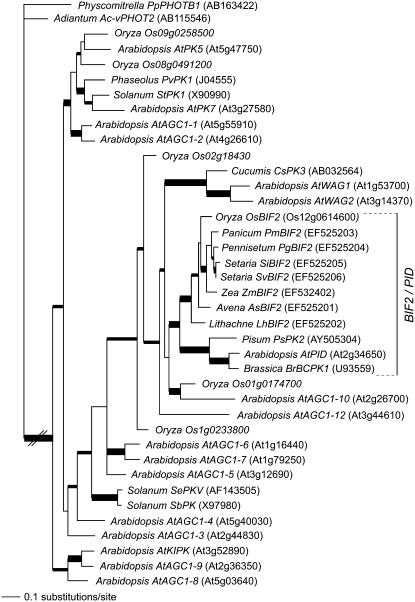
Sequence analysis indicated that bif2 encodes a Ser/Thr protein kinase with high sequence similarity to PID in Arabidopsis (Christensen et al., 2000). To determine the evolutionary history of the relationship between bif2 and PID, bif2-like genes were cloned from seven additional phylogenetically disparate grasses. PCR was performed with degenerate oligos on DNA extracted from common millet (Panicum miliaceum), foxtail millet (Setaria italica), green millet (Setaria viridis), herbaceous bamboo (Lithachne humilis), oats (Avena sativa), pearl millet (Pennisetum glaucum), and rice and the products sequenced. These sequences were aligned with 30 similar kinase sequences obtained from BLAST searches at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and analyzed using Bayesian phylogenetic software. Arabidopsis kinases were named according to Bogre et al. (2003) and Zegzouti et al. (2006b). The phylogeny was rooted using similar kinases from Physcomitrella (moss) and Adiantum (a fern). Phylogenetic analysis of 39 bif2-like protein kinases estimated a well-supported (100% clade credibility [CC]) grass bif2 clade that is sister to a well-supported (100% CC) eudicot PID clade, supporting the hypothesis that bif2 and PID are co-orthologs (Fig. 2). The presence of both grass and eudicot sequences within the bif2/PID clade indicates that the clade dates to at least the separation of the eudicot and monocot lineages approximately 125 million years ago. To date, no bif2/PID orthologs have been isolated from nonangiosperm species.
Figure 2.
Phylogenetic analysis of bif2 homologs from grasses and dicots. Bayesian consensus phylogram of 39 bif2-like protein kinases from diverse angiosperms rooted using Physcomitrella patens PHOTOTROPIN1 (PpPHOTB1) and Adiantum capillus-veneris PHOTOTROPIN2 (Ac-vPHOT2). Thick branches are supported by 1.00 CC and medium thickness branches are supported by >0.95 CC. At = Arabidopsis (Brassicaceae); As = Avena sativa (Poaceae); Br = Brassica rapa (Brassicaceae); Cs = Cucumis sativus (Curcubitaceae); Lh = Lithachne humilus (Poaceae); Os = Oryza sativa (Poaceae); Pg = Pennisetum glaucum (Poaceae); Pm = Panicum miliaceum (Poaceae); Ps = Pisum sativum (Fabaceae); Pv = Phaseolis vulgaris (Fabaceae); Sb = Solanum berthaultii (Solanaceae); Se = Solanum (Lycopersicon) esculentum (Solanaceae); Si = Setaria italica (Poaceae); St = Solanum tuberosum (Solanaceae); Sv = Setaria viridis (Poaceae); Zm = Zea mays (Poaceae).
Sequence alignment of bif2/PID co-orthologs from maize (bif2), rice (Osbif2), Arabidopsis (PID), and pea (Pisum sativum; PsPK2) indicates that similarity is highest in the 11 subdomains that make up the catalytic core of Ser/Thr kinases (Fig. 1E). These proteins belong to subfamily VIIIa of the AGC supergroup of kinases (Hanks et al., 1988; Christensen et al., 2000; Bogre et al., 2003; Bai et al., 2005). This subfamily of kinases is characterized by DFD instead of DFG in subdomain VII and an insertion domain of approximately 50 amino acids between subdomains VII and VIII (Fig. 1E). BIF2 and PID share 55% amino acid identity overall, with 67% amino acid identity in the kinase domain. BIF2 and OsBIF2 have an extended hydrophobic N terminus relative to PID and PsPK2. Outside the kinase domains, these proteins share little sequence conservation except they are Ser and Thr rich with many conserved Ser/Thr positions. The insertion domain between kinase subdomains VII and VIII, which has been shown to be important for subcellular localization of PID in Arabidopsis, shares very little sequence similarity between monocots and eudicots (Zegzouti et al., 2006b). However, the amino acids shown to be important for activation of PID by PDK1 in Arabidopsis (Zegzouti et al., 2006a) are present in BIF2 and OsBIF2.
bif2 Is Transiently Expressed in Axillary Meristems and Lateral Organs in Maize
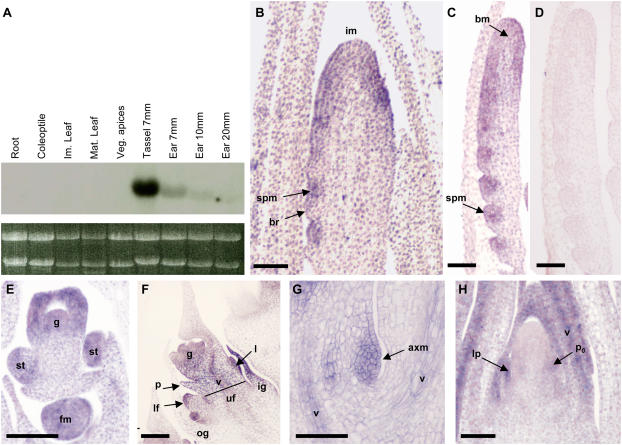
To determine the pattern of bif2 expression, RNA gel-blot and RNA in situ hybridization were performed. RNA gel-blot analysis shows that bif2 is strongly expressed in young tassels and ears, with expression declining in older tassels and ears (Fig. 3A; data not shown). Using 10 μg of total RNA, bif2 expression was below the level of detection in roots, coleoptiles, leaves, and vegetative apices (Fig. 3A).
Figure 3.
bif2 is expressed in axillary meristems and lateral primordia in maize. A, RNA gel-blot analysis using 10 μg total RNA probed with the 0.7-kb EcoRI-NotI restriction fragment. bif2 is highly expressed in immature tassels (7 mm) and ears (7 mm) and declines in expression in older ears (10 and 20 mm). bif2 is not detectable in roots, coleoptile, immature pale green leaf (im leaf), mature dark green leaf (mat leaf), and vegetative apices (veg) at this level of detection. Ethidium bromide-stained gel is shown as a loading control. B to H, RNA in situ hybridization using DIG-labeled antisense (B,C, E–H) or sense (D) probe of bif2. B, bif2 expression in an immature tassel in spikelet pair meristems (spm) in the axil of bracts (br) and on the flanks of the inflorescence meristem (im) before spm arise. C, Tassel branch showing bif2 expression in spikelet pair meristems (spm) on the flanks of the branch meristem (bm). D, Control section showing a tassel branch probed with the sense probe of bif2 has no detectable signal. E, Ear spikelet showing bif2 expression in floral meristem (fm) of lower floret and floral organs (gynoecium [g]; stamens[st]) of upper floret. F, Older ear spikelet showing bif2 expression in developing floral organs of lower floret (lf) and in lemma (l), palea (p), gynoecium (g), and vasculature (v) of upper floret (uf). Note expression in younger inner glume (ig), which is absent from older outer glume (og). G, bif2 expression in a vegetative axillary meristem (axm) and vasculature (v) of the stem and leaves. H, Vegetative apical meristem showing bif2 expression in leaf primordia (lp), including the next arising leaf primordium at plastochron 0 (P0) and vasculature (v) of young leaves. Scale bars in B to H = 100 μm.
Expression of bif2 was monitored using a digoxigenin (DIG)-labeled antisense RNA probe from the 3′ end of the gene. Control hybridizations using the sense probe produced no detectable signal (Fig. 3D). bif2 is detected in the peripheral zone of the inflorescence meristem before axillary meristems arise (Fig. 3B). Subsequently, bif2 is expressed in branch meristems, spikelet pair meristems (Fig. 3, B and C), and each of the spikelet meristems and floral meristems (Fig. 3E). Expression is transient in floral meristems and becomes concentrated in floral organ primordia as they develop (Fig. 3, E and F). Expression is also transient in floral organs and fades as they grow out (see Fig. 3F, outer glume). During vegetative development, bif2 is expressed in axillary meristems that give rise to tillers (Fig. 3G) and is weakly expressed in leaf primordia (Fig. 3H). Expression is also detected in vasculature (Fig. 3, F–H). To conclude, bif2 is expressed in axillary meristems and lateral primordia during both vegetative and inflorescence development in maize.
Expression of Osbif2 in Rice
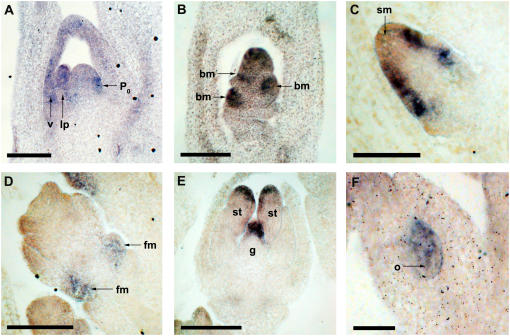
To determine whether the expression of Osbif2 was conserved in rice, RNA in situ hybridization was performed using a probe from the 3′ end of Osbif2. Expression of Osbif2 was detected in leaf primordia and vascular tissue of vegetative meristems in rice (Fig. 4A). Osbif2 expression was detected transiently in all stages of inflorescence development in branch meristems of young inflorescences (Fig. 4B), on the flanks for the spikelet meristem prior to floral organ initiation (Fig. 4C), in the proposed floral meristems of the two sterile, lower florets (Fig. 4D), in stamens and gynoecial tissue (Fig. 4E), and within the ovule (Fig. 4F). Expression was absent when sense probes were used (data not shown). Expression of Osbif2 in vegetative apical meristems and inflorescence branch meristems is supported by a recent report (Morita and Kyozuka, 2007). Therefore, expression of Osbif2 in rice in axillary meristems and lateral primordia is similar to expression of bif2 in maize.
Figure 4.
Osbif2 is expressed in axillary meristems and lateral primordia in rice. RNA in situ hybridization expression patterns for Osbif2. A, Vegetative meristem showing Osbif2 expression in leaf primordia (lp) including the next arising leaf primordia at plastochron 0 (P0) and vascular (v) tissue. B, Young inflorescence showing Osbif2 expression in branch meristems (bm). C, Young spikelet meristem (sm) showing Osbif2 expression on the flanks of the meristem where organs of the fertile upper floret will arise. D, Older spikelet showing Osbif2 expression in floral meristems (fm) of two sterile lower florets. E, In florets, Osbif2 expression is visible in the tips of the stamens (st) in the gynoecium (g). F, In older florets, Osbif2 expression is visible in the ovule (o). Scale bars in A to E = 100 μm; F = 50 μm.
bif2 Plays a Role in Vegetative Meristems
Previous analysis had not identified a vegetative phenotype in bif2 plants (McSteen and Hake, 2001). However, in Pennsylvania growing conditions, we detected a statistically significant reduction in the number of leaves produced in bif2 mutants backcrossed to B73 (normal siblings = 20.08 ± 0.12 leaves, n = 36; bif2 = 18.29 ± 0.27 leaves, n = 14; P < 0.001). In addition, a small, but statistically significant, reduction in plant height was observed (normal siblings = 207.28 ± 2.08 cm; bif2 = 196.88 ± 2.16 cm; P = 0.009). These results contrast with Arabidopsis pid mutants, which have an increased number of leaves, although pin1 mutants have fewer leaves (Bennett et al., 1995).
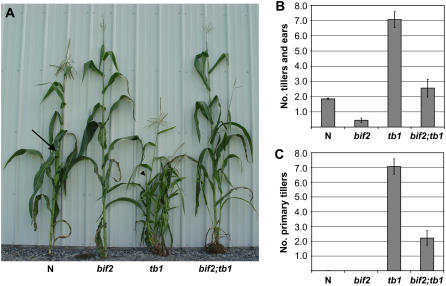
We tested whether bif2 functioned in vegetative axillary meristems by constructing double mutants with tb1-ref (Doebley et al., 1997). In normal maize, axillary meristems arise in the axil of every leaf, but arrest, except for the one or two that will grow out to form ear shoots (Fig. 5A, arrow). In tb1 mutants, however, all axillary buds grow out to form vegetative branches, called tillers, giving the plant a bushy appearance (Fig. 5A; Doebley et al., 1997; Hubbard et al., 2002). In addition, the branches at the ear node are masculinized, forming short tillers (Fig. 5A, arrowhead).
Figure 5.
bif2 plays a role in vegetative axillary meristems. A, bif2;tb1 double-mutant analysis. Normal (N) plant with no tillers and one visible ear (arrow), bif2 mutant plant with no tillers and no ears, tb1 mutant plant with many primary and secondary tillers (the ear shoot that is masculinized is indicated with an arrowhead), bif2;tb1 double mutant with two tillers and no ears. B, Quantitative analysis of all primary branches produced from the main stem (both primary tillers and ear shoots). bif2;tb1 double mutants have a statistically significant (P < 0.001) reduction in the number of tillers and ear shoots compared to tb1. C, Quantitative analysis showing primary tiller number only. bif2;tb1 mutants have a statistically significant (P < 0.001) reduction in the number of tillers compared to tb1. [See online article for color version of this figure.]
bif2;tb1 double mutants produced a reduced number of axillary branches compared to tb1 mutants (Fig. 5). To quantify the defects, the total number of primary tillers and ear shoots were counted in two F2 families segregating for both bif2 and tb1 (Fig. 5B). bif2;tb1 double mutants had significant reduction in the total number of axillary shoots compared to tb1 mutants (Fig. 5B; P < 0.001). This difference was mainly due to a difference in the number of tillers, which were significantly reduced in bif2;tb1 double mutants compared to tb1 mutants (Fig. 5C; P < 0.001). bif2 mutants have a reduced number of ear shoots compared to normal plants (Fig. 5B; McSteen and Hake, 2001). When occasional ear shoots formed in bif2;tb1 double mutants, they were masculinized like tb1 mutants. Because the number of secondary tillers is dependent on the number of primary tillers, these numbers were not included in the graphs shown. However, the number of secondary tillers was also reduced (tb1 = 5.13 ± 0.92; bif2;tb1 = zero), showing that bif2 functions in the formation of both primary and secondary tillers. Therefore, bif2 plays a role in production of vegetative axillary meristems as well as axillary meristems in tassel and ear inflorescences.
bif2 Mutants Have Defects in Auxin Transport and Vasculature
Because PID plays a role in regulating auxin transport in Arabidopsis, we tested whether bif2 mutants had auxin transport defects in maize (Bennett et al., 1995; Benjamins et al., 2001; Friml et al., 2004; Lee and Cho, 2006). To compare our data to that of pid and pin mutants in Arabidopsis, we used the same protocol (Okada et al., 1991; Bennett et al., 1995), except we used [3H]indole acetic acid (IAA) instead of [14C]IAA, because it can be labeled to a higher specific activity.
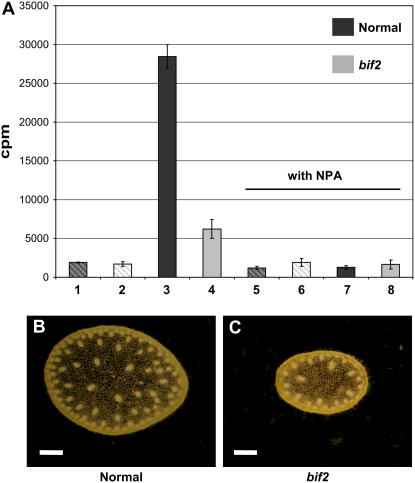
Inflorescence stem pieces from normal and bif2 tassels were placed in upward or downward orientation in tubes containing 1.5 μm [3H]IAA and incubated overnight in the dark. The top 0.5 cm was then counted in a liquid scintillation counter (Fig. 6A). At this stage of development, neither normal siblings nor bif2 had an appreciable level of acropetal transport (Fig. 6A, lanes 1 and 2) and this level was not further reduced by coincubation with N-1-naphthylphthalamic acid (NPA), an auxin transport inhibitor (lanes 5 and 6). However, normal plants displayed a very high level of basipetal transport (lane 3), which was totally abolished by NPA (lane 7). bif2 mutants, on the other hand, had significant reduction in the amount of basipetal transport of [H3]IAA compared to wild type (lane 4; P < 0.001) and the level was further reduced by NPA (lane 8). Therefore, bif2 mutants display a comparable magnitude of reduction of auxin transport in inflorescence stems in maize (21% of wild type) compared to the defects in auxin transport that occur in pid and pin1 (7%–40% of wild type) inflorescence stems in Arabidopsis (Okada et al., 1991; Bennett et al., 1995; Oka et al., 1998).
Figure 6.
Auxin transport and vasculature defects in bif2 inflorescence stems. A, Transport of [3H]IAA in normal and bif2 inflorescence stems. Bars showing transport in normal inflorescences are in dark gray and in bif2 are light gray. Acropetal transport is signified with hatched lines and basipetal transport with solid fill. B, Cross section of a normal inflorescence stem photographed in dark field. This is representative of the inflorescence stem tissue used in the auxin transport assay (below the first branch before the insertion of the stem into the flag leaf node). C, Cross section of bif2 inflorescence stem photographed in dark field. Because bif2 usually has no branches, sections used for the auxin transport assay were taken from the basal part of the inflorescence stem, 8 cm above the insertion of the stem into the flag leaf node. Scale bar in B and C = 1 mm. [See online article for color version of this figure.]
However, a possible explanation for the reduction in auxin transport could be differences in stem anatomy. Inflorescence stems from bif2 mutants were significantly narrower than normal siblings (Fig. 6, B and C; diameter across widest point: normal siblings = 5.08 ± 0.15 mm, n = 18; bif2 = 4.15 ± 0.17 mm, n = 16; P < 0.001) and had significantly fewer vascular bundles (normal siblings = 113.25 ± 4.03, n = 10; bif2 = 47.33 ± 1.57, n = 9; P < 0.001). Therefore, some of the reduction in transport (4.57-fold) could be due to a reduction in the number of vascular bundles (2.39-fold).
DISCUSSION
We cloned bif2 from maize and seven evolutionarily disparate grasses and determined that bif2 is a co-ortholog of PID from Arabidopsis. Sequence and expression patterns of bif2/PID genes are conserved between grasses and eudicots, attesting to the important role of bif2/PID in development. However, analysis of the bif2 loss-of-function phenotype in maize uncovers additional roles of bif2/PID.
bif2/Osbif2 Are Co-Orthologs of PID
Given that there have been at least three rounds of whole-genome duplication since Arabidopsis and grasses last shared a common ancestor (Bowers et al., 2003; Paterson et al., 2004; Yu et al., 2005), orthologous relationships between maize/rice and Arabidopsis genes are not possible with a strict definition of orthology (Sonnhammer and Koonin, 2002). Frequently, especially with transcription factor families, a clade of grass genes is found to be sister to a clade of Arabidopsis genes and these clades are referred to as being co-orthologous (Sonnhammer and Koonin, 2002; Malcomber et al., 2006). Therefore, we refer to bif2/Osbif2 as co-orthologs of PID. Because the maize genome has not been fully sequenced, it is not known how many PID co-orthologs are in the maize genome. However, the fact that grass genes are co-orthologous to a single Arabidopsis gene suggests that there has been extensive loss of other paralogs. Another example where there is correspondence in a signaling pathway between maize and Arabidopsis is thick tassel dwarf1 (td1) and fasciated ear2 (fea2) from maize, which are co-orthologs of CLAVATA1 (CLV1) and CLAVATA2 of Arabidopsis, respectively (Taguchi-Shiobara et al., 2001; Bommert et al., 2005a). However, despite the phylogenetic relationship, the td1/fea2/CLV pathway has diverged between maize and Arabidopsis (Taguchi-Shiobara et al., 2001; Bommert et al., 2005a).
bif2 and Osbif2 share the highest similarity with PID in the 11 subdomains that make up the catalytic domain of Ser/Thr kinases, implying that they also function as kinases (Hanks et al., 1988; Christensen et al., 2000). In addition, amino acids identified as being important for activation of PID by PDK1 in Arabidopsis are also conserved, implying that bif2/Osbif2 may be activated by a PDK1 co-ortholog in maize and rice as in Arabidopsis (Zegzouti et al., 2006a). Outside the kinase domain, there is less sequence conservation, but, interestingly, the position of many Ser/Thr sites is conserved, implying that these positions may be important for autophosphorylation of bif2/PID. On the other hand, both bif2 and Osbif2 have an extended hydrophobic domain at the N terminus that is not present in eudicots. In addition, the insertion domain shown to be important for subcellular localization of PID in Arabidopsis is not conserved (Zegzouti et al., 2006b). Whether these differences in amino sequence cause a difference in the biochemical function of BIF2/OsBIF2 remains to be determined.
Role of bif2/PID in Development
bif2 is expressed in branch, spikelet pair, spikelet, and floral meristems and all floral organs in both maize and rice, and bif2 mutants have fewer branches, spikelets, florets, and floral organs (McSteen and Hake, 2001). In Arabidopsis, PID and PIN are both expressed in floral meristems and floral organs, and pid and pin mutants make fewer florets and floral organs (Okada et al., 1991; Bennett et al., 1995; Christensen et al., 2000; Benjamins et al., 2001; Reinhardt et al., 2003). Because floral meristems are axillary meristems produced by the inflorescence meristem in the axils of cryptic bracts (Long and Barton, 2000), the defects in pid and pin floral meristem initiation are analogous to the defects caused by bif2 in axillary meristem initiation in maize. Therefore, the function of bif2 and PID appears to be conserved in floral meristem and floral organ initiation during inflorescence development. However, because maize has additional types of axillary meristems in the inflorescence compared to Arabidopsis, the phenotype of bif2 in maize extends the role of bif2/PID to all axillary meristems in the inflorescence.
bif2 is also expressed in axillary meristems during vegetative development in maize and rice. Introduction of the bif2 mutant into the tb1 mutant background allowed its effects on vegetative branches (tillers) to be evaluated in maize. The bif2 mutation causes a reduction in tiller number in the tb1 mutant background, showing that bif2 plays a role in vegetative axillary meristems. This is similar to the genetic interaction between ba1 and tb1, except that no tillers form in ba1;tb1 double mutants (Ritter et al., 2002). Mutants that cause a reduction in tiller number, as well as branch and spikelet number, have been identified in rice and other cereals (Babb and Muehlbauer, 2003; Komatsu et al., 2003a; Li et al., 2003). This contrasts with Arabidopsis, in which defects in the production of vegetative branches in pid or pin mutants have not been reported. The additional defects in bif2 mutants reported here show that, similar to the role of bif2 in inflorescence development, bif2 also plays a role in the formation of lateral organs and axillary meristems during vegetative development.
Finally, bif2/Osbif2 and PID (Christensen et al., 2000; Benjamins et al., 2001) are all expressed in vasculature. Loss of bif2, however, has different effects on vasculature than in Arabidopsis. pid mutants have defects in the vasculature of flowers (Christensen et al., 2000), whereas bif2 mutants have no defects in the vasculature of the spikelets (P. McSteen, unpublished data). bif2 mutants have reduced numbers of vascular bundles in the inflorescence stem, whereas pid mutants have no apparent defects in the vasculature of the inflorescence stem (Christensen et al., 2000). This may be due to differences in the vascular pattern of monocots and eudicots (Esau, 1967). In maize, vascular bundles are scattered in the inflorescence stem and a reduction in density is easily detected compared to Arabidopsis, in which the vascular bundles coalesce into a ring. Alternatively, there may be differences in the function of bif2 and PID in vasculature.
Osbif2 Expression Pattern Supports the Hypothesis That Rice Has a Three-Flowered Spikelet
Rice is considered to have either a single- or a three-flowered spikelet, depending on whether the two structures beneath the fertile lemma are interpreted as being glumes or sterile lemmas, the sole remaining organs of two sterile lower florets. The three-flowered spikelet hypothesis was first proposed by Stapf (1917) and this interpretation has recently been supported by gene expression patterns (Prasad et al., 2001; Komatsu et al., 2003b; Malcomber and Kellogg, 2004). Our expression analyses show that, within the spikelet, Osbif2 is expressed in the fertile upper floret. In addition, the distinct expression of Osbif2 on either side of the region subtending the fertile lemma and palea coincides with where the two lower floral meristems are predicted to initiate. This result adds support to the hypothesis that rice has a three-flowered, basipetally maturing, spikelet rather than a single-flowered spikelet.
Role of bif2 in Auxin Transport
Using the standard auxin transport assay on inflorescence stems, pid, pin, and bif2 mutants all have comparable reduction in the level of auxin transport relative to wild-type levels (Okada et al., 1991; Bennett et al., 1995; Oka et al., 1998). bif2 mutants, however, have a significant reduction in the number of vascular bundles and inflorescence stem girth, unlike pid, which does not have apparent defects in vasculature in the inflorescence stem. It is possible that the reduction in auxin transport could be indirectly caused by the reduction in vasculature; however, a recent report of the overexpression of a rice co-ortholog of PID (Morita and Kyozuka, 2007) supports a role for grass co-orthologs in auxin transport.
Expression of bif2 and PID overlaps with expression of the auxin efflux carrier PIN1/ZmPIN1 in vasculature as well as during lateral primordia and axillary meristem initiation (Christensen et al., 2000; Reinhardt et al., 2003; Carraro et al., 2006). PID is proposed to affect auxin transport by regulating PIN1 (Friml et al., 2004; Lee and Cho, 2006). In the pid inflorescence in Arabidopsis, the polarity of PIN1 localization is switched from the apical to the basal end of the cell (Friml et al., 2004). In maize, it has been reported that ZmPIN1 protein is not detected in strong bif2 mutant tassels (Carraro et al., 2006). However, this is likely to be due to the absence of spikelets in strong bif2 mutants because ZmPIN1 is detected when spikelets form in tassels (P. McSteen, unpublished data) and ears (Carraro et al., 2006). Occasional reversals of ZmPIN1 polarity in bif2 mutant ears support the contention that bif2 and PID have conserved roles in regulation of PIN1 (Carraro et al., 2006).
CONCLUSION
To conclude, the sequence and expression of bif2 appears to be conserved between Arabidopsis, maize, and rice. Phylogenetic analysis was aided by identification of homologs from seven grass species, which demonstrated a well-supported co-orthologous relationship between bif/Osbif2 and PID. Conservation of sequence and expression among grasses and Arabidopsis, which last shared a common ancestor 125 million years ago, attests to the important role of bif2/PID in development. Analysis of the bif2 phenotype in maize uncovers additional roles of bif2 in axillary meristem and lateral primordia initiation during both inflorescence and vegetative development.
MATERIALS AND METHODS
Maize Material and Growth
bif2 alleles were isolated as described (McSteen and Hake, 2001). An additional allele, bif2-RM, was identified in maize (Zea mays) Mu active lines generated by the Maize Gene Discovery Project (http://www.maizegdb.org/rescuemu-phenotype.php). For leaf number, plant height, auxin transport assays, and vasculature analysis, bif2-77 was backcrossed four or more times into the B73 genetic background. For bif2;tb1 double-mutant analysis, bif2-1606 was backcrossed four times into the A188 genetic background and tb1-ref (obtained from the Maize Coop) was backcrossed three times into the A188 genetic background. A188 was used for these experiments because bif2 has a strong effect on ear number in this genetic background (McSteen and Hake, 2001) and tb1 does not exhibit a heterozygous effect in this background (Hubbard et al., 2002). For RNA in situ hybridization, meristems were dissected from B73 plants grown in the greenhouse for 5 weeks (tassels) or in the field for 8 weeks (ears). For phenotype and double-mutant analysis, plants were grown during the summer in the field at Penn State Agronomy Research Farm (Rock Springs, PA). Quantitative analysis of single- and double-mutant phenotypes was reported as mean ± se and statistical significance was determined using Student's two-tailed t test.
bif2 Cloning
Genomic DNA of mutants and normal cousins (normal plants from sibling families that did not segregate the mutant phenotype) was extracted (Chen and Dellaporta, 1994), digested with EcoRI, separated on a 0.8% agarose gel, blotted, and probed with Mu1. An 8.5-kb Mu1 hybridizing band was found to cosegregate with the bif2 mutant phenotype in allele bif2-47330 (0 recombinants/59 chromosomes). DNA from the 8.5-kb size range was cut from a preparative gel, purified, and ligated into EcoRI cut ZapExpress vector (Stratagene). The subgenomic DNA library was screened with Mu1 and the 8.5-kb restriction fragment in vector pBK-CMV (pFS3bif2) was purified and restriction mapped. The 8.5-kb insert was sequenced in its entirety by subcloning multiple restriction fragments and primer walking using the Big Dye sequencing kit on an ABI sequencer (Applied Biosystems). This clone contains the entire coding region of bif2 plus 4 kb of promoter sequence (accession no. EF532824). To obtain a cDNA clone, 106 plaques of a cDNA library made from immature tassels (R. Schmidt, University of California, San Diego) were screened with the 0.7-kb EcoRI-NotI restriction fragment. To obtain the 5′ end of the gene, 5′-RACE was performed using the First-Choice RLM-RACE kit (Ambion) on 10 μg total RNA from normal tassels, according to the manufacturer's recommendations, except with the following modifications because bif2 is GC rich: annealing of oligo(dT) was performed at 65°C and SuperScript II enzyme (Invitrogen) was used at 50°C. PCR was performed with RNA adaptor outer primer (Ambion) and 5′-specific outer primer (5′-CAGGTGGTCCGGGAAGCAGGACG-3′) with 1 m betaine and 10% dimethyl sulfoxide (DMSO) in the reaction. Nested PCR was performed with RNA adaptor inner (Ambion) and 5′-specific inner primer (5′-CGACGGCGTCGACGTGCACTG-3′). To obtain a full-length cDNA clone from B73, RT-PCR was performed on 1 μg tassel total RNA according to the SuperScript III (Invitrogen) protocol with the following modifications: annealing of oligo(dT) primer was at 70°C and SuperScript III reverse transcriptase was used at 50°C. bif2 was amplified by PCR using bif2 5′-untranslated region (UTR) forward primer (5′-TCGTCCTCGCTCGGAGACACGGC-3′) and bif2 3′-UTR reverse primer (5′-ACAATGGCGAGTGATGCAACTAGGTC-3′) with the addition of 1 m betaine and 10% DMSO in the reaction. PCR products were cloned into pGEMTeasy (Promega) and sequenced at Penn State Nucleic Acid Facility (accession no. EF532402).
The molecular defect in alleles bif2-47330, bif2-1504, and bif2-RM was determined by PCR with a Mu degenerate primer (5′-AGAGAAGCCAACGCCAWCGCCTCYATTTCGTC-3′) and a gene-specific primer (bif2-3064, 5′-GTCGGCATCACCGGCGCC-3′), including 1 m betaine and 10% DMSO in the reaction mix. Products were sequenced on an ABI sequencer using the Big Dye kit with DMSO in the reaction (Applied Biosystems). bif2-77 was amplified using bif2-57 forward primer (5′-CAGCCTGCCGCGCTGCTCCAGC-3′) and the bif2-250 reverse primer (5′-CGGCGCAGCAGCCTGAAGTCC-3′) and products sequenced at the Penn State Nucleic Acid Facility (the accession no. of the putative transposon insertion in bif2-77 is EF532403). Of the other alleles previously reported (McSteen and Hake, 2001), bif2-1512 came from the same source and had the same insertion as bif2-47330, bif2-70 came from the same source and had the same insertion as bif2-77, and the mutation in bif2-1606 could not be determined because the progenitor is unknown.
DNA Extraction, PCR Amplification, and Sequencing from Other Grasses
Total DNA was isolated from common millet (Panicum miliaceum), foxtail millet (Setaria italica), green millet (Setaria viridis), Lithachne humilis, maize, oat (Avena sativa), pearl millet (Pennisetum glaucum), and rice (Oryza sativa) using an SDS extraction protocol (Dellaporta, 1997). PCR products were amplified using the primers bif2-566F (5′-CCAACCGCCGCTTCCCGCTCCCSTC-3′) and bif2-1277R (5′-CGACGGGCGGGCGSGASGAMCGGAG-3′), including 1 m betaine and 5% DMSO in the PCR reaction. PCR fragments were purified, subcloned, sequenced, and assembled as described (Malcomber and Kellogg, 2004). All sequences were submitted to GenBank (accession nos. EF525201–EF525206).
Phylogenetic Analysis
A total of 39 BIF2-like protein kinase sequences were examined from diverse green land plants. Seven sequences were cloned from disparate grass species (see above) and the remaining 32 genes were identified by BLAST (Altschul et al., 1997) searches at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Nucleotide sequences were aligned based on the conceptual amino acid translation, using MacClade 4 (Maddison and Maddison, 2003) and ClustalX (Jeanmougin et al., 1998). Bayesian phylogenetic analyses were conducted using MrBayes 3.1 (Huelsenbeck and Ronquist, 2001) on the Beowulf parallel-processing cluster at the University of Missouri, using two separate searches of 5 million Monte Carlo Markov Chain generations using default flat priors and the GTR + I + Γ model of sequence evolution (as estimated by MODELTEST [Posada and Crandall, 1998]). Trees were sampled every 500 generations and burn in was determined empirically by plotting the likelihood score against generation number. After burn-in trees had been removed, CC values and the 95% credible set of trees were estimated using MrBayes (Huelsenbeck and Ronquist, 2001).
Expression Analysis in Maize
For RNA gel-blot analysis, total RNA was extracted with TRIzol (Invitrogen). Ten micrograms of total RNA were separated on a 1% agarose gel according to the northernMax-Gly protocol (Ambion), blotted, and hybridized using the 0.7-kb EcoRI-NotI probe at 68°C in 10% dextran sulfate, 1 m NaCl, and 1% SDS.
For RNA in situ hybridization analysis, a 0.6-kb PstI fragment (which overlaps with the 0.7-kb EcoRI-NotI clone) was subcloned into SK+ Bluescript (Stratagene). This plasmid was linearized with XhoI and transcribed with T3 RNA polymerase to produce a DIG-labeled antisense probe. For sense probe, this plasmid was linearized with BamHI and transcribed with T7 RNA polymerase. Although this probe includes part of the conserved kinase domain, high-stringency DNA gel-blot hybridization shows that it detects a single major band in the maize genome and that there is no difference in the hybridization pattern compared to a probe that does not contain the kinase domain (Supplemental Fig. S1). RNA in situ hybridization was performed on developing maize inflorescences as described (Jackson et al., 1994).
Expression Analysis in Rice
Total RNA was extracted from developing rice inflorescences using RNAwiz solution (Ambion) according to the manufacturer's instructions. The template for RNA in situ hybridization was generated using the SuperScript one-step RT-PCR kit (Invitrogen) with the primers Osbif2-566F and Osbif2-1705R (5′-AACAGCAAGGTGATTAGCAG-3′) with a 65°C annealing temperature following the manufacturer's instructions, except 4% DMSO and 1 m betaine were included. The amplified fragment included 500 bp from the C-terminal region of the gene and 161 bp of the 3′-UTR. This probe is proposed to be specific for Osbif2 because the next closest homolog in the rice genome has 34% nucleotide difference in the coding region and is unalignable in the 3′-UTR. RNA in situ hybridization was conducted on developing rice inflorescences, as described (Malcomber and Kellogg, 2004).
Analysis of bif2;tb1 Double Mutants
bif2;tb1 double mutants were identified by phenotype analysis for the bif2 locus and DNA analysis for the tb1 locus. Genomic DNA was extracted (Chen and Dellaporta, 1994), digested with HindIII, separated on a 0.8% agarose gel, blotted, and probed with the 3.4-kb HindIII genomic fragment from the tb1 locus (Doebley et al., 1997). Two F2 families totaling 122 individuals were grown in the field and tiller and ear number counted.
Auxin Transport Assays
Tassels were excised from normal and mutant bif2 plants (segregating 1:1 in the B73 genetic background) before they had fully extended from the leaf whorl. The inflorescence stem between the lowest tassel branch and the point of insertion of the flag leaf were used. Auxin transport assays were performed using the method of Okada et al. (1991) with the following modifications: 2-cm pieces of inflorescence stem were placed in normal or inverted orientation into tubes containing 100 μL 0.5× Murashige and Skoog medium, 1.5 μm 3-[5(n)-3H]indolyl acetic acid (specific activity 25 Ci/mmol; GE Healthcare), plus or minus 15 μm NPA (Chemservice) overnight in the dark. The following day, the pieces were blotted and 0.5 cm from the end that was not immersed in the solution was placed in scintillation fluid (Ready safe; Beckman-Coulter) and counted in a liquid scintillation counter (LSC6000; Beckman-Coulter). Statistical significance was determined using Student's two-tailed t test.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EF525201 to EF525206, EF532402, EF532403, and EF532824.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. DNA gel-blot hybridization with four probes from the bif2 locus.
Supplementary Material
Acknowledgments
We thank Steve Briggs, Guri Johal, Richard Schneeberger, Paul Chomet, Gerry Nueffer, The Maize Gene Discovery Project, and the Maize Coop for genetic stocks. For plant care, we thank David Hantz at the Plant Gene Expression Center greenhouse, Tony Omeis at the Biology Department greenhouse, Penn State, and Bob Oberheim, Ron Shuey, and W. Scott Harkom at Penn State Agronomy Research Farm, Rock Springs, PA. We thank George Chuck and Bob Schmidt for the tassel cDNA library and Jeffrey Buterbaugh for assistance with tb1 genotyping.
This work was supported by the National Science Foundation (grant no. DBI–0110189 to S.H. and E.K. and grant no. IBN–0416616 to P.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Paula McSteen (pcm11@psu.edu).
Some figures in this article are displayed in color online but in black and white in print.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babb S, Muehlbauer GJ (2003) Genetic and morphological characterization of the barley uniculm2 (cul2) mutant. Theor Appl Genet 106 846–857 [DOI] [PubMed] [Google Scholar]
- Bai F, Watson JC, Walling J, Weeden N, Santner AA, DeMason DA (2005) Molecular characterization and expression of PsPK2, a PINOID-like gene from pea (Pisum sativum). Plant Sci 168 1281–1291 [Google Scholar]
- Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R (2001) The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128 4057–4067 [DOI] [PubMed] [Google Scholar]
- Bennett SRM, Alvarez J, Bossinger G, Smyth DR (1995) Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J 8 505–520 [Google Scholar]
- Beveridge CA (2006) Axillary bud outgrowth: sending a message. Curr Opin Plant Biol 9 35–40 [DOI] [PubMed] [Google Scholar]
- Bogre L, Okresz L, Henriques R, Anthony RG (2003) Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci 8 424–431 [DOI] [PubMed] [Google Scholar]
- Bommert P, Lunde C, Nardmann J, Vollbrecht E, Running M, Jackson D, Hake S, Werr W (2005. a) thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 132 1235–1245 [DOI] [PubMed] [Google Scholar]
- Bommert P, Satoh-Nagasawa N, Jackson D, Hirano HY (2005. b) Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol 46 69–78 [DOI] [PubMed] [Google Scholar]
- Bonnett OT (1948) Ear and tassel development in maize. Ann Mo Bot Gard 35 269–287 [Google Scholar]
- Bowers JE, Chapman BA, Rong JK, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422 433–438 [DOI] [PubMed] [Google Scholar]
- Carraro N, Forestan C, Canova S, Traas J, Varotto S (2006) ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol 142 254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler VL, Hardeman KJ (1992) The Mu elements of Zea mays. Adv Genet 30 77–122 [DOI] [PubMed] [Google Scholar]
- Chen J, Dellaporta SL (1994) Urea-based plant DNA miniprep. In M Freeling, V Walbot, eds, The Maize Handbook. Springer-Verlag, New York
- Cheng PC, Greyson RI, Walden DB (1983) Organ initiation and the development of unisexual flowers in the tassel and ear of Zea mays. Am J Bot 70 450–462 [Google Scholar]
- Cheng YF, Dai XH, Zhao YD (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Dagenais N, Chory J, Weigel D (2000) Regulation of auxin response by the protein kinase PINOID. Cell 100 469–478 [DOI] [PubMed] [Google Scholar]
- Clifford HT (1987) Spikelet and floral morphology. In TR Soderstrom, KW Hilu, CS Campbell, ME Barkworth, eds, Grass Systematics and Evolution. Smithsonian Institution Press, Washington, DC, pp 21–30
- Coe EH, Neuffer MG, Hoisington DA (1988) The genetics of corn. In GF Sprague, JW Dudley, eds, Corn and Corn Improvement, Ed 3, Vol 18. ASA-CSSA-SSSA, Madison, WI, pp 81–258
- Dellaporta SL (1997) Plant DNA miniprep and microprep: versions 2.1–2.3. In M Freeling, V Walbot, eds, The Maize Handbook. Springer-Verlag, New York
- Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386 485–488 [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Kent B (1995) Suppressor of sessile spikelets1 (Sos1)—a dominant mutant affecting inflorescence development in maize. Am J Bot 82 571–577 [Google Scholar]
- Esau K (1967) Plant Anatomy, Ed 3. John Wiley & Sons, New York
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PBF, Ljung K, Sandberg G, et al (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306 862–865 [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Zhao Q, Kyozuka J, Meeley RB, Ritter M, Doebley JF, Pe ME, Schmidt RJ (2004) The role of barren stalk1 in the architecture of maize. Nature 432 630–635 [DOI] [PubMed] [Google Scholar]
- Galweiler L, Guan CH, Muller A, Wisman E, Mendgen K, Yephremov A, Palme K (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282 2226–2230 [DOI] [PubMed] [Google Scholar]
- Grbic V (2005) Comparative analysis of axillary and floral meristem development. Can J Bot 83 343–349 [Google Scholar]
- Hanks SK, Quinn AM, Hunter T (1988) The protein-kinase family—conserved features and deduced phylogeny of the catalytic domains. Science 241 42–52 [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17 1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard L, McSteen P, Doebley J, Hake S (2002) Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162 1927–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17 754–755 [DOI] [PubMed] [Google Scholar]
- Irish EE (1997) Class II tassel seed mutations provide evidence for multiple types of inflorescence meristems in maize (Poaceae). Am J Bot 84 1502–1515 [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S (1994) Expression of maize knotted1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120 405–413 [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with ClustalX. Trends Biochem Sci 23 403–405 [DOI] [PubMed] [Google Scholar]
- Komatsu K, Maekawa M, Ujiie S, Satake Y, Furutani I, Okamoto H, Shimamoto K, Kyozuka J (2003. a) LAX and SPA: major regulators of shoot branching in rice. Proc Natl Acad Sci USA 100 11765–11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J (2003. b) FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130 3841–3850 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Maekawa M, Shimamoto K, Kyozuka J (2001) The LAX1 and FRIZZY PANICLE2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev Biol 231 364–373 [DOI] [PubMed] [Google Scholar]
- Lee SH, Cho HT (2006) PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells. Plant Cell 18 1604–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Qian Q, Fu ZM, Wang YH, Xiong GS, Zeng DL, Wang XQ, Liu XF, Teng S, Hiroshi F, et al (2003) Control of tillering in rice. Nature 422 618–621 [DOI] [PubMed] [Google Scholar]
- Lisch D (2002) Mutator transposons. Trends Plant Sci 7 498–504 [DOI] [PubMed] [Google Scholar]
- Long J, Barton MK (2000) Initiation of axillary and floral meristems in Arabidopsis. Dev Biol 218 341–353 [DOI] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP (2003) MacClade: Analysis of Phylogeny and Character Evolution. Sinauer Associates, Sunderland, MA
- Malcomber ST, Kellogg EA (2004) Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell 16 1692–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber ST, Preston JC, Reinheimer R, Kossuth J, Kellogg EA (2006) Developmental gene evolution and the origin of grass inflorescence diversity. Adv Bot Res 44 425–481 [Google Scholar]
- McSteen P, Hake S (2001) barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 128 2881–2891 [DOI] [PubMed] [Google Scholar]
- McSteen P, Laudencia-Chingcuanco D, Colasanti J (2000) A floret by any other name: control of meristem identity in maize. Trends Plant Sci 5 61–66 [DOI] [PubMed] [Google Scholar]
- McSteen P, Leyser O (2005) Shoot branching. Annu Rev Plant Biol 56 353–374 [DOI] [PubMed] [Google Scholar]
- Morita Y, Kyozuka J (2007) Characterization of OsPID, the rice ortholog of PINOID, and its possible involvement in the control of polar auxin transport. Plant Cell Physiol 48 540–549 [DOI] [PubMed] [Google Scholar]
- Oka M, Ueda J, Miyamoto K, Okada K (1998) Activities of auxin polar transport in inflorescence axes of flower mutants of Arabidopsis thaliana: relevance to flower formation and growth. J Plant Res 111 407–410 [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y (1991) Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov IA, Teale WD, Trebar M, Blilou K, Palme K (2005) The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci 10 170–177 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA 101 9903–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, et al (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312 914–918 [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14 817–818 [DOI] [PubMed] [Google Scholar]
- Prasad K, Sriram P, Kumar CS, Kushalappa K, Vijayraghavan U (2001) Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev Genes Evol 211 281–290 [DOI] [PubMed] [Google Scholar]
- Przemeck GKH, Mattsson J, Hardtke CS, Sung ZR, Berleth T (1996) Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200 229–237 [DOI] [PubMed] [Google Scholar]
- Reinhardt D (2005) Phyllotaxis—a new chapter in an old tale about beauty and magic numbers. Curr Opin Plant Biol 8 487–493 [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Mandel T, Kuhlemeier C (2000) Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260 [DOI] [PubMed] [Google Scholar]
- Ritter MK, Padilla CM, Schmidt RJ (2002) The maize mutant barren stalk1 is defective in axillary meristem development. Am J Bot 89 203–210 [DOI] [PubMed] [Google Scholar]
- Scanlon MJ (2003) The polar auxin transport inhibitor N-1-naphthylphthalamic acid disrupts leaf initiation, KNOX protein regulation, and formation of leaf margins in maize. Plant Physiol 133 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz G, Theres K (2005) Shoot and inflorescence branching. Curr Opin Plant Biol 8 506–511 [DOI] [PubMed] [Google Scholar]
- Shimamoto K, Kyozuka J (2002) Rice as a model for comparative genomics of plants. Annu Rev Plant Biol 53 399–419 [DOI] [PubMed] [Google Scholar]
- Sonnhammer ELL, Koonin EV (2002) Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet 18 619–620 [DOI] [PubMed] [Google Scholar]
- Stapf O (1917) Gramineae. Flora of Tropical Africa. Lowell Reeve & Co, London
- Steeves T, Sussex I (1989) Patterns in Plant Development. Cambridge University Press, Cambridge, UK
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D (2001) The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev 15 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Zhu L, Shou HX, Wu P (2005) A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol 46 1674–1681 [DOI] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li SG, Li H, Zhou J, Ni PX, Dong W, Hu SN, Zeng CQ, et al (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3 266–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegzouti H, Anthony RG, Jahchan N, Bogre L, Christensen SK (2006. a) Phosphorylation and activation of PINOID by the phospholipid signaling kinase 3-phosphoinositidedependent protein kinase 1 (PDK1) in Arabidopsis. Proc Natl Acad Sci USA 103 6404–6409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegzouti H, Li W, Lorenz TC, Xie MT, Payne CT, Smith K, Glenny S, Payne GS, Christensen SK (2006. b) Structural and functional insights into the regulation of Arabidopsis AGC VIIIa kinases. J Biol Chem 281 35520–35530 [DOI] [PubMed] [Google Scholar]
- Zhao YD, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 306–309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.