Abstract
Peanut (Arachis hypogaea) seed proteins Ara h 1, Ara h 2, and Ara h 3 are considered to be the major peanut allergens. However, little is known about their temporal and spatial expression during seed development and upon germination and seedling growth. In this study, transcript levels of the three major peanut allergen genes, ara h 1, ara h 2, and ara h 3, and their corresponding proteins were found in all cultivars. Expression patterns were heterogeneous depending on the specific peanut allergen gene and the cultivars tested. However, ara h 3 expression patterns among the cultivars were more variable than ara h 1 and ara h 2. Transcripts were tissue specific, observed in seeds, but not in leaves, flowers, or roots, and were undetectable during seed germination. In situ hybridizations and immunotissue prints revealed that both embryonic axes and cotyledons expressed the allergens. However, more ara h 1 and ara h 3 messenger RNA was detected in cotyledons relative to embryonic axes. Allergen polypeptide degradation patterns were different in embryonic axes compared with cotyledons during germination and seedling growth, with levels of Ara h 1 and Ara h 2 dramatically reduced compared to the Ara h 3 polypeptides in embryonic axes. These characterization studies of major peanut allergen genes and their corresponding seed storage proteins can provide the basic information needed for biochemical and molecular approaches to obtain a hypoallergenic peanut.
Cultivated peanut (Arachis hypogaea) is a valuable oilseed and food crop containing 45% to 53% oil and 24% to 29% protein in its seed. However, ingestion of peanut seeds is one of the most serious causes of fatal food-induced anaphylaxis (Yocum and Khan, 1994). Peanut allergy is mediated by IgE (Pumphrey et al., 1999), and the major peanut allergens are seed storage proteins. Although about 11 peanut allergens have been reported (www.allergome.org), Ara h 1, Ara h 2, and Ara h 3 are classified as the major peanut allergens because they are generally recognized by more than 50% of peanut-allergic patients (Koppelman et al., 2001). Specifically, Ara h 1 and Ara h 2 are recognized by 70% to 90% of patients with peanut allergy (Burks et al., 1995b, 1998; Clarke et al., 1998), and Ara h 3 is recognized by serum IgE from approximately 44% to 54% of different patient populations with a history of peanut sensitivity (Kleber-Janke et al., 1999; Rabjohn et al., 1999).
Ara h 1 is a 63.5-kD glycoprotein that has significant homology with vicilin seed storage proteins in other legume plants (Burks et al., 1991, 1995a). Ara h 1 has 40% similarity with soybean (Glycine max) and pea (Pisum sativum) vicilins at the protein level (Lycett et al., 1983), and 64% similarity with broad bean (Vicia faba) and pea vicilins at the DNA level (Burks et al., 1995b). The linear IgE-binding epitopes of Ara h 1 have been mapped and shown to contain 23 independent binding sites, which are evenly distributed along the linear sequence of the molecule (Burks et al., 1997). Based on a molecular model of the tertiary structure of Ara h 1, it is a homotrimer with IgE-binding epitopes located in the area of monomer-monomer contact mediated primarily through hydrophobic interactions (Shin et al., 1998). Northern-blot analysis showed that the ara h 1 transcript is 2.3 kb and is abundant in mature peanut cotyledons (Burks et al., 1995b).
Ara h 2 is a member of the conglutin family (Stanley et al., 1997). Ara h 2 is composed of two glycoproteins containing eight Cys residues that migrate closely on SDS-PAGE (Burks et al., 1992; Sen et al., 2002). An ara h 2 clone of 741 bp was identified from a peanut cDNA library, and this clone hybridizes to an approximately 0.7-kb mRNA (Stanley et al., 1997). Two complete isoform cDNAs of ara h 2, named ara h 2.01 and ara h 2.02, were isolated and characterized (Chatel et al., 2003). The difference between these isoforms is that Ara h 2.02 contains a 12-amino acid insertion. These additional 12 amino acids in Ara h 2.02 corresponded to the difference in molecular mass of the two Ara h 2 bands found on SDS-PAGE (Chatel et al., 2003). Recently, it was shown that these isoforms are homeologous genes that are orthologs, each derived from one of the diploid ancestors of peanut (Ramos et al., 2006).
Ten IgE-binding epitopes have been identified by peptide analysis for Ara h 2. Three of these epitopes are considered immunodominant epitopes, and two regions within these three epitopes contain the amino acid sequence, DPYSPS, which is considered to be necessary for IgE binding. The additional 12 amino acids of Ara h 2.02 contain a third repeat of this major linear IgE epitope motif (Stanley et al., 1997; Chatel et al., 2003). The allergenicities of Ara h 1 and Ara h 2 are resistant to heat and several proteinases due to protein structure and modifications to these proteins that occur following these treatments (Koppelman et al., 1999; Maleki et al., 2000a, 2000b, 2003). In particular, disulfide bonds contribute significantly to the overall structure and stability of Ara h 2 (Sen et al., 2002).
The nucleotide sequence of ara h 3 encodes 507 amino acid residues, and the calculated size of the deduced amino acid is approximately 57 kD. It is a legumin-like seed storage protein that has high sequence similarity to glycinin, which is the major 11S globulin seed storage protein in soybean (Rabjohn et al., 1999). Database searches for sequence similarity revealed that there are several ara h 3-related genes, including ara h 4, ara h 3/ara h 4, and gly1. These are common storage proteins in legumes that function as a source of nitrogen for the developing plant and are initially synthesized as 60-kD premature globulins. However, a 14-kD protein was originally identified as Ara h 3 (Eigenmann et al., 1996; Burks et al., 1998). It was reported that there are several posttranslational processing sites such as Asn-Gln, Asn-Gly, and Cys residues that can form disulfide bonds in Ara h 3 (Rabjohn et al., 1999). Recently, protein extracts from peanut seed run on SDS-PAGE revealed a series of Ara h 3 polypeptides ranging in size from 14 to 45 kD (Koppelman et al., 2003). Also, based on their N-terminal sequences, the observed set of polypeptides is the result of posttranslational proteolysis (Burks et al., 1998; Koppelman et al., 1999, 2003; Boldt et al., 2005).
Correctly formed and assembled mature storage proteins are stably accumulated in developing seeds and degraded during germination and seedling growth to small peptides or amino acids that are subsequently transported to the growing seedling (Goldberg et al., 1989; Shewry et al., 1995). Degradation occurs only after a period of rest and imbibition of water, when seeds germinate and seedlings start to grow (Shutov et al., 2003). Synthesis and degradation, the antagonistic processes of protein turnover, occur during different developmental stages.
Although the major peanut allergens have been studied by many research groups, information on their temporal and spatial expression during seed development and upon germination and seedling growth is limited. Therefore, in this study, we characterized and compared the expression patterns of these three major peanut allergens during seed maturation and germination. Also, spatial relationships between peanut allergen mRNAs and their corresponding proteins in seeds were examined. Understanding the expression of peanut allergens will allow for a better understanding of mechanisms to control their synthesis and degradation for future production of a hypoallergenic peanut.
RESULTS
Expression of Peanut Allergens during Seed Development in Selected Cultivars
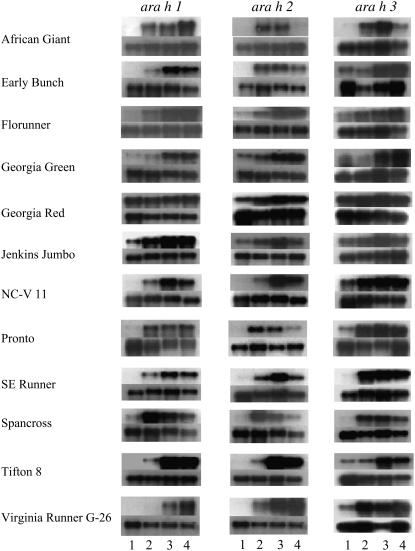
All 12 peanut cultivars analyzed expressed the three major allergen genes: ara h 1, ara h 2, and ara h 3. Transcripts of the three allergens were detected by northern-blot analysis as single bands of 2.3 kb (ara h 1), 740 bp (ara h 2), and 1.5 kb (ara h 3) that are differentially expressed during seed development within and between cultivars (Fig. 1). For ‘Florunner,’ ‘Georgia Green,’ ‘Jenkins Jumbo,’ and ‘Tifton 8,’ transcripts of all three genes were either not detected or were at low levels in stage 1, the earliest stage of seed development tested, and their levels peaked at the more mature stages, 3 or 4. In ‘African Giant,’ ara h 2 and ara h 3 transcript levels were dramatically lower at stage 4 compared to ara h 1 transcript levels, which were highest at stage 4. Interestingly, all three genes were highly expressed at all four seed stages of ‘Georgia Red.’ For ‘NC-V 11’ and ‘SE Runner,’ transcripts of ara h 1 and ara h 2 reached maximum levels in stage 3 seed, whereas ara h 3 transcript levels peaked earlier (stage 2). In ‘Spancross,’ transcript levels of the three genes were highest at stage 2 and decreased as the seed matured. The transcripts of ara h 1 and ara h 3 accumulated after stage 2 in ‘Pronto,’ whereas the expression of ara h 2 resembled that of ‘Spancross.’ ‘African Giant’ and ‘Virginia Runner G-26’ displayed higher transcript levels of ara h 1 and ara h 2, while ‘Pronto’ and ‘Spancross’ had substantially higher transcript levels of ara h 3 compared with the other cultivars examined, respectively. Additionally, transcripts of ara h 3 were much higher than those of ara h 1 and ara h 2 within ‘Pronto’ and ‘Spancross’ compared to the other two allergen gene transcripts, whereas ara h 1 and ara h 2 transcripts were higher than that of ara h 3 in ‘Virginia Runner G-26.’ These results were consistent for each cultivar regardless of the planting location.
Figure 1.
Northern-blot analysis for the three major peanut allergen genes in 12 genotypes during seed maturation. The top segment for each genotype is ara h 1, ara h 2, or ara h 3 transcripts, as indicated. The bottom segment for each genotype is 25S rRNA transcript used as an internal control. 1, 2, 3, and 4 represent seed developmental stages from immature to mature.
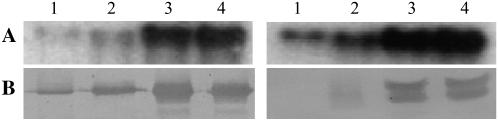
Western-blot analyses were carried out to examine expression patterns of Ara h 1 and Ara h 2 allergens at the protein level during seed maturation, and the results were compared with their corresponding transcript patterns (Fig. 2). For this study, two specific polyclonal antibodies against Ara h 1 and Ara h 2 were prepared and hybridized with seed proteins of ‘Georgia Green,’ a widely grown cultivar in the southern United States. The results show that these proteins accumulate to higher levels in mature seeds than in immature seeds. The accumulation of protein was similar to the transcriptional gene expression patterns in ‘Georgia Green.’ Ara h 1 could be detected easily, although at low levels at stages 1 and 2, whereas Ara h 2 was not observed at stage 1 and at very low levels at stage 2. Therefore, it appears that Ara h 1 accumulated sooner in developing peanut seed than Ara h 2.
Figure 2.
Expression of ara h 1 and ara h 2 during seed maturation. A, Transcript levels of ara h 1 or ara h 2 were analyzed by northern blots. B, Protein levels of Ara h 1 and Ara h 2 were analyzed by western blots. 1, 2, 3, and 4 represent the seed developmental stages of ‘Georgia Green.’
Seed-Specific Expression of Peanut Allergens
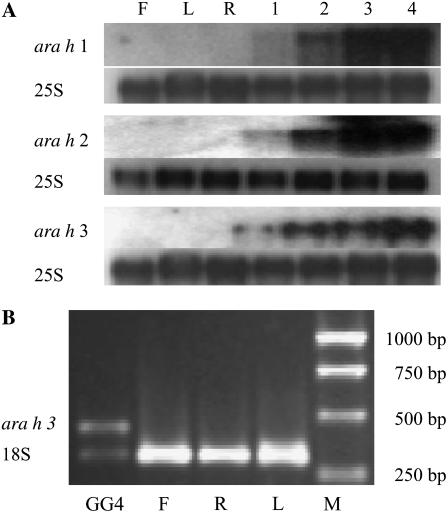
To determine tissue specificity of gene expression, total RNA was isolated from flowers, leaves, roots, and developing seeds of ‘Georgia Green.’ Transcripts accumulated in developing seed, as earlier described, but transcripts were not detected for any of the allergens in flowers, leaves, and roots (Fig. 3A). However, because there are several ara h 3-related genes that cannot be distinguished with the probe used, it was necessary to confirm tissue specificity using reverse transcription (RT)-PCR with ara h 3 gene-specific primers. Based on RT-PCR specific for ara h 3, there were no amplified PCR products from flower, root, and leaf, whereas an ara h 3 band was amplified from the synthesized seed cDNA (Fig. 3B).
Figure 3.
Seed-specific expression of the three major peanut allergen genes. A, Northern blots for tissues including flowers (F), leaves (L), roots (R), and four seed developmental stages of ‘Georgia Green’ (1–4) for ara h 1, ara h 2, and ara h 3 genes. ara h 1, ara h 2, and ara h 3 indicates transcript signals, respectively. As a control, the transcript levels of 25S rRNA (25S) were used. B, Transcript level of ara h 3 was analyzed by RT-PCR from mRNA extracted from flowers (F), leaves (L), roots (R), and stage 4 seed of ‘Georgia Green’ (GG4). The transcript level of 18S rRNA (18S) was used as an internal control with an amplified band of 315 bp. A 1-kb DNA marker (M; Promega) was used as a size standard.
Peanut Allergens Are Expressed and Accumulate in Both Embryonic Axes and Cotyledons
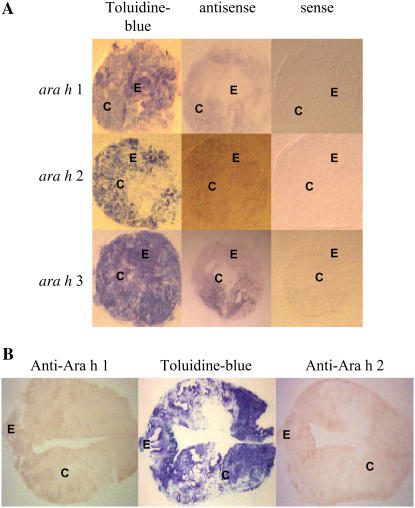
Localization of allergen transcripts was examined to determine whether there were differences observed between embryonic axes and cotyledons (Fig. 4A). In cross sections of stage 3 seeds, transcripts were detected in both embryonic axes and cotyledons for all three allergen genes; however, more transcript could be detected in cotyledons than in embryonic axes. Transcripts were not detected in stem or root sections, confirming that the expression of these genes is seed specific (data not shown).
Figure 4.
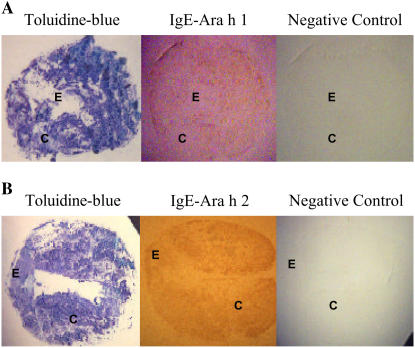
Localization of ara h 1, ara h 2, and ara h 3 transcripts and Ara h 1 and Ara h 2 in seed by tissue print hybridization. A, Localization of transcripts of ara h 1, ara h 2, and ara h 3 in seed cross sections (stage 3). To verify the specificity of the signal from the antisense RNA probe, a sense RNA probe was used as a negative control. B, Longitudinal section of seed hybridized with anti-Ara h 1 and anti-Ara h 2 antibodies. Embryonic axis (E) and cotyledon (C) are indicated. As a control, seed section blots were stained with toluidine blue. [See online article for color version of this figure.]
When tissue print blots were probed with anti-Ara h 1 or anti-Ara h 2, signals were detected in the entire seed, including embryonic axes and cotyledons (Fig. 4B). Immunotissue prints produced from hybridization with sera from peanut-allergic patients containing peanut-specific IgE for Ara h 1 (Fig. 5A) or Ara h 2 (Fig. 5B) confirmed that allergenic proteins accumulated in both embryonic axes and cotyledons.
Figure 5.
Tissue print immunoblotting for Ara h 1 and Ara h 2 in seed using peanut-specific IgE. Seed cross sections hybridized with human serum containing peanut-specific IgE for Ara h 1 (A) and Ara h 2 (B). Negative control was performed with human serum containing nonspecific IgE to peanut allergens. Embryonic axis (E) and cotyledon (C) are indicated. As a control, seed section blots were stained with toluidine blue. [See online article for color version of this figure.]
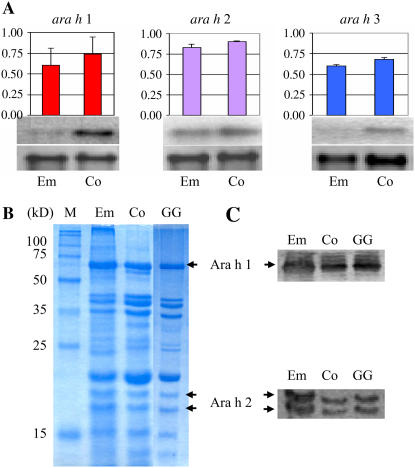
To further examine allergen expression levels within the seed, northern and western blots were performed (Fig. 6). Transcript levels of ara h 1 and ara h 3 were higher in the cotyledons, but mRNA of ara h 2 accumulated to similar levels in both the embryonic axes and cotyledons (Fig. 6A). However, protein levels of Ara h 1 and Ara h 2 were higher on a dry weight basis in the embryonic axes compared to the cotyledons (Fig. 6, B and C).
Figure 6.
Expression of the three major peanut allergen genes in embryonic axes and cotyledons. A, Northern blots were performed for ara h 1, ara h 2, and ara h 3 with mRNA extracted from embryonic axes (Em) and cotyledons (Co). The top segment is ara h 1, ara h 2, or ara h 3 transcripts as indicated. The lower segment is 25S rRNA transcript used as an internal control. Signal values obtained from each gene were normalized using the 25S rRNA signal value and analyzed. Units on the y axis: allergen transcripts/25S r RNA transcript. Each bar represents the sd. B and C, SDS-PAGE (B) and western blots (C) hybridized with anti-Ara h 1 and anti-Ara h 2 antibodies to proteins from embryonic axes (Em), cotyledons (Co), and stage 3 whole seed of ‘Georgia Green’ (GG). Protein size marker (M; Novagen) is indicated. [See online article for color version of this figure.]
Allergen Expression and Content during Seed Germination and Seedling Growth
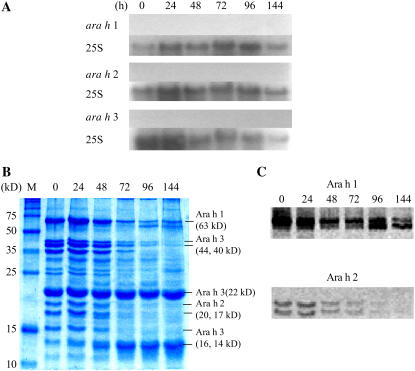
Expression of the allergen genes was examined during seed germination and seedling growth to determine how their corresponding seed storage proteins were degraded to supply nutrients for early seedling development. Twenty seeds were harvested at each time point, and the length of the seedling was measured after radicle emergence. More than 75% of the imbibed seeds showed radicle emergence after 24 h. Therefore, germination is complete following 24 h of imbibition. After 24 h, growth continues producing a seedling that is more than 7 cm in length, with primary leaves at 96 h after imbibition. Allergen transcripts were not detected at any stage of germination and seedling growth (Fig. 7A). However, SDS-PAGE indicated that there were large changes in protein composition in the samples during germination (Fig. 7B). Ara h 1 and Ara h 2 levels are dramatically reduced during germination and seedling growth. In the case of Ara h 1, protein degradation was easily observed beginning at 48 h, with degradation products clearly detectable at the 96- and 144-h time points. Also, Ara h 2 levels began to decline at 48 h but were undetectable at 144 h. SDS-PAGE showed that the 44-, 40-, and 16-kD Ara h 3 polypeptides degraded during the time course beginning at 48 h. However, interesting results were observed for two Ara h 3 polypeptides. Levels of the Ara h 3 22-kD basic chain were unchanged over the time course, remaining high even 144 h after imbibition, and the Ara h 3 14-kD polypeptide increased along with seedling growth (Fig. 7B). Western-blot analysis provided more detailed results for Ara h 1 and Ara h 2 (Fig. 7C). Ara h 1 degradation products were dominant at 96 h, and each of the two Ara h 2 polypeptides degraded at similar rates over the time points.
Figure 7.
Expression of the three major allergen genes during peanut seed germination and seedling growth. A, Northern blots were performed for ara h 1, ara h 2, and ara h 3 during peanut seed germination and seedling growth. ara h 1, ara h 2, and ara h 3 indicate transcript signals, respectively. As a control, the transcript levels of 25S rRNA (25S) were used. B and C, SDS-PAGE (B) and western blots (C) for Ara h 1 and Ara h 2. Ara h 1, Ara h 2, and Ara h 3 polypeptides are indicated as the major allergens along with their corresponding sizes. 24, 48, 72, 96, and 144, Harvesting times after imbibition (hours). For the 0 time point, seeds were harvested after 20 min of imbibition. Protein size marker (M; Novagen) is indicated. [See online article for color version of this figure.]
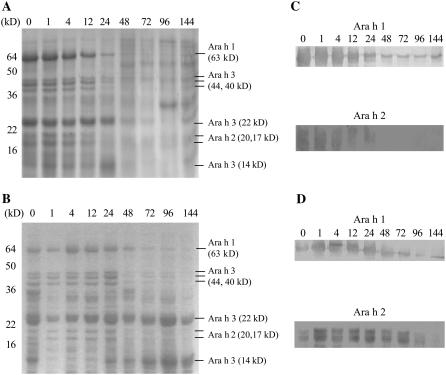
To examine the germination process in greater detail, germinated seeds were collected at 1, 4, 12, 24, 48, 72, 96, and 144 h after imbibition. The fresh weight of the cotyledons and embryonic axes were unchanged before radicle protrusion at the 24-h time point. However, the fresh weight of embryonic axes increased dramatically, while that of the cotyledons increased slowly after 24 h. Because seedling growth begins after 24 h, fresh weight changes in embryonic axes were more profound. The percent of soluble protein in the fresh weight of embryonic axes decreased sharply up until 96 h, while it decreased slowly in cotyledons over time. These results reveal that the degradation of soluble proteins can occur earlier in embryonic axes than in cotyledons. SDS-PAGE showed similar results (Fig. 8). Ara h 1, Ara h 2, and Ara h 3 could be detected only up to 24 h in embryonic axes (Fig. 8A). However, they were detected over the entire time course in cotyledons, even though their levels decreased slowly after 24 h (Fig. 8B). Western-blot results were similar to the results from SDS-PAGE, with the degradation of Ara h 1 and Ara h 2 being more pronounced in embryonic axes than in cotyledons (Fig. 8, C and D). Ara h 2 was nearly completely degraded in embryonic axes after 48 h (Fig. 8C).
Figure 8.
SDS-PAGE and western blots of embryonic axes and cotyledons during germination and seedling growth. Peanut seed proteins were isolated from embryonic axes (A), and cotyledons (B) and their polypeptide patterns are shown on SDS-PAGE. Western blots were performed with proteins from embryonic axes (C) and cotyledons (D) to detect levels of Ara h 1 and Ara h 2 with anti-Ara h 1 and anti-Ara h 2 polyclonal antibodies. 1, 4, 12, 24, 48, 72, 96, and 144, Harvesting times after imbibition (hours). For the 0 time point, seeds were harvested after 20 min of imbibition. Protein size marker (M; Novagen) is indicated.
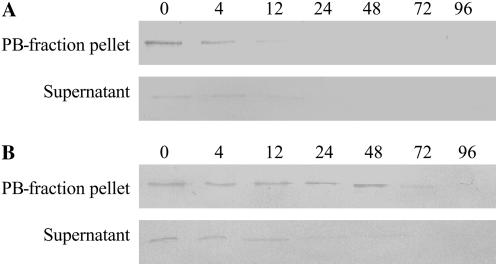
It has been shown that seed storage proteins accumulate in protein bodies (PBs) after synthesis and that they can be broken down in the PBs subsequently. Therefore, Ara h 1 degradation in PBs from embryonic axes and cotyledons was analyzed at different times during germination and seedling growth (Fig. 9). It was observed that PBs containing Ara h 1 from embryonic axes rapidly disappeared (Fig. 9A), but cotyledon PBs containing Ara h 1 remained stable up until 48 h (Fig. 9B).
Figure 9.
Ara h 1 levels in PBs during germination and seedling growth. Western blots of Ara h 1 were performed on PBs from embryonic axes (A) and cotyledons (B). Western blots were performed with proteins from the PB-fraction pellet and supernatant after PB isolation. 4, 12, 24, 48, 72, and 96, Harvesting times after imbibition (hours). For the 0 time point, seeds were harvested after 20 min of imbibition.
DISCUSSION
All peanut cultivars examined expressed the three major allergen genes, ara h 1, ara h 2, and ara h 3, in seed. In most of these cultivars, the expression of these allergen genes was similar to those of other seed storage protein genes in that their transcripts were abundant in the embryo, and their accumulation is spatially and temporally controlled during seed development (Kroj et al., 2003). However, their expression patterns were heterogenous depending on the specific peanut allergen gene and on the properties of the cultivars tested. In particular, ara h 3 expression patterns among the cultivars were more variable than ara h 1 and ara h 2 (Fig. 1). These variations may be due to the existence of homologous, multiple ara h 3 genes that are differentially regulated.
Protein accumulation of Ara h 1 and Ara h 2 during seed development was similar to transcript accumulation, indicating that these genes are most likely under transcriptional control (Fig. 2). Additionally, the transcripts were found only in seeds (Fig. 3). Based on their function as major seed storage proteins, these results are not surprising. However, peanut allergens have been reported to possess functions other than storage. For example, it was reported that ara h 2 and ara h 3 have functional and sequence homologies with trypsin inhibitor (Maleki et al., 2003; Dodo et al., 2004).
The tissue print hybridization studies clearly showed that transcripts for all three genes are found in both embryonic axes and in the cotyledons (Fig. 4), and northern-blot analysis revealed that higher transcript levels were detected in the cotyledons compared to the embryonic axes for ara h 1 and ara h 3 (Fig. 6A). Previous studies have reported differential patterns of transcriptional activity of seed protein genes in legume seeds (Hauxwell et al., 1990). In pea, the onset of vicilin and legumin gene expression was synchronous throughout the cotyledons with mRNAs found in regions of reduced mitotic activity (Harris et al., 1989; Hauxwell et al., 1990). In soybean, a wave of transcriptional activity of storage protein genes was detected from the outer surface to the inner surface of the cotyledons (Perez-Grau and Goldberg, 1989).
Ara h 1 and Ara h 2 that were IgE reactive were observed in both the embryonic axis and cotyledons (Fig. 5). However, protein levels for both allergens were higher in the embryonic axes (Fig. 6, B and C). These results agree with previous research that indicates that during legume seed maturation, protein reserves are deposited in the embryonic axis (Müntz, 1998).
Although Ara h 1, Ara h 2, and Ara h 3 were present in germinating seeds and seedlings, the transcripts encoding these proteins could not be detected (Fig. 7A). Therefore, it may be concluded that transcription of ara h 1, ara h 2, and ara h 3 does not occur during germination or that their mRNA abundance levels are not high enough to detect. Hydrolysis of seed storage proteins provides amino acids for protein synthesis in the growing seedling. Germination is regarded as finished when the radicle breaks through the seed coat, approximately 24 h after imbibition in legumes (Bewley and Black, 1994). Based on these facts, our study also showed that the allergen proteins are degraded after 24 h (Fig. 7, B and C). Levels of Ara h 1 and Ara h 2 gradually declined and eventually were undetectable during the course of seedling development.
Most Ara h 3 isoforms, such as the 44-, 40-, and 36-kD polypeptides, followed the same degradation pattern as Ara h 1 and Ara h 2; however, the 22-kD band, corresponding to the basic chain of Ara h 3, was present throughout seedling development. Furthermore, levels of the 14-kD polypeptide of Ara h 3 increased as the other seed storage protein levels decreased (Fig. 7B). Ara h 3 was originally identified as a 14-kD protein (Eigenmann et al., 1996; Burks et al., 1998), and this polypeptide is the result of posttranslational processing products of Ara h 3 isoforms (Rabjohn et al., 1999). Therefore, the increase seen for the 14-kD polypeptide band is most likely the result of an accumulation of the processed Ara h 3 isoforms. Figure 7B shows that both the 22- and 14-kD Ara h 3 bands appear predominantly in cotyledons during seedling growth, especially after 72 h.
In legumes, Cys proteinases are thought to be the major proteinases responsible for storage globulin breakdown (Wilson et al., 1986; Shutov and Vaintraub, 1987; Müntz, 1998), and multiple mechanisms modulate proteinase activity during seed germination. PBs containing storage proteins have been found in the embryonic axes of taxonomically distant plants, and protein mobilization in the axes precedes storage protein breakdown in cotyledons (Schlereth et al., 2000; Tiedemann et al., 2000). Consequently, the decline in most seed storage proteins in cotyledons and embryonic axes is not synchronous. In this work, the level of allergen proteins remained relatively high in cotyledons for 48 h but was already reduced in embryonic axes at that time (Fig. 8). This result may be a consequence of a slower turnover of these proteins in cotyledons than in embryonic axes. Therefore, degradation of soluble proteins can occur much earlier in embryonic axes than in cotyledons. Schlereth et al. (2000) observed that globulin proteins, including vicilin and legumin, are restricted to PBs in dry vetch seed, and PBs from the embryonic axes are more fragile than cotyledon PBs that are stable for at least 72 h after imbibition. Similarly, Ara h 1 degradation in PBs of peanut embryonic axes occurs prior to that in the peanut cotyledons (Fig. 9). Therefore, the results indicate that the degradation of globulin proteins, including Ara h 1, occurs inside PBs in peanut.
CONCLUSION
In this study, we characterized the timing and localization of expression of the three major peanut allergen genes, ara h 1, ara h 2, and ara h 3, and their corresponding proteins during seed development and upon germination and seedling growth. Seed developmental expression patterns of the allergens at the transcript level were cultivar dependent. Therefore, generalizations regarding the pattern of allergen gene expression at the mRNA level cannot be made and must be examined for each peanut genotype of interest. To reduce the allergenicity of peanut, these major allergens must be altered, minimized, or eliminated in the seed. Because their expression was limited to the seed, such alterations should not adversely affect plant growth or function. Transcription factors and amino acid transporters are being investigated in many systems to manipulate the regulation of seed protein composition (Tegeder et al., 2000; Kwong et al., 2003). Therefore, the characterization of expression patterns of the major peanut allergen genes in cultivars provides fundamental information that can be utilized in the future to produce a hypoallergenic peanut.
MATERIALS AND METHODS
Plant Material
Twelve peanut (Arachis hypogaea) cultivars (‘African Giant,’ ‘Early Bunch,’ ‘Georgia Green,’ ‘Georgia Red,’ ‘Jenkins Jumbo,’ ‘NC-V 11,’ ‘Pronto,’ ‘SE Runner,’ ‘Spancross,’ ‘Tifton 8,’ ‘Virginia Runner G-26,’ and ‘Florunner’) were collected at four stages of seed development from two locations (Tifton, GA and Citra, FL). Other peanut plant tissue samples (‘Georgia Green’), including leaf, flower, stem, and root, were grown and harvested at the Plant Science Research Unit (Citra, FL). Seed stage determination and harvesting were conducted according to Pattee et al. (1974), where stage 1 is the most immature seed stage followed by stages 2, 3, and 4, which is fully mature. Seed coats were left intact for the immature stages 1 and 2, but were removed prior to RNA extraction for the more mature stage 3 and the mature stage 4 seed.
Germination
Dry seeds of ‘Georgia Green’ were surface sterilized in 10% commercial bleach for 30 min, subsequently rinsed three times with distilled water, and imbibed in sterilized distilled water for 2 h. The imbibed seeds were spread onto two layers of moistened Whatman No. 1 filter paper on a mesh tray and germinated under aseptic conditions at 25°C in the dark. Samples were taken at 24, 48, 72, 96, and 144 h following imbibition. Dry seeds imbibed for 20 min were used as a control and considered to be the 0-h time point. Fresh weight and soluble protein content were measured for seeds from each time point.
Northern-Blot Analysis
Total RNA was isolated using the modified method of De Vries et al. (1988). Gel electrophoresis and northern-blot membrane transfer of total RNA were performed according to Sambrook et al. (1989) with slight modification. Total RNA (5 μg) was electrophoresed on a 1.0% (w/v) agarose/formaldehyde denaturing gel for 3 h 30 min at 90 V in 1× MOPS running buffer (0.2 m MOPS, 20 mm sodium acetate, 10 mm EDTA). After electrophoresis, the gel was stained in a 0.5 μg/mL ethidium bromide (Sigma) solution for 5 min at RT and destained in diethyl pyrocarbonate-treated distilled water. Stained gel images were taken with the Gel Doc 1000/2000 gel documentation system using Quantity One software (Bio-Rad). The gel and Hybond-N membrane (Amersham-Pharmacia Biotech) were soaked in 6× SSC solution for 15 min at RT followed by capillary transfer overnight. The membrane was removed from the transfer plate and UV cross linked at 120 J/cm2 for 5 min (SpectroLinker XL-10000; Spectronics).
32P-labeled probes were synthesized using the Prime-a-Gene Labeling system (Promega), as suggested by the manufacturer. Probes for ara h 1 and ara h 2 were made from their corresponding 1.7-kb and 0.5-kb cDNAs, respectively (provided by Dr. G. Bannon, University of Arkansas). A partial ara h 3 cDNA fragment (576 bp) was amplified from an aliquot of a peanut cDNA library constructed from developing seeds (provided by Dr. A. Abbott, Clemson University) by PCR with gene-specific primers (P1, 5′-GTGCAAAACCTAAGAGGCGAG-3′; P2, 5′-CCTTGAGTCTGTGTTGAATGC-3′).
Northern blots were prehybridized for 2 h at 65°C in hybridization buffer (0.5 m sodium phosphate, pH 7.2). Hybridization was in the same buffer for 16 to 18 h at 65°C. The membranes were then washed two times for 15 min at 65°C in 20 mm sodium phosphate, pH 7.2, with 5% SDS and 1% SDS, separately. Radioactive bands from the gel blots were directly quantified using an IMAGEQUANT PhosphorImager (version 3.0, Molecular Dynamics World Headquarters). For internal control experiments, blots were stripped for 2 h at 65°C in 50% formamide. After detecting no signal, the stripped blots were rehybridized with α-32P dCTP-labeled 25S radish ribosomal RNA (rRNA) gene fragments to confirm loading amounts. All signals were normalized to the 25S rRNA signals obtained from each blot. Northern-blot analysis was repeated three times for each experiment.
RT-PCR
cDNA was synthesized using total RNA from mature seed (stage 4), flowers, leaves, and roots of ‘Georgia Green.’ Total RNA (1 μg) was treated with 1 unit of RQ1 DNase (Promega) for 15 min at room temperature prior to RT-PCR to remove residual DNA contamination, and the DNase was inactivated using RQ1 DNase Stop Solution provided by the manufacturer following the protocol provided. Aliquots of total RNA were reverse transcribed into cDNA with 50 mm of a random hexamer (Promega). A gene-specific primer set (P1, 5′-TGCCCAGTTCCAGCGCCTC-3′; and P2, 5′-TGTCGTGGTCGTTGTAGA-3′) was designed to produce an ara h 3 product (400 bp). For each RT-PCR reaction, a pair of plant 18S rRNA internal standard primers (Ambion) was included as a loading control with a pair of gene-specific primers for ara h 3 in the same tube. The pair of 18S rRNA-specific primers and a pair of competitive primers were mixed at a ratio of 2:8, respectively, to generate unsaturated RT-PCR signals over the concentration range of total RNA used in this study. A range of 2 to 256 ng of total RNA was tested, and 64 ng of total RNA was found to generate unsaturated RT-PCR product accumulation through 28 cycles of PCR. For the analysis, RT-PCR was conducted twice with two independently isolated total RNA samples.
A total of 20 μL from each PCR reaction was fractionated on a 1.5% agarose gel in 1× Tris-borate/EDTA buffer and stained with ethidium bromide (0.5 μg/mL; Sigma). After destaining, the gels were digitally photographed and quantified with the Gel Doc 1000/2000 gel documentation system using the Quantity One software (Bio-Rad).
Peanut Protein Extraction and SDS-PAGE
Peanut proteins were extracted from seeds by a modification of the method by Koppelman et al. (2001). Peanut protein extracts were made by mixing 100 mg of seed ground in liquid nitrogen with 1 mL of 20 mm Tris-HCl, pH 8.2. After 2 h of stirring at RT, the aqueous fraction was collected by centrifugation (3,000g) for 5 min at RT. The aqueous phase was subsequently centrifuged (10,000g) for 15 min at RT to remove residual traces of oil and insoluble particles; extracts were stored at −20°C until use. Soluble protein concentration was determined using the Dc Protein Assay kit according to the manufacturer's instructions (Bio-Rad).
SDS-PAGE was performed essentially according to Laemmli (1970) with a Mini Protean II system (Bio-Rad). Peanut protein extract (10 μg) was mixed with an equal volume of 2× SDS-PAGE sample buffer (0.09 m Tris-HCl, pH 6.8; 20% glycerol; 2% SDS; 0.02% bromphenol blue; 0.1 m dithiothreitol). The mixture was boiled 10 min for denaturation and then spun for 5 to 10 s. Electrophoresis was performed on 12% SDS-PAGE gels in running buffer (Gly, 14.4 g/L; Tris-base, 3.03 g/L; 20% SDS, 5 mL/L) for 1 h 10 min at 150 V. Gels not used for immunoblotting were stained with 0.1% Coomassie Brilliant Blue R-250 and then destained. Blue Plus2 Pre-Stained Standard (Invitrogen) with molecular mass of 4, 6, 16, 22, 36, 50, 64, 98, 148, and 250 kD and Perfect Protein Markers (Novagen) with molecular mass of 10, 15, 25, 35, 50, 75, 100, 150, and 225 kD were used as references.
Preparation of Polyclonal Antibodies against Ara h 1 and Ara h 2
cDNAs corresponding to each allergen were amplified with gene-specific primer sets by PCR and cloned into pGEM-T (Promega). The cloned inserts were confirmed by sequencing and were subsequently isolated by double digestion using NheI and SalI and were ligated into pET-21b(+) vector (Novagen).
The pET-21b(+) plasmids containing ara h 1 and ara h 2 cDNA inserts were transformed into Escherichia coli BL21-Codon PLUS (DE3)-RIL cells (Stratagene). After the induction with 0.4 mm isopropyl-B-d-thiogalactopyranoside, the recombinant proteins were purified using Ni-NTA Agarose (Qiagen) as suggested by the manufacturer. The purified proteins were electrophoresed on SDS-PAGE, and concentrations were analyzed by the Dc Protein Assay kit (Bio-Rad). The purified proteins were sent to Lampire Biological Laboratories (Pipersville) and injected into rabbits (two rabbits per each antigen) for production of polyclonal antibodies. Their protocols were used for bleeds and screening to obtain reliable antibodies, and the sera containing specific polyclonal antibodies against Ara h 1 and Ara h 2 were used for our studies without purification.
Western-Blot Analysis
After SDS-PAGE, the separated proteins were electrophoretically transferred to polyvinylidene-difluoride membranes (Bio-Rad), as described by Towbin et al. (1979). Blotting was performed in transfer buffer (10 mm Tris-HCl, 100 mm Gly, 10% methanol) using a Mini-Trans Blot system (Bio-Rad). A modified method of Xiang et al. (2002) was used for immunoblotting. Membranes were blocked overnight with Tris-buffered saline plus Tween 20 (TBST; 25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.05% Tween 20) containing nonfat 5% dry milk and subsequently incubated overnight at 4°C with the specific polyclonal antibodies for Ara h 1 and Ara h 2 diluted 1:100 and 1:10 in TBST, respectively. After three washes of 10 min each with TBST, membranes were incubated with the secondary antibody (goat anti-rabbit IgG-whole molecule peroxidase conjugate, Sigma) that was diluted 1:10,000 in TBST. Membranes were washed as above with TBST, and antigen-antibody complexes were detected by chemiluminescence using the ECL plus reagent (Amersham-Pharmacia Biotech) and then imaged using Biomax ML film (Eastman Kodak).
To study the immunoreactivity of the proteins with human sera, transferred membranes were blocked in TBST and subsequently incubated overnight at RT with patient serum diluted 1:10 in TBST. After washes with TBST, membranes were incubated with horseradish peroxidase conjugated to goat anti-human IgE (Bethyl Labs) diluted 1:1,000 in TBST. The bound IgE membranes were detected as above.
Tissue Print RNA Hybridization
After harvest from the field, seeds, stems, and roots of ‘Georgia Green’ were cut into 1-cm cross sections and 2-cm longitudinal sections with a new razor blade, and the cut surfaces were pressed onto a positively charged nylon membrane (Roche Molecular Chemicals) for approximately 10 to 15 s. Duplicate membranes were cross linked using UV light at 120 J/cm2 for 5 min (SpectroLinker XL-10000; Spectronics) and stored at 4°C until use.
Prehybridization and hybridization was performed with digoxigenin (DIG) DNA Labeling and Detection kit (Roche Molecular Chemicals). The linearized DNAs were used as templates for DIG-labeled sense and antisense RNA probes by in vitro transcription using T7 or SP6 RNA polymerase (Promega). After hybridization, washing and detection of DIG-labeled probes were performed by manufacturer's instructions.
Tissue Print Immunoblots
Tissue print immunoblotting was performed as described by Cassab and Varner (1987). Nitrocellulose membranes (Bio-Rad) were soaked in 0.2 m CaCl2 for 30 min, and dried on Whatman 3MM paper. Once dried, three layers of Whatman 3MM paper were put on a plastic plate, and nitrocellulose membrane was laid on top of them. The cut seeds were pressed onto the membrane for 15 to 30 s using a gloved fingertip.
The primary and secondary antibodies for Ara h 1 and Ara h 2 were prepared according to the same protocols used for western blotting. Two different peanut-allergic sera (nos. 18500-DB and 18140-KT) were used to detect the allergenic proteins in peanut seeds. Both patient sera (diluted 1:10 in TBST) were used as the primary antibodies; horseradish peroxidase conjugated to goat anti-human IgE (Bethyl Labs) was diluted 1:1,000 in TBST and used as the secondary antibody. After incubation with antibodies, antigen-antibody complexes were detected by ImmunoPure Metal enhanced DAB substrate kit (Pierce), a colorimetric detection system in the presence of horseradish peroxidase. After detection, photographs of tissue prints were taken under the dissecting microscope.
Protein Body Isolation
Protein bodies were isolated according to the method of Schlereth et al. (2000) with slight modifications. Peanut seeds were homogenized in a buffer containing 100 mm MES, pH 5.5, 1 mm EDTA, and 600 mm mannitol. After filtration using Miracloth (Calbiochem), the extract was centrifuged (100g) for 4 min at RT. The supernatant was removed and placed onto a solution of 5% Ficoll (Sigma) in the same buffer and centrifuged (100g) for 20 min at RT. The resulting pellet was washed twice and resuspended in 100 mm Tris-HCl, pH 8.0, containing 150 mm NaCl. Protein bodies and supernatants containing protein from the cytosol and other cellular compartments were used for immunoblot analysis.
This work was supported by the U.S. Department of Agriculture (Cooperative State Research, Education, and Extension Service-administered special grant no. 00–34420–9178), by the Georgia Peanut Commodity Commission, by the Florida Peanut Producers Association, and by the Institute of Food and Agricultural Sciences at the University of Florida.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Maria Gallo (mgm@ufl.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Bewley JD, Black M (1994) Seeds: Physiology of Development and Germination, Ed 2. Plenum Press, New York
- Boldt A, Fortunato D, Conti A, Petersen A, Ballmer-Weber B, Lepp U, Reese G, Becker WM (2005) Analysis of the composition of an immunoglobulin E reactive high molecular weight protein complex of peanut extract containing Ara h 1 and Ara h 3/4. Proteomics 5 675–686 [DOI] [PubMed] [Google Scholar]
- Burks AW, Cockrell G, Connaughton C, Karpas A, Helm RM (1995. a) Epitope specificity of the major peanut allergen, Ara h II. J Allergy Clin Immunol 95 607–611 [DOI] [PubMed] [Google Scholar]
- Burks AW, Cockrell G, Stanley JS, Helm RM, Bannon GA (1995. b) Recombinant peanut allergen Ara h I expression and IgE binding in patients with peanut hypersensitivity. J Clin Invest 96 1715–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burks AW, Sampson HA, Bannon GA (1998) Review Article Series II. Peanut allergens. Allergy 53 725–730 [DOI] [PubMed] [Google Scholar]
- Burks AW, Shin D, Cockrell G, Stanley JS, Helm RM, Bannon GA (1997) Mapping and mutational analysis of the IgE binding epitopes on Ara h I, a legume storage protein and a major peanut allergen. Eur J Biochem 245 334–339 [DOI] [PubMed] [Google Scholar]
- Burks AW, Williams LW, Connaughton C, Cockrell G, O'Brien TJ, Helm RM (1992) Identification and characterization of a second major peanut allergen, Ara h II, with use of the sera of patients with atopic dermatitis and positive peanut challenge. J Allergy Clin Immunol 90 962–969 [DOI] [PubMed] [Google Scholar]
- Burks AW, Williams LW, Helm RM, Connaughton C, Cockrell G, O'Brien TJ (1991) Identification of a major peanut allergen, Ara h I, in patients with atopic dermatitis and positive peanut challenges. J Allergy Clin Immunol 88 172–179 [DOI] [PubMed] [Google Scholar]
- Cassab GI, Varner JE (1987) Immunocytolocalization of extensin in developing soybean seed coats by immunogold-silver staining and by tissue printing on nitrocellulose paper. J Cell Biol 105 2581–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel JM, Bernard H, Orson FM (2003) Isolation and characterization of two complete Ara h 2 isoforms cDNA. Int Arch Allergy Immunol 131 14–18 [DOI] [PubMed] [Google Scholar]
- Clarke MC, Kilburn SA, Hourihane JO, Dean KR, Warner JO, Dean TP (1998) Serological characteristics of peanut allergy. Clin Exp Allergy 28 1251–1257 [DOI] [PubMed] [Google Scholar]
- De Vries S, Hoge H, Bisseling T (1988) Isolation of total and polysomal RNA from plant tissues. In SB Gelvin, RA Schilperoort, DPS Verma, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–13
- Dodo HW, Viquez OM, Maleki SJ, Konan KN (2004) cDNA clone of a putative peanut (Arachis hypogaea L.) trypsin inhibitor has homology with peanut allergens Ara h 3 and Ara h 4. J Agric Food Chem 52 1404–1409 [DOI] [PubMed] [Google Scholar]
- Eigenmann PA, Burks AW, Bannon GA, Sampson HA (1996) Identification of unique peanut and soy allergens in sera adsorbed with cross-reacting antibodies. J Allergy Clin Immunol 98 969–978 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Barker SJ, Perez-Grau L (1989) Regulation of gene expression during plant embryogenesis. Cell 56 149–160 [DOI] [PubMed] [Google Scholar]
- Harris N, Grindley H, Mulchrone J, Croy RRD (1989) Correlated in situ hybridization and immunocytochemical studies of legumin storage protein deposition in pea (Pisum sativum L.). Cell Biol Int Rep 13 23–35 [Google Scholar]
- Hauxwell AJ, Corke FMK, Hedley CL, Wang TL (1990) Storage protein gene expression is localized to regions lacking mitotic activity in developing pea embryos. An analysis of seed development in Pisum sativum XIV. Development 110 283–289 [DOI] [PubMed] [Google Scholar]
- Kleber-Janke T, Crameri R, Appenzeller U, Schlaak M, Becker WM (1999) Selective cloning of peanut allergens, including profilin and 2S albumins, by phage display technology. Int Arch Allergy Immunol 119 265–274 [DOI] [PubMed] [Google Scholar]
- Koppelman SJ, Bruijnzeel-Koomen CA, Hessing M, de Jong HH (1999) Heat-induced conformational changes of Ara h 1, a major peanut allergen, do not affect its allergenic properties. J Biol Chem 274 4770–4777 [DOI] [PubMed] [Google Scholar]
- Koppelman SJ, Knol EF, Vlooswijk RAA, Wensing M, Knulst AC, Hefle SL, Gruppen H, Piersma S (2003) Peanut allergen Ara h 3: isolation from peanuts and biochemical characterization. Allergy 58 1144–1151 [DOI] [PubMed] [Google Scholar]
- Koppelman SJ, Vlooswijk RA, Knippels LM, Hessing M, Knol EF, van Reijsen FC, Bruijnzeel-Koomen CA (2001) Quantification of major peanut allergens Ara h 1 and Ara h 2 in the peanut varieties Runner, Spanish, Virginia, and Valencia, bred in different parts of the world. Allergy 56 132–137 [DOI] [PubMed] [Google Scholar]
- Kroj T, Savino G, Valon C, Giraudot J, Parcy F (2003) Regulation of storage protein gene expression in Arabidopsis. Development 130 6065–6073 [DOI] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ (2003) Leafy cotyledon 1-like defines a class of regulators essential for embryo development. Plant Cell 15 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Lycett GW, Delauney AJ, Gatehouse JA, Gilroy J, Croy RR, Boulter D (1983) The vicilin gene family of pea (Pisum sativum L.): a complete cDNA coding sequence for preprovicilin. Nucleic Acids Res 11 2367–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleki SJ, Chung SY, Champagne ET, Raufman JP (2000. a) The effects of roasting on the allergenic properties of peanut proteins. J Allergy Clin Immunol 106 763–768 [DOI] [PubMed] [Google Scholar]
- Maleki SJ, Kopper RA, Shin DS, Park CW, Compadre CM, Sampson H, Burks AW, Bannon GA (2000. b) Structure of the major peanut allergen Ara h 1 may protect IgE-binding epitopes from degradation. J Immunol 164 5844–5849 [DOI] [PubMed] [Google Scholar]
- Maleki SJ, Viquez O, Jacks T, Dodo H, Champagne ET, Chung SY, Landry SJ (2003) The major peanut allergen, Ara h 2, functions as a trypsin inhibitor, and roasting enhances this function. J Immunol 112 190–195 [DOI] [PubMed] [Google Scholar]
- Müntz K (1998) Deposition of storage proteins. Plant Mol Biol 38 77–99 [PubMed] [Google Scholar]
- Pattee HE, Johns EB, Singleton JA, Sanders TH (1974) Composition of peanut fruit parts during maturation. Peanut Sci 1 57–62 [Google Scholar]
- Perez-Grau L, Goldberg RB (1989) Soybean seed protein genes are regulated spatially during embryogenesis. Plant Cell 1 1095–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumphrey RSH, Wilson PB, Faragher EB, Edwards SR (1999) Specific immunoglobulin E to peanut, hazelnut and brazil nut in 731 patients: similar patterns found at all ages. Clin Exp Allergy 29 1256–1259 [DOI] [PubMed] [Google Scholar]
- Rabjohn P, Helm EM, Stanley JS, West CM, Sampson HA, Burks AW, Bannon GA (1999) Molecular cloning and epitope analysis of the peanut allergen Ara h 3. J Clin Invest 103 535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos M, Fleming G, Chu Y, Akiyama Y, Gallo M, Ozias-Akins P (2006) Chromosomal and phylogenetic context for conglutin genes in Arachis based on genomic sequence. Mol Genet Genomics 275 578–592 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schlereth A, Becker C, Horstmann C, Tiedemann J, Müntz K (2000) Comparison of globulin mobilization and cysteine proteinases in embryonic axes and cotyledons during germination and seedling growth of vetch (Vicia sativa L.). J Exp Bot 51 1423–1433 [PubMed] [Google Scholar]
- Sen M, Kopper R, Pons L, Abraham EC, Burks AW, Bannon GA (2002) Protein structure plays a critical role in peanut allergen stability and may determine immunodominant IgE-binding epitopes. J Immunol 169 882–887 [DOI] [PubMed] [Google Scholar]
- Shewry PR, Napier JA, Tatham AS (1995) Seed storage proteins: structures and biosynthesis. Plant Cell 7 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DS, Compadre CM, Maleki SJ, Kopper RA, Sampson H, Huang SK, Burks AW, Bannon GA (1998) Biochemical and structural analysis of the IgE binding sites on Ara h1, an abundant and highly allergenic peanut protein. J Biol Chem 273 13753–13759 [DOI] [PubMed] [Google Scholar]
- Shutov AD, Baümlein H, Blattner FR, Müntz K (2003) Storage and mobilization as antagonist functional constraints on seed storage globulin evolution. J Exp Bot 54 1645–1654 [DOI] [PubMed] [Google Scholar]
- Shutov AD, Vaintraub IA (1987) Degradation of storage proteins in germinating seeds. Phytochemistry 26 1557–1566 [Google Scholar]
- Stanley JS, King N, Burks AW, Huang SK, Sampson H, Cockrell G, Helm RM, West CM, Bannon GA (1997) Identification and mutational analysis of the immunodominant IgE binding epitopes of the major peanut allergen Ara h 2. Arch Biochem Biophys 342 244–253 [DOI] [PubMed] [Google Scholar]
- Tegeder M, Offler CE, Frommer WB, Patrick JW (2000) Amino acid transporters are localized to transfer cells of developing pea seeds. Plant Physiol 122 319–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedemann J, Neubohn B, Müntz K (2000) Different functions of vicilin and legumin are reflected in the histopattern of globulin mobilization during germination of vetch (Vicia sativa L.). Planta 211 1–12 [DOI] [PubMed] [Google Scholar]
- Towbin HK, Staehelin TH, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets. Proc Natl Acad Sci USA 76 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KA, Rightmire BR, Chen JC, Tan-Wilson A (1986) Differential proteolysis of glycinin and β-conglycinin polypeptides during soybean germination and seedling growth. Plant Physiol 82 71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang P, Beardslee TA, Zeece MG, Markwell J, Sarath G (2002) Identification and analysis of a conserved immunoglobulin E-binding epitope in soybean G1a and G2a and peanut Ara h 3 glycinins. Arch Biochem Biophys 408 51–57 [DOI] [PubMed] [Google Scholar]
- Yocum MW, Khan DA (1994) Assessment of patients who have experienced anaphylaxis: a 3-year survey. Mayo Clin Proc 69 16–23 [DOI] [PubMed] [Google Scholar]