Abstract
Expression of two Arabidopsis (Arabidopsis thaliana) apyrase (nucleoside triphosphate-diphosphohydrolase) genes with high similarity, APY1 and APY2, was analyzed during seedling development and under different light treatments using β-glucuronidase fusion constructs with the promoters of both genes. As evaluated by β-glucuronidase staining and independently confirmed by other methods, the highest expression of both apyrases was in rapidly growing tissues and/or tissues that accumulate high auxin levels. Red-light treatment of etiolated seedlings suppressed the protein and message level of both apyrases at least as rapidly as it inhibited hypocotyl growth. Adult apy1 and apy2 single mutants had near-normal growth, but apy1apy2 double-knockout plants were dwarf, due primarily to reduced cell elongation. Pollen tubes and etiolated hypocotyls overexpressing an apyrase had faster growth rates than wild-type plants. Growing pollen tubes released ATP into the growth medium and suppression of apyrase activity by antiapyrase antibodies or by inhibitors simultaneously increased medium ATP levels and inhibited pollen tube growth. These results imply that APY1 and APY2, like their homologs in animals, act to reduce the concentration of extracellular nucleotides, and that this function is important for the regulation of growth in Arabidopsis.
Apyrases (nucleoside triphosphate-diphosphohydrolases) are enzymes that can hydrolyze nucleoside triphosphates (NTPs) and/or diphosphates, but not nucleoside monophosphates or nonnucleoside phosphates. They are found in all eukaryotes and are far more efficient in removing phosphates from NTP/nucleoside diphosphate than other phosphatases. They are characterized by conserved motifs (Handa and Guidotti, 1996) and by their relative insensitivity to specific inhibitors of P-type, F-type, and V-type ATPases and to most inhibitors of alkaline and acid phosphatases (Zimmermann, 2001; Steinebrunner et al., 2003).
The majority of characterized apyrases are ectoapyrases (i.e. enzymes that are anchored in the plasma membrane with their active site pointing out into the extracellular matrix [ECM] of cells). In animal cells, where a signaling role for extracellular ATP (eATP) and ADP has been established for over two decades (Burnstock and Knight, 2004), ectoapyrases play a crucial role in terminating signal transduction initiated by extracellular nucleotides (Zimmermann, 2001). Of the apyrases characterized in plants, some are plasma membrane associated (Thomas et al., 1999; Day et al., 2000), but the subcellular locale of most of them has not been determined. Plasma membrane-associated apyrases in plants could, in principle, function as ectoapyrases because plant cells, like animal cells, release significant quantities of ATP into their ECM when they are mechanically stimulated (Jeter et al., 2004), when they are wounded (Song et al., 2006), and when they are engaged in activities that involve active secretion, such as growth (Kim et al., 2006). Moreover, control of this eATP could be important because plant cells have significant signaling responses to submicromolar ATP (Demidchik et al., 2003; Song et al., 2006) and extensive depletion of eATP can result in loss of cell viability (Chivasa et al., 2005).
Arabidopsis (Arabidopsis thaliana) has seven apyrases, two of which, APY1 and APY2, are most similar to the pea (Pisum sativum) ectoapyrase NTP9 (Steinebrunner et al., 2000). These two apyrases are 87% identical in protein sequence and, like the pea enzyme, both have signal peptides (Steinebrunner et al., 2000). Overexpression of APY2 lowers the sensitivity of Arabidopsis leaves to applied ATP (Song et al., 2006), consistent with the hypothesis that it can function as an ectoapyrase. An initial genetic investigation of the role of APY1 and APY2 revealed that they were needed for pollen germination (Steinebrunner et al., 2003).
Here we report three lines of evidence that APY1 and APY2 play an important role in the control of plant cell growth: Transcript abundance for APY1 and APY2 is highest in tissues and cell types that are growing rapidly, constitutive expression of one of these genes results in enhanced growth of hypocotyls and pollen tubes, and suppression of both genes in Arabidopsis or chemical suppression of apyrase enzyme activity results in impaired growth. We also show that the same light signal that suppresses the growth of hypocotyls simultaneously induces a loss of transcripts and protein of APY1 and APY2 in this tissue and provide evidence that a key function of the two apyrases is, like their vertebrate counterparts (Zimmermann, 2001), to reduce the concentration of eATP. These results reveal that expression of APY1 and APY2 is closely correlated with growth and we discuss ways their enzymatic function could participate in growth control.
RESULTS
Expression of APY1 and APY2 Is Strongest in Cells That Are Rapidly Expanding and/or Accumulate Auxin
In the primary roots of 7-d-old seedlings, promoter:GUS analysis shows that both APY1:GUS and APY2:GUS are expressed highly in the root-hypocotyl junction (Fig. 1A) and root tip, mainly the root cap and the columella cells (Fig. 1, B and C), but with some staining also in the more proximal meristematic zone. However, in the distal elongation zone, expression of the two constructs differs, with APY2:GUS but not APY1:GUS showing strong expression there (Fig. 1, B and C).
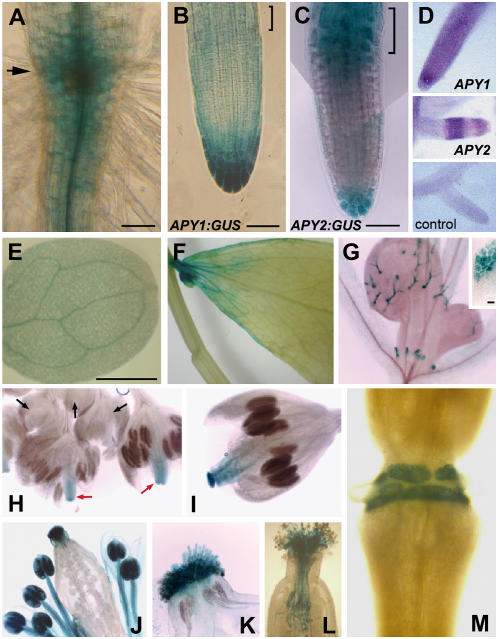
Figure 1.
Promoter:GUS or in situ assays of apyrase expression in various tissues. A, Representative staining for APY1:GUS or APY2:GUS in the region close to the root-hypocotyl junction (arrow) and in the more apical region of the differentiation zone of the primary root. B and C, Promoter:GUS expression in the apical region of primary root, including the elongation zone (brackets). Bar = 50 μm in A to C. D, In situ hybridization of digoxigenin-labeled antisense APY1 in primary root (top), and APY2 in lateral root (middle). Control (bottom) shows the lack of staining in a lateral root when the reverse transcriptase is left out of the PCR step of the sample preparation. E, Representative staining for APY1:GUS or APY2:GUS in the cotyledon. Bar = 100 μm. F, Representative staining for APY1:GUS or APY2:GUS in the mature cauline leaf. G, APY2:GUS expression in young leaf trichomes and the upper region of stipules (inset). H to L, Representative promoter:GUS staining for either APY1 or APY2 at various stages of flower development. There is no expression in flowers younger than stage 8 (H, black arrows), but some stigma staining between stages 9 and 12 (H, red arrows, I). J and K, Staining in flowers at stages 13 to 15. L, Staining of pollen tubes growing through style. M, Staining in abscission zone of sepal at the end of stage 15.
The pattern in apical roots was verified independently by in situ localization (Fig. 1D). The in situ staining pattern for APY1 in primary roots (top) was the same as its pattern in lateral roots (data not shown) and the in situ staining pattern for APY2 in lateral roots (shown in Fig. 1D, middle) is identical to that of the promoter:GUS staining pattern for APY2 in lateral roots found by Sun (2003). These findings are in accord with prior results that show that expression patterns of transcripts in equivalent tissues of lateral and primary roots are similar (Masucci et al., 1996).
In the apical or early maturation zone just basal to the elongation zone, APY1:GUS and APY2:GUS are expressed mostly in the vascular tissue, with less staining in the surrounding cortex and epidermal layers (Fig. 1A, bottom). As the root elongates, expression of APY1:GUS and APY2:GUS in the tissue between the apical and basal regions of the zone of maturation totally disappears (data not shown).
Among aerial vegetative tissues, staining for APY1:GUS and APY2:GUS is evident in the veins of light-grown cotyledons (Fig. 1E), is weak or nonexistent throughout mature leaves and stems, except in some leaf veins and near the leaf base (Fig. 1F), and is readily detectable in young, but not mature, trichomes and in stipules (Fig. 1G). Stipule staining is restricted to the upper part of this tissue (Fig. 1G, inset), where auxin and flavonoids also accumulate (Aloni et al., 2003; Schwalm et al., 2003).
Among flowering tissues, promoter:GUS fusions showed that expression of these two constructs varied with the stage of floral development. Development of Arabidopsis flowers has been divided into 12 stages, each characterized by landmark events (Smyth et al., 1990). Expression of APY1:GUS and APY2:GUS was examined in these different stages. APY1:GUS and APY2:GUS staining did not appear in flowers younger than stage 8 (Fig. 1H, black arrows). Between stages 8 and 12, APY1:GUS and APY2:GUS were expressed only in the stigma and in vascular tissue of carpels (Fig. 1H, red arrows, and 1I). No expression was observed in developing pollen. In flowers between stages 13 and 15, APY1:GUS and APY2:GUS were expressed strongly in the stamen, especially in pollen (Fig. 1J) and stigma tissue (Fig. 1K). Staining in the stamen first appeared in pollen, then less strongly in the whole stamen, including the anther wall and filaments (Fig. 1J), by which time expression in the carpel vascular tissue had disappeared (Fig. 1J). Pollen tubes growing through the style were also strongly stained (Fig. 1L).
Postpollination, both APY1:GUS and APY2:GUS were also expressed strongly in the separation layer of the abscission zone of flower organs, where petal, sepal, and stamen fall off after anthesis. Figure 1M shows that after abscission of these organs, staining exists only at the former area of attachment to those flower structures. When pollination was prevented by removing the stigma before anthesis, staining in this area remained strong (data not shown), indicating that expression of apyrases in the separation layer is pollination independent.
Light Down-Regulates Apyrase Expression in Parallel with Growth Inhibition in Hypocotyls
Light rapidly inhibits elongation of hypocotyls in Arabidopsis, where growth changes become apparent in about 8 min (Parks and Spalding, 1999). We tested whether expression of Arabidopsis apyrases in hypocotyls is also light regulated. Figure 2A shows that APY1:GUS and APY2:GUS are expressed in etiolated wild-type hypocotyls, but when the seedling is grown in light, expression of APY1:GUS and APY2:GUS in hypocotyls is not detectable.
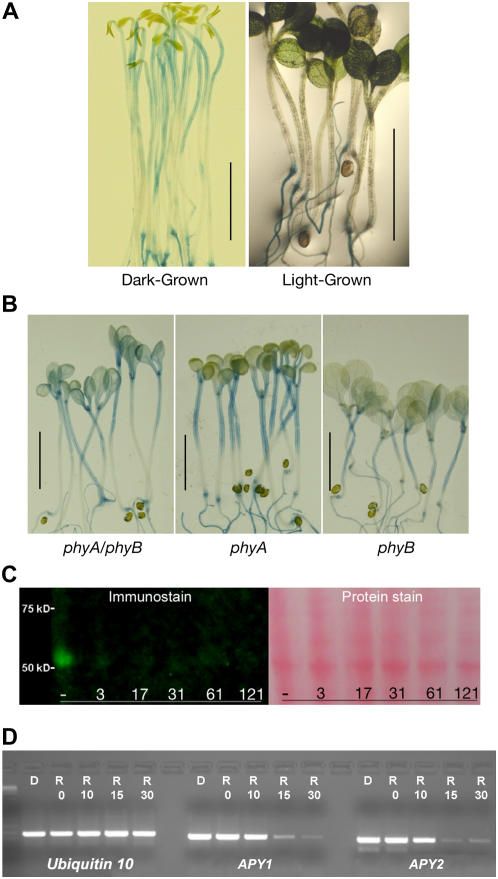
Figure 2.
Light suppresses APY:GUS staining and the expression of apyrase protein. A, Representative staining for APY1:GUS in 7-d-old dark-grown seedlings and in seedlings grown in white light. Staining for APY2:GUS was identical (data not shown). B, Light does not suppress APY1:GUS staining in 7-d-old phy mutants grown in light. Identical results were observed for APY2:GUS staining in 7-d-old phy mutants (data not shown). Scale bars in A and B = 0.5 cm. C, Immunoblot analysis showing red-light-induced rapid loss of apyrase protein from crude extracts of 4-d-old etiolated seedlings. The 7-min time point is 3 min after the 4-min irradiation was over. The protein stain is Ponceau S. D, RT-PCR assay of transcript abundance of APY1 and APY2 in unirradiated seedlings (D) and in seedlings 0, 10, 15, and 30 min after 4-min red-light (R) irradiation.
The ability of white light to inhibit hypocotyl growth is reduced in phytochrome A (phyA), phyB, and phyAphyB mutants (Neff and Chory, 1998). When these phy mutants were transformed with APY1:GUS and APY2:GUS, GUS expression was detected in the hypocotyls of light-grown seedlings (Fig. 2B). This indicates that both phyA and phyB are needed for down-regulation of APY1 and APY2 expression in the hypocotyls of light-grown seedlings.
Detailed immunoblot analysis of the kinetics of red-light-induced changes in apyrase protein content in hypocotyls, using a polyclonal antibody that recognizes both APY1 and APY2, revealed that the apyrase protein level decreased dramatically within 3 min after the end of 4-min irradiation and then remained barely detectable for 125 min after (Fig. 2C). Similarly, but with a slightly longer lag time, transcript levels for both APY1 and APY2 declined significantly by 15 min after 4-min red-light irradiation, as revealed by semiquantitative reverse transcription (RT)-PCR assay (Fig. 2D).
Suppression of Expression of APY1 and APY2 Also Suppresses Root and Shoot Growth
To reveal APY1 and APY2 gene function in whole plants, it would be important to characterize the phenotype of double-knockout (DKO) plants. However, DKO progeny could not be produced easily because DKO pollen cannot germinate (Steinebrunner et al., 2003). Therefore, a complementation strategy with an inducible promoter was chosen: Double-heterozygous plants (apy1/+; apy2/+) for a T-DNA insertion in the APY1 (apy1) and APY2 (apy2) genes were transformed with cDNA for either APY1 or APY2, each under the dexamethasone (DEX)-inducible promoter (Aoyama and Chua, 1997). The resulting lines were called DEX:APY1 and DEX:APY2 lines, respectively. The DEX promoter provided a means to reverse the DKO phenotype by reenabling pollen germination when flowering T-DNA mutants complemented with the DEX:APY1 or DEX:APY2 construct were sprayed with DEX. Progeny without intact wild-type genes for APY1 and APY2 (apy1/apy1; apy2/apy2) were identified by PCR (see “Materials and Methods”) and titled DKOs. Although these DKO plants carried no wild-type APY1 and APY2 genes, they did contain a gene construct with the wild-type cDNA for APY1 or APY2 under the DEX-inducible promoter.
Growth phenotypes of these DKO plants were compared to wild type and plants carrying the transformation vector alone (+vector) as controls. Some DKO plants from transgenic lines DEX:APY1 and DEX:APY2 expressed the transgene even in the absence of DEX (data not shown) as determined by Southern-blot analysis of RT-PCR products (see “Materials and Methods”). These DKO mutants were termed leaky and their phenotype was indistinguishable from the wild type and vector control. In nonleaky DKO mutants, on the other hand, no APY1 and APY2 message was detectable by Southern-blot analysis of RT-PCR products in the absence of DEX (data not shown). Of six DEX:APY1 and four DEX:APY2 lines tested, only four and two were nonleaky, respectively.
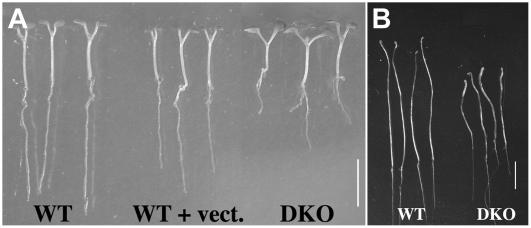
Seed germination was carried out on agar and the germination rate was the same in DKO and wild-type plants (data not shown). The phenotype of light-grown aerial tissue, cotyledons, and hypocotyls was also identical for all genotypes during the first 7 d after germination (Fig. 3A); however, primary roots were already severely affected by the absence of APY1 and APY2 transcripts after 7 d of growth (Fig. 3A). Roots of wild-type, vector-only control, and leaky DKO seedlings were 14.5 ± 1.40 mm in length, whereas those of nonleaky DKO plants were only 4.76 ± 1.56 mm long. In etiolated seedlings, the hypocotyls of DKO plants (length 18.1 ± 1.0 mm) were shorter than those of wild-type plants (length 23.4 ± 0.2 mm) and this difference was statistically significant (P ≤ 0.01; Fig. 3B).
Figure 3.
Dwarf phenotype of DKO (apy1apy2; DEX: APY2) seedlings. A, Wild-type plants, plants with vector-only control, and DKO plants (apy1apy2) containing APY2 under the steroid-inducible promoter grown 7 d in light. B, 9-d-old wild-type and DKO (apy1apy2; DEX: APY2) seedlings grown in darkness. Scale bars = 0.5 cm.
As DKO seedlings continued to grow in the light on agar, their hypocotyl and root size differences compared to wild-type plants became greater. To evaluate the basis of the size differences in these tissues, quantitative analysis comparing the lengths of hypocotyl and root cortex cells in wild-type and DKO plants was carried out. This analysis revealed that, after 10- and 13-d growth in the light, the length of hypocotyl cortex cells was significantly smaller in DKO plants (Table I), approximately enough to account for the size difference in wild-type and DKO hypocotyls. In contrast, after 10-d growth, the length of root cortex cells in DKO plants was not significantly different from these cells in wild-type plants (Table I). Thus, the significantly smaller root length of DKO plants at this stage should be attributed to reduced cell number. However, in 13-d-old plants, the lengths of root cortex cells, like those of hypocotyl cells, were significantly smaller in DKO plants. Note that measurements in 10- and 13-d-old seedlings were carried out separately.
Table I.
Hypocotyl and root cell lengths in DKO and wild-type plants
| Characters Measured | DKO Plantsa
|
Wild-Type Plants
|
||
|---|---|---|---|---|
| 10-d-Old | 13-d-Old | 10-d-Old | 13-d-Old | |
| Hypocotyl cortex length | 16.7 ± 4.9b | 12.9 ± 4.8 | 27.5 ± 7.9 | 24.0 ± 5.5 |
| Root cortex length | 11.6 ± 2.1 | 8.8 ± 1.7 | 10.3 ± 1.6 | 11.7 ± 3.2c |
DKO = apy1; apy2; DEX: APY2.
All numbers are mean ± sd in micrometers. Numbers in bold indicate DKO values that are significantly smaller than same-age/cell-type values of wild-type plants (P ≤ 0.05; n ≥ 9 for all hypocotyl and root values unless otherwise noted).
n = 3.
The only cellular abnormalities observed in the primary roots of mutant seedlings suppressed in APY1 and APY2 expression were that the root tips of these mutants lacked a well-defined meristematic zone and had a greatly reduced zone of elongation. As a result, the zone of differentiation, marked by the differentiation of root hairs, extended almost all the way to the root tip (data not shown).
Seedlings were transferred to soil after 7 d on agar and 7 d later all genotypes had developed to the same extent (Fig. 4, A–D). After 17 d on soil, however, the growth difference of nonleaky DKO plants to those with APY1 and/or APY2 transcripts had become very apparent. Wild-type plants with vector-only and leaky DKO mutants had grown at least three sets of true leaves (Fig. 4, E–G) and these leaves were several times bigger than 10 d earlier (Fig. 4, A–C). In nonleaky DKO plants (Fig. 4H), true leaves had remained the size of the cotyledons from stage 7 (Fig. 4D) and the third set of true leaves was barely visible. Growth of nonleaky DKO plants arrested in this stage so, at day 24 on soil (Fig. 4L), the number and size of leaves had not changed. Plants with APY1 and/or APY2 transcripts, however, had grown flower stalks after 24 d on soil (Fig. 4, I–K) and their overall size was approximately 10 times the size of nonleaky DKO mutants. Some nonleaky DKO plants grew tiny flower stalks (data not shown), but none ever formed seed-containing siliques.
Figure 4.
Wild-type (WT) and DKO (apy1; apy2; DEX: APY2) seedlings at various stages of growth. A to L, Wild-type, transgenic plants (WT) for APY1 and APY2 containing vector only (WT + vector) and apy1; apy2 (DKO) plants containing APY2 under the steroid-inducible promoter (DEX: APY2) were grown without DEX on an agar surface for 7 d and then transferred to soil. Pictures were taken after 7 d (A–D), 17 d (E–H), and 24 d (I–L) of growth on soil. DKO lines (apy1; apy2; DEX: APY2) producing the APY2 transcript even in the absence of DEX were classified as leaky. Scale bars = 1 cm.
We tested whether induction of the APY1 or APY2 transgene could reverse the dwarf phenotype of nonleaky DKO plants. DEX treatment of these plants started after 7 d on soil. Although transcription of APY1 or APY2 from the DEX promoter clearly reversed the blockage of germination observed in DKO pollen, DEX treatment of intact plants did not usually restore wild-type growth (data not shown). Most plants remained in their arrested stage, similar to the untreated plants (Fig. 4, H and L) and eventually died like the untreated control. In one nonleaky DKO mutant, DEX treatment induced some growth of the flowering stem and production of a small number of seeds.
To further assess the effects of suppression of APY1 and APY2 on growth, we carried out this suppression by inducing an apyrase-directed RNAi construct in apy2 plants that were wild type for APY1, but homozygous for the apy2 knockout mutation. The RNAi construct was made by inserting a sense and an antisense region of APY1 cDNA (132 bp) into the vector with an intron in between. The structure of this construct predicts that when estradiol is applied to transformed plants harboring the RNAi construct, the hormone will induce production of the sense-intron-antisense mRNA, which will form a hairpin structure, making it a target for breakdown by the RNAi machinery of the cell. The small pieces (approximately 23 bp) of double-strand RNA formed from this breakdown would be expected to target and silence APY1, and our results indicate that this happens.
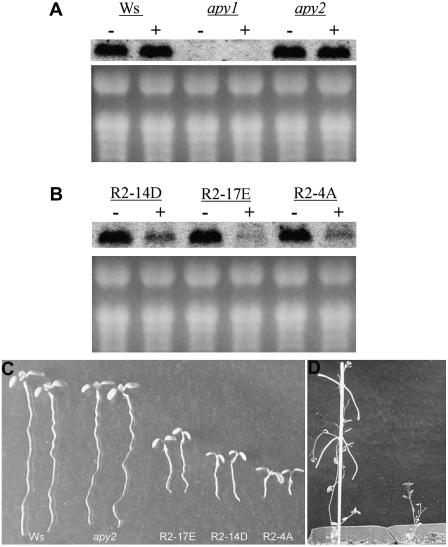
We developed three lines of transgenic plants harboring the RNAi construct and designed a gene-specific probe that could be used to assess transcript levels of APY1 in them by RNA gel-blot analysis (Fig. 5A). After confirming that the induction of the RNAi construct by estradiol in apy2 mutants significantly depressed the expression of APY1 (Fig. 5B), we found that induction also significantly reduced the growth of all three lines, both at the seedling and flowering stages of growth (Fig. 5, C and D). Although all three lines had suppressed growth, the level of growth suppression did not correlate with the level of message reduction.
Figure 5.
Induction of an RNAi construct targeting APY1 in apy2 plants reduces APY1 transcript abundance and suppresses growth in light. A, Level of APY1 transcripts in hypocotyls of wild-type (Ws ecotype), apy1, and apy2 mutants by RNA gel-blot analysis. In both A and B, seedlings were either treated with 4 μm estradiol (+) or not treated with estradiol (−). The membrane was hybridized with a 330-bp APY1-specific probe containing the −350- to −20-bp upstream region of the APY1 start codon. B, Abundance of APY1 transcripts in three lines of apy2 mutants suppressed in APY1 by RNAi. C, Dwarf growth of apy2 seedlings suppressed in APY1 by RNAi (three R2 lines) compared to wild-type (Ws) and apy2 seedlings after 7-d growth. D, Adult apy2 mutants suppressed in APY1 by RNAi (right) or not suppressed (left).
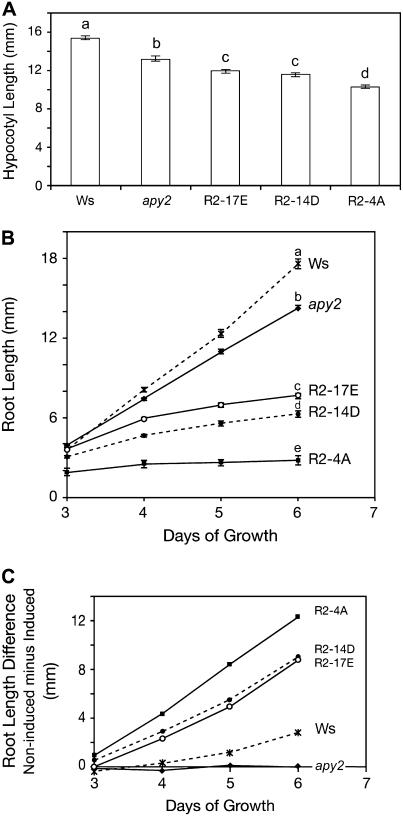
In dark-grown seedlings, the most rapidly growing tissue is the hypocotyl. Shortened hypocotyl length was found in 3.5-d-old etiolated seedlings of all three RNAi lines after they had germinated and grown the entire time in medium containing estradiol. The length of hypocotyls in apy2 mutant plants was about 15% shorter than in the wild-type strain and the average length of etiolated hypocotyls in RNAi lines was about 70% of the wild-type control (Fig. 6A). All three RNAi lines and apy2 mutant plants had significantly shorter hypocotyl lengths compared to wild-type seedlings (P < 0.01; Fig. 6A).
Figure 6.
Suppression of APY1 expression in apy2 plants suppresses growth of hypocotyls and roots. A, Suppression of growth in etiolated hypocotyls of apy2 plants expressing an RNAi construct for APY1. The hypocotyl length was measured in 3.5-d-old seedlings that had been grown the entire time in estradiol to silence APY1. Different letters above the bars indicate mean values that are significantly different from one another (P < 0.01; n > 20). B, Suppression of root growth in light-grown apy2 plants expressing an RNAi construct for APY1. Growth was assayed from days 3 to 6 of RNAi lines (R2-17E, R2-14D, and R2-4A), wild-type (Ws), and apy2 mutant lines. Different letters above the bars indicate mean values that are significantly different from one another (P < 0.01; n > 20). C, Difference in root length between estradiol-treated and not treated RNAi lines, wild type (Ws), and apy2 from days 3 to 6.
In light-grown seedlings, the most rapidly growing tissue is the primary root. Primary root growth of estradiol-treated RNAi seedlings grown in the light was analyzed from day 3 to day 6. Seedlings exhibited a significantly reduced rate of root elongation in all three lines (Fig. 6B), resulting in significantly shorter roots by day 6 (Fig. 6B).
To test whether estradiol itself can inhibit root growth, the difference of root length in estradiol-treated and nontreated seedlings, both apy2 and wild-type plants, was measured. Data showed estradiol had no effect on apy2 mutant plants, which were used as the background plants of the RNAi lines. Although estradiol did slightly reduce the growth of wild-type roots, still it reduced the root growth of the three RNAi lines to a much greater extent than did the wild type and apy2 mutants (Fig. 6C). This demonstrated that the shorter root length of RNAi lines was not due to applying estradiol. Wild-type adult plants treated with estradiol were indistinguishable from RNAi plants that were not treated with estradiol (data not shown). Although estradiol-treated RNAi seedlings (Fig. 5C) resemble the genetic DKO seedlings (Fig. 3A) in being dwarf, there is one statistically significant difference between the two: the former have more radially expanded (larger diameter) primary roots than the latter (diameter of 0.371 ± 0.018 mm compared to 0.323 ± 0.032 mm; P < 0.05, n = 7).
Lines Suppressed in Apyrase Expression Have Fewer Lateral Roots and More Adventitious Roots
Beyond decreased growth, the most notable developmental effects of suppressing the expression of both APY1 and APY2 were decreased formation of lateral roots and increased formation of adventitious roots. In lateral root measurements (Table II), the apy2 mutant used as the control was not different from wild-type plants in this characteristic. In the adventitious root measurements, comparison of a representative 23 wild-type and 23 DKO plants grown on the same plates revealed that only two of the wild-type plants, but 17 of the DKO plants, had adventitious roots, a difference that was statistically significant (P < 0.01). Roots emerging from the root-shoot interface in DKO plants in Figure 3A are adventitious roots, not lateral roots.
Table II.
Lateral root numbers in 14-d-old light-grown seedlings of four different mutant lines with suppressed apyrase expression
| Mutant Line Assayed | Average Lateral Root No.a |
|---|---|
| apy2 | 6.65 ± 1.33 |
| R2-4A | 0.39b ± 0.49 |
| R2-14D | 1.26b ± 0.82 |
| R2-17E | 2.03b ± 1.29 |
All averages are ±sd.
Significantly different from average number in apy2 plants (P < 0.01; n ≥ 28).
Overexpression of Apyrase Enhances Growth
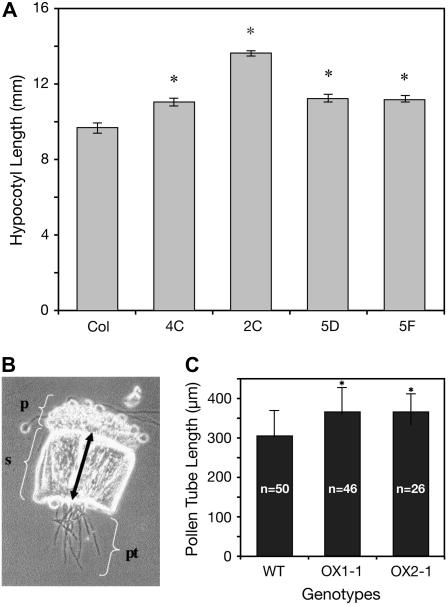
In etiolated seedlings, overexpressing lines of APY1 and APY2 were analyzed by measuring the hypocotyl length. Overexpressing APY1 resulted in a 15% increase in growth over that of wild type (Fig. 7A), but overexpressing APY2 did not increase growth (data not shown). Under continuous light conditions, growth of wild-type plants and plants overexpressing APY1 or APY2 was not different.
Figure 7.
Constitutive expression of an apyrase enhances growth. A, Enhancement of hypocotyl growth in 3.5-d-old 35S:apy1 plants overexpressing APY1. Hypocotyl length was measured in 3.5-d-old seedlings of four different lines (4C, 2C, 5D, and 5F) of 35S:apy1 plants and in Columbia ecotype wild-type plants (Col). The mean values marked with an asterisk are significantly different from mean wild-type hypocotyl lengths as determined by Student's t test (P < 0.01). B, Bright/dark-field image of a wild-type (wt) style (s) with wild-type pollen tubes (pt). Pollen tube lengths were determined by adding the length of the style (double-headed arrow) to the emerged length of a particular pollen tube. p, Papillae. C, Lengths of wild-type (WT) pollen tubes were compared to those from transgenic lines overexpressing APY2 (OX1-1, OX2-1). The mean values marked with an asterisk are significantly (P ≤ 0.05) different from mean wild-type pollen tube lengths as determined by Student's t test. The measurements of one of two independent experiments are shown, both of which had identical statistical results. n, Number of pollen tubes measured.
Growth of APY2-overexpressing (Song et al., 2006) and wild-type pollen tubes was compared in vitro, but no difference was detected (data not shown). Therefore, a semi-in vivo pollen tube growth assay was conducted, which allows pollen tubes to grow much faster than in vitro (Wilhelmi and Preuss, 1997), thereby making differences in growth rate more apparent. For each genotype, pollen grains were allowed to germinate on a wild-type stigma of a style removed from its ovary. Growing through the transmission tract of the style aligned pollen tubes before emerging into the growth medium (Fig. 7B). Lengths of emerged tubes were determined after 3 h and the distance from the base of the papillae to the cut end of the style was added. In this assay, pollen tubes from two overexpressing lines (OX1-1 and OX2-1) were significantly longer than the wild-type control (Fig. 7C). Pollen tubes from single-knockout lines, however, showed no difference in length to wild-type tubes, which averaged 304 ± 63 μm long (data not shown).
Inhibition of Apyrase Activity in Pollen Tubes Inhibits Their Growth
Polyclonal antibodies to Arabidopsis apyrase were tested for their effects on apyrase activity and on pollen tube growth. As expected, antiapyrase immune serum (but not preimmune serum) strongly inhibited the apyrase activity from Arabidopsis pollen germination medium (PGM; Supplemental Fig. S1). Immune serum applied in amounts of 0.4 μg or above significantly inhibited pollen tube growth, but preimmune serum at 0.6 μg did not (Fig. 8A). The level of inhibition was increased with the increasing concentration of immune serum (Fig. 8A). Consistent with these results, two selective chemical inhibitors of apyrase previously characterized (Windsor, 2000; Windsor et al., 2002, 2003) also inhibited pollen tube growth (Supplemental Fig. S2A).
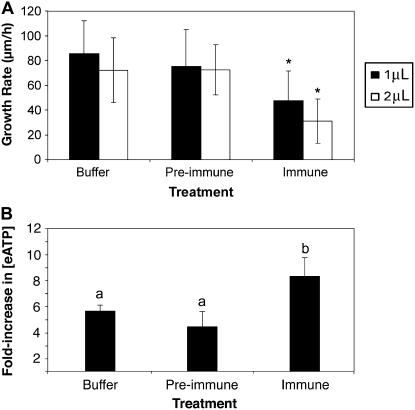
Figure 8.
Inhibition of apyrase activity in pollen tubes by antibodies decreases tube growth rate and increases the [eATP]. A, Treatment of growing pollen tubes with polyclonal antiapyrase antibodies rapidly decreases their growth rate. Error bars are ±sd and asterisk marks growth rates that are significantly different from that of the buffer control (P ≤ 0.02; n ≥20). The differences in growth rates of tubes treated with preimmune serum and of tubes treated with buffer are not statistically different (P > 0.6; n ≥ 20). Protein concentration of the preimmune sera was 0.3 μg/μL, and of the immune sera was 0.4 μg/μL. B, Treatment of growing pollen tubes with polyclonal antiapyrase antibodies rapidly increases the concentration of eATP in the medium. Medium [ATP] was measured at 1 and 15 min after addition of the treatment solution (buffer control, preimmune sera, or immune sera as in A), and the bar height gives the average fold increase in [eATP] ±sd between the two time point comparisons. The average [eATP] at 1 min was 6.1 ± 0.41 nm for buffer controls, 8.22 ± 1.81 nm for samples treated with preimmune sera, and 6.77 ± 1.38 nm for samples treated with immune sera. Different letters above the bars indicate mean values that are significantly different from one another (P < 0.05; n = 4).
For pollen growth experiments in which the volume of growth medium applied was 150 μL, the [eATP] in the medium was measured by luciferin-luciferase assay (Jeter et al., 2004) at 1 min after it was placed over the germinated grains and then again at 15 min after being applied. In all trials, the [eATP] of the medium rose between the first and second measurements and these increases were normalized for all experiments by expressing them as fold increases between the first and second time points assayed. The average fold increase in [eATP] after control treatments with buffer only was 5.66 (Fig. 8B) and, in all cases, the [eATP] of the bulk medium was at least 5 nm for the first measurement and more than 33 nm for the second measurement, indicating that ATP was being released during the growth of the tubes.
When apyrase antibodies were applied to germinated pollen, they consistently increased the [eATP] of the pollen growth medium compared to medium without immune sera. Expressed as fold increases, they were statistically significantly higher in the samples treated with immune serum than those recorded in the preimmune and buffer control samples (Fig. 8B). Apyrase inhibitors also consistently increased the [eATP] of the pollen growth medium (Supplemental Fig. S2B). The inhibitor/antibody-induced increase in medium [ATP] was evident within the same time frame (15 min) that these agents inhibited pollen tube growth.
DISCUSSION
Pollen tubes, etiolated hypocotyls, and root tips are among the fastest growing tissues in plants. All three show high expression of APY1:GUS and APY2:GUS. In contrast, nongrowing tissue, such as mature, fully expanded leaves, have little or no expression of the GUS constructs. Moreover, expression in hypocotyls is drastically reduced by the same light signal that induces their decreased growth. The promoter:GUS results are consistent with PCR assays in hypocotyls, which showed that red light induces a rapid decrease in the level of apyrase transcripts. Even more pertinent to the argument that apyrase expression is correlated with growth is the observation that the red-light signal that activates phytochromes induces the disappearance of the apyrase protein from etiolated hypocotyls within 3 min after the end of 4-min irradiation, or less than one-half the time it reportedly takes for red light to noticeably reduce the growth of hypocotyls, which is about 8 min (Parks and Spalding, 1999). Primary roots grow faster in light than in darkness and promoter:GUS signals appear generally more intense in light-grown roots than in dark-grown roots (Sun, 2003). However, the effect of light on the transcript abundance of APY1 and APY2 in roots was not quantitatively evaluated in this study.
Regulation of apyrase transcripts by light could involve both mRNA turnover and down-regulation of transcription. The 3′-untranslated region of APY2, but not of APY1, has a AUUUA motif that can serve as an instability domain (Zhang and Mehdy, 1994) and the cis-regulatory elements of both genes contain some motifs that are light responsive (Table III). Consistent with the fact that auxin also plays a major role in the regulation of hypocotyl growth, there are also auxin-responsive cis-regulatory elements in the promoters of both apyrase genes (Table III). Light-induced loss of apyrase transcripts by itself probably cannot account for the light-induced loss of apyrase protein, which is so rapid that it almost certainly requires proteolytic destruction, although additional experiments would be needed to confirm this.
Table III.
cis-Regulatory elements in the upstream promoter regions of APY1 (AP1) and APY2 (AP2) that are light or auxin responsive
| Motifs | Sequence | Function | AP1 | AP2 |
|---|---|---|---|---|
| Light responsive | ||||
| -10PEHVPEBD | TATTCT | Light regulated | X | X |
| CCA1ATLHCB1 | AAMAATCT | Phytochrome regulation | X | |
| CGACGOSAMY3 | CGACG | Coupling element for G box | X | X |
| GATA box | GATA | Conserved in LHCII-type I cab genes | X | X |
| GT1CORE | GGTTAA | Critical GT-1 binding site | X | |
| GT1 consensus | GRWAAW | GT-1 binding site | X | |
| IBox/IBox core | GATAA(G) | Conserved in light-regulated genes | X | X |
| INRNTPSADB | YTCANTYY | Light-responsive without TATA box | X | X |
| REALPHALGLHCB21 | AACCAA | Phytochrome regulation | X | |
| Auxin responsive | ||||
| ARFAT | TGTCTC | Binding site, auxin-responsive genes | X | |
| CATATGGMSAUR | CATATG | Auxin responsive | X | X |
Other regions of high expression of APY1:GUS and APY2:GUS are associated either with cell growth or differentiation. These include the root-shoot interface, the stigma papilla cells, veins of cotyledons, and the abscission zone of floral tissues. All of these sites are also regions of auxin accumulation and/or transport and most show high expression of the pin-formed (PIN) proteins associated with auxin transport (Leyser, 2005; Aloni et al., 2006).
Promoter:GUS results suggest the possibility that apyrases may play a central role in growth control. Strong support for this hypothesis comes from the observation that transgenic plants suppressed in apyrase expression have a dwarf phenotype with drastically reduced root and shoot growth. This phenotype was confirmed by two independent genetic approaches: (1) generation of apy1/apy2 DKO lines and (2) suppression of APY1 in apy2 mutants by the estradiol-inducible expression of double-stranded APY1 RNA.
In light-grown seedlings of both DKO plants and RNAi-suppressed plants, growth inhibition is more evident in roots, which grow more rapidly in wild-type seedlings than in hypocotyls, which grow only slowly in the light. Although hypocotyl cortical cells are shorter in DKO plants than in wild type at both ages tested (10 and 13 d), 10-d-old root cortical cells are not. However, Figure 3 reveals that the roots of 7-d-old DKO seedlings grown in the light already are dramatically shorter than wild-type roots. This would suggest that APY1 and APY2, which both have some expression in the meristematic region of root tips (Fig. 1), may play a role in cell division control as well as in cell expansion control, and that both roles contribute to overall growth control in roots. Consistent with this hypothesis, root tips of DKO seedlings have a poorly defined meristematic zone and a greatly reduced zone of elongation (data not shown).
The estradiol treatment that induced the expression of double-strand APY1 RNA clearly suppressed normal growth in those mutants, but DEX treatment only rarely reversed the growth defects in DEX-inducible DKO mutants, even though induction of the transgene after DEX application was confirmed by RT-PCR (data not shown). One explanation could be that the chimeric DEX-regulated steroid receptor that is part of the inducible system (Aoyama and Chua, 1997) can exert toxic effects, as described by Kang et al. (1999). Upon binding to DEX, the receptor translocates to the nucleus, where it functions as a transcription factor. Kang et al. (1999) documented that there are severe growth defects in transgenic lines carrying the chimeric transcription factor only, possibly due to its binding to low-affinity sites in the wild-type genome. Thus, DEX usage on those plants may have indirectly caused a phenotype similar to that of nonleaky DKO mutants. Nonetheless, the DEX-inducible system effectively reversed the male-sterile phenotype of DKO mutants and thus allowed us to generate DKO plants.
In estimating background expression of apyrase even in the so-called nonleaky DKO mutants, there is always the possibility that there is some low expression that is undetectable by RT-PCR. That this may be the case in the nonleaky DKO mutants is suggested by the fact that DKO lines generated without an inducible system display an even more severe dwarf phenotype than the DKO mutants presented here (C. Wolf, M. Hennig, and I. Steinebrunner, unpublished data).
If DKO mutants have some low level of expression of APY1 and/or APY2, it is unlikely that this expression is identical to that in the mutants silenced by the RNAi constructs. This may be one explanation as to why the seedlings of these two mutant types do not have identical phenotypes, with the hypocotyls and roots of the mutants expressing RNAi constructs being thicker. A BLAST search of the whole genome of Arabidopsis revealed that, except for APY1 and APY2, there are no other genes that have sequences similar to that used in the RNAi construct, including other apyrases, so it is unlikely that the phenotypic differences between the two types of mutants can be attributed to suppression of other apyrase genes in RNAi-silenced mutants. Nonetheless, this and other possible explanations would have to be tested.
Although the ineffectiveness of DEX in rescuing the DKO lines will make it more difficult to propagate DKO lines, the dwarf growth of multiple lines of DKO plants and plants suppressed in apyrase expression by double-stranded RNA induction underscores a key role for apyrase in growth. Obviously, neither cell division nor even the growth of newly divided cells is totally blocked in plants mutated in APY1 and APY2. Nonetheless, combination of the expression data in Figures 1 and 2 and the suppression results of Figures 3 to 6 strongly support the conclusion that apyrase expression is critical for normal, full cell expansion in Arabidopsis.
To the extent that APY1 and APY2 are expressed in locales accessible to chemical inhibitors of their activity, these inhibitors could mimic the growth effects of suppressing the APY1 and APY2 genes. Polyclonal antibodies that recognize both APY1 and APY2 would be the most specific agents to block the activity of these enzymes and, as would be expected, they do inhibit the apyrase enzyme activity released by Arabidopsis pollen tubes as they grow. The observation that they also inhibit pollen tube growth reinforces genetic evidence linking APY1 and APY2 expression to growth and points to the likelihood that these enzymes are functioning as ectoapyrases. Small-molecule inhibitors of apyrase enzyme activity have been described (Windsor et al., 2002) and, although the two chosen for the growth assays differ significantly in their structure, they have similar strong potency in inhibiting Arabidopsis apyrase activity. The fact that inhibition of apyrase activity by antibodies or other chemical agents results in a rapid rise in the [ATP] of the growth medium indicates that this activity plays a significant role in controlling the equilibrium concentration of ATP outside the plasma membrane.
Although APY1 and APY2 are already strongly expressed in rapidly growing hypocotyls and pollen tubes, the constitutive (and increased) expression of APY1 in hypocotyls and of APY2 in pollen further enhances their growth. These growth-promoting effects are not as dramatic as the growth suppression that results from reduced apyrase expression, suggesting that in wild-type hypocotyls and pollen cells apyrase expression is near optimal for growth. Still, the overexpression data further support the hypothesis that the expression of APY1 and APY2 is closely linked to growth control.
Based on apyrase studies in vertebrates and yeast (Saccharomyces cerevisiae), at least two different functions could account for why apyrases exert such dramatic effects on growth. One function is that of an ectophosphatase to reduce the signaling activity of [eATP] and thus turn off nucleotide activation of purinoceptors. The other is as a Golgi enzyme that regulates glycoprotein synthesis.
Regarding ectophosphatase function, APY1 and APY2 could potentially play this role because they both have signal peptides (Steinebrunner et al., 2000) and overexpression of APY2 is correlated with decreased sensitivity of Arabidopsis leaves to wound-released ATP (Song et al., 2006). As remarked above, more direct proof that some of the APY1 and APY2 enzymes are ectoapyrases is the fact that apyrase activity is released by growing pollen into the pollen growth medium and this activity is blocked by polyclonal antibodies that bind both APY1 and APY2 (Supplemental Fig. S1).
In animal cells, ATP is released to the outside of cells through secretory activity because secretory vesicles enclose high levels of ATP and they release this into the ECM when they fuse with the plasma membrane (Lazarowski et al., 2003). Of course, growth of plant cells is necessarily accompanied by the delivery of Golgi contents to the ECM (Clark et al., 2005). That ATP is released from regions of cell growth in plant cells was recently demonstrated by Kim et al. (2006). They used a novel luciferase reporter bound to wall cellulose to visualize eATP in roots of Medicago truncatula and they found that the activity of this bound luciferase was closely correlated with regions of cell expansion. Notably, luciferase-detected eATP was particularly abundant at root hair tips. If a parallel phenomenon occurs in pollen, one would expect that most of the ATP released during the growth of tubes would be from their tip.
Consistent with the findings of Kim et al. (2006), our measurements of the change in [ATP] in the bulk medium during pollen tube growth indicate that this growth may result in the release of ATP. To the extent that ectoapyrases play a role in reducing the [eATP], one would predict that blocking the activity of these apyrases in growing cells would result in at least a transient increase in the [eATP]. We observed this in growing pollen tubes and, coincident with the antibody- and inhibitor-induced rise in the [eATP], there was a decrease in growth rate. The coincidence of ectoapyrase inhibition and growth inhibition in pollen tubes growing in vitro is consistent with genetic data linking apyrase suppression with growth suppression.
The nanomolar levels of ATP measured in the bulk medium in which pollen is growing is likely a vast underestimate of the level of ATP at least transiently present at the cell surface when nucleotides are released from the cell. A recent report by Yegutkin et al. (2006) assayed the pericellular ATP pool on the surface of lymphocytes using a novel intrinsic sensor of [ATP] and found that levels at the membrane surface were often 1,000-fold higher than that measured in the bulk medium in which the cells were growing, reaching over 40 μm. Similar measurements of the pericellular [ATP] in pollen tubes with inhibited apyrase activity would be needed to reveal what eATP concentrations in the vicinity of the membrane surface are correlated with growth inhibition.
Kim et al. (2006) correctly note that the level of [eATP] accumulating at the growing points of cells could be linked to the production of superoxide, which, in turn, is generally linked to growth promotion. Our data suggest that if the equilibrium between eATP accumulation at growth points and the removal of this eATP is disturbed by inhibition of ectoapyrase activity, the [eATP] rises to the point where growth becomes inhibited. On the other hand, extensive removal of eATP by application of high concentrations of potato (Solanum tuberosum) apyrase can lead to diminished growth (Kim et al., 2006) or even cell death (Chivasa et al., 2005). Taken together, these data predict that there is an optimal level of eATP that can stimulate superoxide production leading to growth promotion, but significantly higher or lower levels would turn on alternative pathways leading to growth inhibition. Consistent with this interpretation, we previously showed that there are low levels of applied nucleotides that can stimulate hypocotyl growth, whereas higher levels inhibit growth (Roux et al., 2006).
How could an increase in [eATP] be linked to a decrease in growth? As yet there are insufficient data to answer this question. The fact that submicromolar levels of eATP can induce signaling changes in plants (Demidchik et al., 2003; Song et al., 2006) would suggest that plants, like animals, have purinoceptors that can bind eATP with a high affinity and transduce that binding into transduction pathways. However, as discussed by Demidchik et al. (2003), Roux et al. (2006), and others, as yet no purinoceptor has been identified in plants. Thus, in parallel with the history of auxin, gibberellin, and other growth regulators in plants, evidence that eATP can influence growth is preceding the discovery of how its growth influence is initiated, whether that be by a receptor or by some other mechanism.
Apart from receptor considerations, one downstream effect initiated by eATP that could help explain its influence on growth is on auxin transport. Increased [eATP] (by external application) can block auxin transport and promote auxin accumulation in tissues (Tang et al., 2003), most likely through its inhibitory effects on the multidrug resistance transporters that facilitate auxin efflux from cells (Lin and Wang, 2005; Geisler and Murphy, 2006). That high levels of auxin inhibit growth has been demonstrated repeatedly in the literature, including recently by Hu et al. (2005), who showed that the asymmetric accumulation of auxin on the lower side of roots induces nitric oxide (NO) production there, leading to asymmetric growth inhibition and gravitropic bending.
To the extent that high [eATP] can lead to the accumulation of growth-inhibitory levels of auxin (and/or of NO), maintenance of lower [eATP] through apyrase activity at growth points may be needed to maintain growth. Observations consistent with this interpretation are that APY1:GUS and APY2:GUS assays show the highest expression in cells where the PIN auxin efflux facilitators are also highly expressed, and plants suppressed in apyrase expression have decreased lateral root formation, just like both mutants in aux1 (Marchant et al., 2002) and wild-type plants that have been treated with auxin transport inhibitors (Casimiro et al., 2001).
The considerations above do not exclude the possibility that some fraction of APY1 and APY2 function in the endoplasmic reticulum (ER) or Golgi, for certainly apyrases would move through the ER-Golgi pathway on the way to the plasma membrane. There is a well-developed model for apyrase function in the Golgi of yeast related to control of protein glycosylation (Hirschberg et al., 1998). However, in animal cells, the ectoapyrase CDC39 is inactive until it arrives at the plasma membrane (Zhong et al., 2001) and evidence of APY1/APY2 activity in ER or Golgi would be needed to make this role for them seem probable.
The two apyrases do not have to play identical roles in Arabidopsis. The amino acid sequences of APY1 and APY2 are 87% identical and these two apyrases at least partially complement each other's function (Steinebrunner et al., 2003). However, the tissue expression patterns of these two apyrases are different, at least in roots, so we cannot assume their subcellular distribution is identical. Moreover, the polyclonal antibodies used to inhibit apyrase activity in pollen growth medium do not distinguish whether both apyrases are present or only one. Nonetheless, the results presented here do make it clear that wherever APY1 and APY2 function, their combined activities play a central role in growth control in Arabidopsis.
In summary, our data reveal the finding that the expression of two closely related apyrase enzymes that can lower the [eATP] of plant cells is closely correlated with growth and, in fact, is needed for normal growth in Arabidopsis. Because cells release ATP as a consequence of growth (Kim et al., 2006; Fig. 8) and high levels of eATP can inhibit growth (Roux et al., 2006), our results point to the likelihood that plant cells must control their [eATP] to sustain growth and that APY1 and APY2 function as key players in the mechanisms whereby cells achieve this control. Tests of this hypothesis will require developing methods to quantify how much the concentration of eATP changes during growth and how increasing or decreasing ectoapyrase expression modulates these changes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Unless otherwise noted, Arabidopsis (Arabidopsis thaliana) ecotypes Columbia (CS907) and Wassilewskija (Ws) were used as wild types in this study. Seeds were planted directly on autoclaved Metro-Mix 200 soil or surface sterilized and planted on solidified Murashige and Skoog medium (4.3 g/L Murashige and Skoog salts [Sigma], 0.5% [w/v] MES, 1% [w/v] Suc, and 0.8%, 1.0%, or 1.2% [w/v] agar, raised to pH 5.7 with 5 m KOH). The apy1 and apy2 mutants were isolated previously (Steinebrunner et al., 2003). For root and hypocotyl growth assays, seeds were sown on the surface of solidified Murashige and Skoog medium. Plates were placed upright in a culture chamber and grown at 23°C under 24-h fluorescent light. For the root growth assay, images were taken every 24 h on days 3 through 6 with a Nikon Coolpix 990 digital camera. For measurements of the growth of etiolated hypocotyls, plates with sown seeds were wrapped in aluminum foil and placed in a growth chamber for 3.5 d, then unwrapped under white light and photographed immediately.
Phytochrome mutants phyA-201, phyB-5, and phyA201/phyB-5 and Ws wild-type plants were used for plant transformation. All types of plants were grown at 22°C under continuous light. For etiolated seedlings, seeds were put on solidified Murashige and Skoog medium and grown in the dark for 7 to 10 d.
In assays of DKO plants complemented with a wild-type gene, to induce the transgene, plants were sprayed with water containing 30 μm DEX and 0.01% (v/v) Tween 20 or watered with 30 nm DEX.
Measurements of Hypocotyl and Root Cortex Cells
Wild-type and DKO plants used for microscopic analysis were grown together on the same agar plate. Plants were processed using protocols from Ruzin (1999). After fixation and embedding in paraffin, 10-μm sections of tissue were cut, mounted on glass slides, and analyzed using an ocular micrometer and standard light microscopy techniques. Measurements of hypocotyl cells were taken from just below the cotyledons to just above the root junction and of root cells from the root hypocotyl junction to just above the zone of maturation. All cell lengths from each hypocotyl or root were averaged.
Primers Used
In the text below, more than a score of different primers were used, defined as follows: AAR566 (5′-CACAGCGTAATTCTTCGGACC-3′), AP1F (5′-CCCAAGCTCTCTCCGCTACCTTTGGAATTCAGACG-3′), AP1R (5′-GCGTCGACTCGATAGACACAAGTCCCTGATGAGAGTC-3′), AP2F (5′-ACGCGTCGACATGGTCATTTGAGGTGGCAGAGAATATG-3′), AP2R (5′-GCTCTAGACGTCAACAGAGTCGGATGTAGGAGAATGG-3′), APT1_for (5′-TCCCAGAATCGCTAAGATTGCC-3′), APT1_rev (5′-CCTTTCCCTTAAGCTCTG-3′), APY1-NF (5′-TAGAAGCAGTATCCTCACC-3′), APY1-NR (5′-ACAGAGGTTACGTATGCGG-3′), APY2-NF (5′-CATAGTTGGGAGTTACCCATCTCCC-3′), APY2-NR (5′-TACCAGACTCCAGGAGCTCAGTGG-3′), Apy1-SalI (5′-ATAGTCGACGTATTTCACCTTCTT-3′), Apy1-XhoI (5′-ATACTCGAGAAACCAACCTGTGGC-3′), Apy-a (5′-ATAGAATTCATGACGGGGAAGGGA-3′), Apy-b (5′-ATCGATACCGTCGACCTCGAGTGGTGAGGATACTGCTTCT-3′), AraF172 (5′-GCAGCCGTAACTTGCAATC-3′), AraF172 (5′-GCAGCCGTAACTTGCAATC-3′), Arapy2F (5′-GCTTTCCCAAATTCACCGT-3′), DEXF (5′-GCCGCCAGTGTGATGGATATCTGC-3′), Myc-c (5′-AGAAGCAGTATCCTCACCATCTCGAGGTCGACGGTATCGA-3′), Myc-e (5′-GTATCATTCATTCAGTCAAAAGTCCTC-3′), RNAiI-EcoRI (5′-ATAGAATTCGTATTTCACCTTCTT-3′), RNAiI-SpeI (5′-ATAACTAGTAAACCAACCTGTGGC-3′), UBQ1 (5′-GATCTTTGCCGGAAAACAATTGGAGGATGGT-3′), and UBQ2 (5′-CGACTTGTCATTAGAAAGAAAGAGATAACAGG-3′).
Construction of APY1 Promoter:GUS Fusion and Plant Transformation
The Arabidopsis genomic bacterial artificial chromosome (BAC) clone T6K12, which contains the APY1 promoter region, was obtained from the Arabidopsis Biological Resource Center (ABRC). MRG7, which contains the APY2 promoter region, was kindly provided by Kazusa DNA Research Institute. Both promoter regions used include the 5′-untranslated region sequences of APY1 and APY2 genes. The promoter of APY1 was first amplified by PCR using primers AP1F and AP1R and subcloned into TOPO vector pCR2.1 (Invitrogen). Then the 3-kb HindIII-SalI fragment was subcloned into binary vector pBI101 (provided by Dr. Mark Estelle, University of Indiana), which contains a promoterless GUS gene, to create pBI-APY1:GUS. To construct pBI-APY2:GUS, the APY2 promoter was amplified using primers AP2F and AP2R by PCR and subcloned into vector pCR2.1. Then a 2.8-kb SalI-XbaI fragment was subcloned into vector pBI101. The constructs pBI-APY1:GUS and pBI-APY2:GUS were transformed into Agrobacterium tumefaciens strain GV3101(pMP90) and the vacuum infiltration method (Clough and Bent, 1998) was used to transform the Arabidopsis plants. Several independent APY1:GUS and APY2:GUS transgenic lines were carried to T3 homozygous and stained for GUS activity according to Lehman et al. (1996).
Histochemical GUS Staining
GUS transformants were grown on regular Murashige and Skoog medium for root and cotyledon staining. For staining of flowers, siliques, and leaves, soil-grown plants were used. GUS staining was performed according to Lehman et al. (1996).The staining was performed for 1 to 4 h with constant monitoring every 30 min until a desired staining intensity was reached.
To make the staining for APY1:GUS and APY2:GUS in pollen tubes stand out more clearly in the pollinated flower, wild-type plant stigmas were used for in vivo germination of pollen from APY1:GUS and APY2:GUS plants. Wild-type flowers were emasculated the day before pollination. The next morning, pollen from fully opened APY1:GUS or APY2:GUS flowers were brushed onto the wild-type stigmas and allowed to germinate for 2 h. A regular GUS-staining procedure was then followed to make APY1:GUS and APY2:GUS expression in pollen tubes clearly visible (Fig. 1L).
In Situ Localization of Apyrase Transcripts in Roots
The method followed was a liquid-phase whole-mount RT-PCR protocol that was a combination of the in situ RT-PCR protocol (Koltai and Bird, 2000) and the whole-mount in situ protocol (Engler et al., 2001). Soil-grown seedlings were fixed in heptane:fixation buffer (0.08 m EGTA, 5% [v/v] formaldehyde, and 10% [v/v] dimethylformamide; 1:1 [v/v]) for 30 min, dehydrated twice for 5 min in absolute methanol, and three times for 5 min in absolute ethanol. Samples were typically stored 1 to 3 d in ethanol at −20°C, then rinsed once in absolute ethanol, and incubated for 30 min in absolute ethanol:xylene (1:1 [v/v]). Samples were then washed twice for 5 min in absolute ethanol, twice for 5 min in absolute methanol, and once for 5 min in methanol:PBT (phosphate-buffered saline + 0.1% [v/v] Tween 20; 1:1 [v/v]). They were postfixed for 30 min in PBT containing 5% (v/v) formaldehyde followed by one rinse with phosphate-buffered saline and two rinses with double-distilled water.
Tissues thus prepared then underwent liquid-phase RT-PCR in a solution containing RNase inhibitor, Moloney murine leukemia virus reverse transcriptase, and either the APY1 or APY2 gene-specific reverse primer to reverse transcribe the APY1 or APY2 message. PCR reactions were performed using Taq polymerase with forward (APY1-NF, APY2-NF) and reverse (APY1-NR, APY2-NR) primers with digoxigenin-labeled dUTP to yield a labeled PCR product of about 250 bp for APY1 and 270 bp for APY2.
Samples were stained immediately after PCR. They were washed twice for 5 min in PBT and blocked for 30 min in PBT containing 3% (w/v) bovine serum albumin, then incubated overnight at 4°C in 1 mL of antibody, which was a preabsorbed, alkaline-phosphatase-conjugated antidigoxigenin monoclonal antibody (Boehringer Mannheim/Hoffmann-La Roche) diluted 1:1,500 in blocking solution. The antibody solution was then replaced by fresh blocking solution and incubated for 10 min. Samples were washed five times in PBT for 15 to 30 min and placed in 35 × 10-mm petri plates with 1 mL of washing buffer (10 mm Tris, 15 mm NaCl, pH 9.5) containing 150 μg/mL 4-nitroblue tetrazolium chloride and 370 μg/mL 5-bromo-4-chloro-3-indolyl-phosphate (Boehringer Mannheim/Hoffmann-La Roche). Color development was monitored by microscopy and stopped by rinsing samples with double-distilled water.
Control samples were treated exactly as above (fixation, permeabilization, washing, etc.), except in some cases only the reverse transcriptase was excluded from the RT reaction or only Taq polymerase was left out of the PCR reactions. Both controls looked essentially the same (negative) and the control in Figure 1D is a no-Taq control.
Antibody Production and Purification
Sequence for the APY1 open reading frame without the first 105 bp coding for a putative transmembrane domain was inserted into the BamHI site following the polyhistidine tag of the pET28a vector (Novagen). The BamHI cleavage site was added to either end of the APY1 sequence by PCR. In-frame insertion of each construct was confirmed by sequencing. The recombinant protein was expressed in the Escherichia coli strain BL21 (Novagen) and purified under denaturing conditions on a nickel-nitrilotriacetic acid agarose column (Qiagen) following the manufacturer's instructions. This column-purified protein was loaded onto 10% SDS-PAGE, and the band containing the protein of the correct size was excised. The gel slice was sent to Pocono Rabbit Farm and Laboratory, Inc., to immunize two guinea pigs, gp18 and gp19, respectively. The affinity of antiserum gp18 was equal to both recombinant denatured APY1 and APY2 proteins. Gp18 serum was affinity purified on CM Affi-Gel Blue according to the manufacturer's instructions and used for the immunoblot analysis, growth assays, and apyrase activity assays.
Immunoblot Analysis
Etiolated 4-d-old seedlings were harvested in a cold room, 4°C, illuminated only by a dim green light-emitting diode. Their roots were removed with a razor blade and their aerial portion placed in a 1.5-mL reaction tube and snap frozen in liquid nitrogen. Before harvesting the light-treated seedlings, the petri dishes containing them were placed vertically in racks, set into light chambers, and exposed to a light-emitting diode source with an emission maximum of 670 nm and an irradiance of 31 μmol m−2 s−1 for 240 s at 22°C.
Frozen tissue was ground in a mortar filled with liquid nitrogen and a pestle. Once the tissue was ground and placed in a 1.5-mL reaction tube, 15 or 25 μL of a buffer containing 125 mm Tris-HCl, pH 8.8, 1% (w/v) SDS, 10% (v/v) glycerol, 50 mm Na2S2O5 were added and tissue immediately boiled for 5 min. Samples were then centrifuged briefly at 4°C, then quantified via Bradford assay with Bio-Rad protein assay dye reagent concentrate. Fifteen micrograms total protein were mixed with 6× sample buffer and proteins separated via SDS-PAGE, transferred to 0.2-μm nitrocellulose membranes (Schleicher & Schuell), and probed with gp18 antibody serum diluted 1:1,000. Secondary antibody was a conjugated affinity-purified anti-guinea pig IgG (goat) coupled to an 800-nm fluorochrome diluted 1:5,000 (Rockland IRDye 800CW). The fluorochrome signals were detected and analyzed using the Odyssey infrared imaging system (LI-COR Biosciences).
RT-PCR Assay of Transcript Abundance
All seeds were sterilized with 20% (v/v) bleach and plated on Murashige and Skoog agarose plates containing 1% (w/v) Suc. Seeds were allowed to vernalize for 4 d, then were grown in darkness for 4 d. Seedlings were given light treatments as follows: D, no light, etiolated tissue; R, 4 min, 30 μmol m−2 s−1 red light, then harvested immediately; R10, 4 min, 30 μmol m−2 s−1 red light followed by 10-min darkness, then harvested immediately; R15, 4 min, 30 μmol m−2 s−1 red light followed by 15-min darkness, then harvested immediately; R30, 4 min, 30 μmol m−2 s−1 red light followed by 30-min darkness, then harvested immediately. Tissue was harvested by cutting the aerial portion of the seedlings away from the roots and freezing them in liquid nitrogen. Total RNA isolation was performed on each sample using the RNeasy mini kit (Qiagen), following the manufacturer's protocol. Two micrograms of RNA were treated with Deoxyribonuclease I (Invitrogen). DNAse-treated RNA was then used to synthesize first-strand cDNA using SuperScript II reverse transcriptase (Invitrogen), following the manufacturer's protocol. Two microliters of the first-strand cDNA reaction were used as template in 25-cycle PCR reactions. The following pairs of gene-specific primers were used separately to amplify each of the cDNA samples: UBQ1 and UBQ2 (Weigl and Glazebrook, 2001) were used to amplify UBQ; apyrase-specific primers AAR566 and AraF172 were used to amplify APY1; and Arapy2F and AAR566 were used to amplify APY2. PCR products were run on 1.5% (w/v) agarose gel.
Generation of Conditional DKOs
Double-heterozygous plants were transformed with either the cDNA for APY1 or APY2 under the steroid-inducible vector pTA7002 (Aoyama and Chua, 1997) or with pTA7002 alone by vacuum infiltration. Cloning of the genetic construct and the screening process for DKOs, including genomic DNA isolation and PCR conditions, were described elsewhere (Steinebrunner et al., 2003). Seven T1 lines complemented with DEX:APY1, 14 lines complemented with DEX:APY2, and seven lines with the vector alone were selected. To induce the transgene, plants were sprayed with water containing 30 μm DEX and 0.01% (v/v) Tween 20 or watered with 30 nm DEX.
Screening for Leakiness in DEX Lines
RNA was isolated from leaves of untreated DEX lines, subjected to DNAseI digestion, and reverse transcribed as published previously (Steinebrunner et al., 2003). The primer combination DEXF and ApyR (see Steinebrunner et al., 2003) produced a DEX-induced specific band of 1.6 kb. Southern-blot analysis was performed of the RT-PCR products using a 603-bp fragment (nucleotides 1,304–1,907 of accession no. AF093604) of the APY1 cDNA. This probe bound to APY1 and APY2 cDNA sequence. As positive control for successful RT, PCR of either adenine phosphoribosyl transferase (APT1_for; APT1_rev; product size: 479 bp) or APY1 (ApyF and ApyR; product size: 1.4 kb) was performed.
Semi-in Vivo Pollen Tube Growth
Pollen and styles were used from flowers from the upper one-third of primary inflorescences from same-age plants. Wild-type styles from emasculated flowers at stage 12 (Smyth et al., 1990) were cut off with a scalpel after pollen from flowers at stages 13 to 14 (Smyth et al., 1990) were dipped onto them. The styles were transferred into 100 μL of PGM (17% [w/v] Suc, 1.6 mm boric acid, 3 mm calcium citrate, pH 7.2) on a cavity slide and incubated as a sitting drop culture for 3 h according to Shivanna and Rangaswamy (1992). Pollen tube growth was terminated by incubation in 70% (v/v) ethanol overnight. After another incubation in 8 m NaOH for 8 h, pollen tubes were stained with aniline blue (as described in Shivanna and Rangaswamy, 1992) for length measurements with the analySIS software (Soft Imaging Systems). Three to 25 pollen tubes were measured per style. Only pollen tubes that could be traced emerging from the cut style end to its tip were evaluated. The length of the trace was determined with the software's polygon selection tool. Because the length of styles was not uniform, it was added to the trace lengths of its pollen tubes to obtain the total pollen tube lengths. Mean pollen tube lengths and sds were calculated per experiment, which consisted of several styles per pollen genotype under investigation.
Assays of in Vitro Pollen Growth and ATP Level in Growth Medium
PGM consisted of 400 μL of 1.6 mm boric acid, 1 mm MgSO4, 1 mm CaCl2, 1 mm Ca(NO3)2, and 5 mm HEPES buffer in 18% (w/v) Suc, 1% (w/v) agar, pH 7.0. Forty microliters of medium were applied to the bottom of each multiple well of a depression slide and after the agar set pollen from a single Arabidopsis flower was deposited, resulting in about 100 pollen grains per well. Then the slide was suspended by wooden sticks in a petri dish containing 14 mL of double-distilled water and two paper filter discs, the dish was covered, placed in a dark incubator set at 26°C for 4 h, and allowed to germinate. Only wells that achieved at least a 60% germination rate were used in the experimentation.
For experiments testing the effects of antiapyrase immune and preimmune sera on pollen tube growth, between 0.5 and 1.5 μg of Affigel-Blue purified gp18 serum or preimmune serum protein (see section below) was added in volumes of 2 μL or less to the 150 μL of PGM solution that was applied to the well on top of the semisolid medium and germinated pollen, and pictures of pollen tubes were taken at 1 and 15 min after solution application, as described above. At least 20 pollen tubes were measured for each treatment to get a representative growth rate of the tubes in that well.
For experiments testing the effects of apyrase inhibitors on pollen tube growth, the inhibitors (NGXT 191 and no. 4, both at 2.5 μg/mL) or control PGM solution (minus agar), all in 0.1% (v/v) dimethylformamide, were applied in 150 μL to the well on top of the semisolid medium within the first hour after the pollen had germinated. Pictures of pollen tubes were taken at 1 and 15 min after solution application using a PixeLINK PL-662 microscopy camera and used to calculate growth rates (rate/h = total micrometer length increase during the 15 min after the treatment was applied/15 × 60). At least 20 pollen tubes were measured in a well for each treatment to get a representative growth rate of the tubes in that well.
For experiments measuring the [ATP] of the PGM, aliquots (30 μL) of medium were removed at 1 and 15 min after the treatments were applied, immediately placed in 1.5-mL graduated plastic vials, labeled, sealed, and frozen in liquid nitrogen until they were analyzed for ATP concentration as described by Jeter et al. (2004).
The ATP concentration of PGM medium in which pollen tubes were growing was measured using the Enliten ATP assay system bioluminescence kit produced by Promega and ATP standard curve solutions. All samples were assayed using a Turner Designs 20/20 luminometer. Three individual 10-μL samples were assayed from each sample to ensure internal consistency of the sample.
Apyrase Activity Assay
PGM used in wells for pollen germination and growth was prepared as described above, with the following modifications: 200 μL of liquid PGM was applied over Arabidopsis pollen on semisolid PGM. Pollen was incubated at 26°C for 4.5 h and allowed to germinate. From those wells where pollen had a germination rate of at least 60%, 100 μL of liquid PGM was siphoned off in such a way as to exclude any pollen. These solutions had significant apyrase activity and were combined into aliquots of 500 μL, frozen in liquid nitrogen, and held at −40°C until ready for use. Prior to the activity assay, 30 μg cytochrome c were added to each thawed 500-μL aliquot, and sample volumes were reduced to 50 μL using Microcon Ultracel YM-10 centrifugal filter devices (Millipore). These concentrated samples were termed PGM apyrase and were used for all apyrase activity assays.
The activity assay was based on the method of Traverso-Cori et al. (1965) with modifications. The reaction volumes were scaled down from 2 mL to 200 μL. Each nucleotide solution consisted of 3.0 mm AMP or ATP dissolved in 60 mm HEPES buffer, 3.0 mm MgCl2, 3.0 mm CaCl2, and four ATPase/phosphatase inhibitors: 1.14 mm ascorbic acid, 0.2 mm Na2MoO4·2H2O, 0.5 mm NaN3, and 1.0 mm Na3VO4, pH adjusted to 6.6 using NaOH. Activity assays were initiated by the addition of 7 μL of PGM apyrase and the amount of phosphate released was measured at 1, 5, and 12 min after addition of apyrase. The amount of phosphate released was typically linear over this period. To stop apyrase activity and measure the phosphate concentration from ATP hydrolysis, 800 μL of a solution containing 0.625% (w/v) (NH4)6Mo7O24·4H2O, 0.625 n sulfuric acid, and 3.12% (w/v) FeSO4·7H2O was added. The samples were developed for 5 min then their absorption at 660 nm was measured (Beckman DU 530 spectrophotometer) and compared to that of phosphate standards.
Generation of the RNAi Construct and Plant Transformation
To generate the RNAi construct, the sense cDNA region containing the 220 bp near the 3′ end of APY1 was amplified by primers Apy1-XhoI and Apy1-SalI. The antisense region was amplified by primers RNAiI-EcoRI and RNAiI-SpeI. The PCR products of sense and antisense fragments were sequenced and subcloned into pSKint in the sense direction by XhoI and SalI and in the antisense direction by EcoRI and SpeI. The fragment containing the sense, an actin 11 intron, and the antisense sequences was cut by XhoI and SpeI. The released fragment was used to replace the original GFP-RNAi fragment in the pX7-GFP binary vector to produce pX7-APY1. The pX7-GFP binary vector and pSKint were provided by Dr. Nam-Hai Chua (Rockefeller University; Guo et al., 2003). The pX7-APY1 binary vector was electroporated into A. tumefaciens strain GV3101. The apy2 mutants were transformed by pX7-APY1 via the floral-dip method (Clough and Bent, 1998). Transgenic plants were selected on Murashige and Skoog plates containing 20 μg/mL hygromycin (Sigma). Homozygous and single-locus insertion lines were selected by examining the resistance for hygromycin in T2 seeds.
To induce expression of the RNAi constructs, the transformed plants were either germinated and grown on agar in medium containing 4 μm estradiol (Sigma) or germinated and grown on soil that was watered at regular intervals with 4 μm estradiol in double-distilled water. The aerial parts of plants grown on soil were also sprayed with 4 μm estradiol whenever they were watered.
Generation of Overexpressing Lines and Plant Transformation
To generate 35S:APY1-Myc lines, the cDNA region of APY1 was amplified by the primers Apy-a and Apy-b to produce PCR fragment Apy1-M (containing the first 21 bp of Myc at the 3′ end). Six copies of the Myc epitope tag were amplified by primers Myc-c and Myc-e to produce a PCR product A-Myc (containing the last 20 bp of APY1 at the 5′ end). APY1-Myc was generated by mixing Apy1-M and A-Myc together and amplifying by primers Apy-a and Myc-e. The PCR product was subcloned into the pCR2.1-TOPO vector (Invitrogen) to generate pTOPO-APY1. The APY1-Myc fragment was sequenced and cut with EcoRI. The released insert was then ligated into the EcoRI site of the pLBJ21 binary vector, which contained the 35S promoter of Cauliflower mosaic virus. This construct was introduced into the A. tumefaciens strain GV3101 that was used to transform Columbia wild type by the vacuum infiltration method (Clough and Bent, 1998). Twenty transgenic lines containing the construct were selected with 50 μg/mL kanamycin on germination plates. Plants with resistance were selected and transplanted to soil. T2 seeds from individual T1 plants were screened to generate homozygous and single-locus insertion lines.
RNA Gel-Blot Analysis
Seven-day-old Arabidopsis seedlings were collected and frozen in liquid nitrogen. Total RNA was isolated using the RNeasy plant mini kit (Qiagen). Ten micrograms of RNA were separated in a 1.2% (w/v) agrose gel with 6% (v/v) formaldehyde. RNA was transferred to a Zeta-Probe GT Membrane (Bio-Rad) and hybridization was performed according to the manufacturer's instructions.
Assay of Lateral Root Formation
Fourteen-day-old light-grown seedlings were used to detect lateral root formation in RNAi lines. As control plants, apy2 single-knockout mutants were used. Both control and RNAi lines were planted in medium containing 4 μm estradiol to induce RNAi silencing. Control and RNAi lines were grown in one 150-mm petri dish to assure identical growth conditions. The experiment was repeated three times. The n value for each RNAi line and control was around 30.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At3g04080 (APY1) and At5g18280 (APY2). Phytochrome mutants, phyA-201 (stock no. CS6219), phyB-5 (stock no. CS6219), and phyA201/phyB-5 (stock no. CS6224) were obtained from the ABRC.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Apyrase activity released from Arabidopsis pollen is inhibited by apyrase inhibitor 4, NGXT191, NGXT1913, and polyclonal antiapyrase antibodies.
Supplemental Figure S2. A, Treatment of growing pollen tubes with apyrase inhibitors rapidly decreases their growth rate. B, Treatment of growing pollen tubes with apyrase inhibitors rapidly increases the concentration of ATP in the pollen growth medium.
Supplementary Material
Acknowledgments
We thank Enamul Huq and Greg Clark for advice and critical reading of the manuscript and Mari Salmi for semiquantitative RT-PCR analyses of APY1 and APY2 transcript levels after red-light irradiation. Figure 1F was contributed by Carolin Wolf. Phytochrome mutants, phyA-201, phyB-5, and phyA201/phyB-5 and the Arabidopsis genomic BAC clone T6K12 were obtained from the ABRC. Arabidopsis BAC clone MRG7 was kindly provided by Kazusa DNA Research Institute (Japan).
This work was supported by the National Science Foundation (grant no. 0344221 to S.R.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Stanley J. Roux (sroux@uts.cc.utexas.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aloni R, Aloni E, Langhans M, Ullrich CI (2006) Role of auxin in regulating Arabidopsis flower development. Planta 223 315–328 [DOI] [PubMed] [Google Scholar]
- Aloni R, Schwalm K, Langhans M, Ullrich CI (2003) Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216 841–853 [DOI] [PubMed] [Google Scholar]
- Aoyama T, Chua N-H (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11 605–612 [DOI] [PubMed] [Google Scholar]
- Burnstock G, Knight GE (2004) Cellular distribution and functions of P2 receptor subtypes in different systems. Int Rev Cytol 240 31–51 [DOI] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inze D, Sandberg G, Casero PJ, et al (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivasa S, Ndimba BK, Simon WJ, Lindsey K, Slabas AR (2005) Extracellular ATP functions as an endogenous external metabolite regulating plant cell viability. Plant Cell 17 3019–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark GB, Lee DW, Dauwalder M, Roux SJ (2005) Immunolocalization and histochemical evidence for the association of two different Arabidopsis annexins with secretion during early seedling growth and development. Planta 220 621–631 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Day RB, McAlvin CB, Loh JT, Denny RL, Wood TC, Young ND, Stacey G (2000) Differential expression of two soybean apyrases, one of which is an early nodulin. Mol Plant Microbe Interact 13 1053–1070 [DOI] [PubMed] [Google Scholar]
- Demidchik V, Nichols C, Oliynyk M, Dark A, Glover BJ, Davies JM (2003) Is ATP a signaling agent in plants? Plant Physiol 133 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler JD, De Groodt R, Van Montagu M, Engler G (2001) In situ hybridization to mRNA of Arabidopsis tissue sections. Methods 23 325–334 [DOI] [PubMed] [Google Scholar]
- Geisler M, Murphy AS (2006) The ABC of auxin transport: the role of p-glycoproteins in plant development. FEBS Lett 580 1094–1102 [DOI] [PubMed] [Google Scholar]
- Guo FQ, Okamoto M, Crawford NM (2003) Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302 100–103 [DOI] [PubMed] [Google Scholar]
- Handa M, Guidotti G (1996) Purification and cloning of a soluble ATP-diphosphohydrolase (apyrase) from potato tubers (Solanum tuberosum). Biochem Biophys Res Commun 218 916–923 [DOI] [PubMed] [Google Scholar]
- Hirschberg CB, Robbins PW, Abeijon C (1998) Transporters of nucleotide sugars, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu Rev Biochem 67 49–69 [DOI] [PubMed] [Google Scholar]
- Hu XY, Neill SJ, Tang ZC, Cai WM (2005) Nitric oxide mediates gravitropic bending in soybean roots. Plant Physiol 137 663–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeter CR, Tang WQ, Henaff E, Butterfield T, Roux SJ (2004) Evidence of a novel cell signaling role for extracellular adenosine triphosphates and diphosphates in Arabidopsis. Plant Cell 16 2652–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HG, Fang YW, Singh KB (1999) A glucocorticoid-inducible transcription system causes severe growth defects in Arabidopsis and induces defense-related genes. Plant J 20 127–133 [DOI] [PubMed] [Google Scholar]
- Kim SY, Sivaguru M, Stacey G (2006) Extracellular ATP in plants: visualization, localization, and analysis of physiological significance in growth and signaling. Plant Physiol 142 984–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltai H, Bird DM (2000) High throughput cellular localization of specific plant mRNAs by liquid-phase in situ reverse transcription-polymerase chain reaction of tissue sections. Plant Physiol 123 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowski ER, Boucher RC, Harden TK (2003) Mechanisms of release of nucleotides and integration of their action as P2X- and P2Y-receptor activating molecules. Mol Pharmacol 64 785–795 [DOI] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR (1996) HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85 183–194 [DOI] [PubMed] [Google Scholar]
- Leyser O (2005) Auxin distribution and plant pattern formation: how many angels can dance on the point of PIN? Cell 121 819–822 [DOI] [PubMed] [Google Scholar]
- Lin RC, Wang HY (2005) Two homologous ATP-binding cassette transporter proteins, AtMDR1 and AtPGP1, regulate Arabidopsis photomorphogenesis and root development by mediating polar auxin transport. Plant Physiol 138 949–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklof J, Casero PJ, Bennett M, Sandberg G (2002) AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW (1996) The homeobox gene GLABRA 2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122 1253–1260 [DOI] [PubMed] [Google Scholar]
- Neff MM, Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Spalding EP (1999) Sequential and coordinated action of phytochromes A and B during Arabidopsis stem growth revealed by kinetic analysis. Proc Natl Acad Sci USA 96 14142–14146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux SJ, Song C, Jeter C (2006) Regulation of plant growth and development by extracellular nucleotides. In F Baluska, S Mancuso, D Volkmann, eds, Communication in Plants. Springer, Berlin, pp 221–234
- Ruzin S (1999) Plant Microtechnique and Microscopy. Oxford University Press, New York
- Schwalm K, Aloni R, Langhans M, Heller W, Stich S, Ullrich CI (2003) Flavonoid-related regulation of auxin accumulation in Agrobacterium tumefaciens-induced plant tumors. Planta 218 163–178 [DOI] [PubMed] [Google Scholar]
- Shivanna KR, Rangaswamy NS (1992) Pollen Biology. Springer-Verlag, Berlin
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CJ, Steinebrunner I, Wang XZ, Stout SC, Roux SJ (2006) Extracellular ATP induces the accumulation of superoxide via NADPH oxidases in Arabidopsis. Plant Physiol 140 1222–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinebrunner I, Jeter C, Song C, Roux SJ (2000) Molecular and biochemical comparison of two different apyrases from Arabidopsis thaliana. Plant Physiol Biochem 38 913–922 [Google Scholar]
- Steinebrunner I, Wu J, Sun Y, Corbett A, Roux SJ (2003) Disruption of apyrases inhibits pollen germination in Arabidopsis. Plant Physiol 131 1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y (2003) Distribution and expression of apyrases in pea and Arabidopsis. PhD thesis. University of Texas, Austin, TX
- Tang WQ, Brady SR, Sun Y, Muday GK, Roux SJ (2003) Extracellular ATP inhibits root gravitropism at concentrations that inhibit polar auxin transport. Plant Physiol 131 147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Sun Y, Naus K, Lloyd A, Roux S (1999) Apyrase functions in plant phosphate nutrition and mobilizes phosphate from extracellular ATP. Plant Physiol 119 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverso-Cori A, Chaimovich H, Cori O (1965) Kinetic studies and properties of potato apyrase. Arch Biochem Biophys 109 173–184 [DOI] [PubMed] [Google Scholar]
- Weigl D, Glazebrook J (2001) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Wilhelmi LK, Preuss D (1997) Blazing new trails—pollen tube guidance in flowering plants. Plant Physiol 113 307–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windsor B (2000) Establishing a role for ecto-phosphatase in drug resistance. PhD thesis. University of Texas, Austin, TX
- Windsor B, Roux SJ, Lloyd A (2003) Multiherbicide tolerance conferred by AtPgp1 and apyrase overexpression in Arabidopsis thaliana. Nat Biotechnol 21 428–433 [DOI] [PubMed] [Google Scholar]
- Windsor JB, Thomas C, Hurley L, Roux SJ, Lloyd AM (2002) Automated colorimetric screen for apyrase inhibitors. Biotechniques 33 1024–1030 [DOI] [PubMed] [Google Scholar]
- Yegutkin GG, Mikhailov A, Samburski SS, Jalkanen S (2006) The detection of micromolar pericellular ATP pool on lymphocyte surface by using lymphoid ecto-adenylate kinase as intrinsic ATP sensor. Mol Biol Cell 17 3378–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SQ, Mehdy MC (1994) Binding of a 50-kD protein to a U-rich sequence in a messenger-RNA encoding a proline-rich protein that is destabilized by fungal elicitor. Plant Cell 6 135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XT, Malhotra R, Woodruff R, Guidotti G (2001) Mammalian plasma membrane ecto-nucleoside triphosphate diphosphohydrolase 1, CD39, is not active intracellularly—the N-glycosylation state of CD39 correlates with surface activity and localization. J Biol Chem 276 41518–41525 [DOI] [PubMed] [Google Scholar]
- Zimmermann H (2001) Ectonucleotidases: some recent developments and a note on nomenclature. Drug Dev Res 52 44–56 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.