Abstract
Since late 2004, the swine industry in the province of Quebec has experienced a significant increase in death rate related to postweaning multisystemic wasting syndrome (PMWS). To explain this phenomenon, 2 hypotheses were formulated: 1) the presence of a 2nd pathogen could be exacerbating the porcine circovirus 2 (PCV-2) infection, or 2) a new and more virulent PCV-2 strain could be infecting swine. In 2005, 13 PMWS cases were submitted to the Quebec provincial diagnostic laboratory and PCV-2 was the only virus that could be found consistently by PCR in all 13 samples. The PCR detection results obtained for other viruses revealed the following: 61.5% were positive for porcine reproductive and respiratory syndrome virus, 30.8% for swine influenza virus, 15.4% for porcine parvovirus, 69.2% for swine torque teno virus (swTTV), 38.5% for swine hepatitis E virus (swHEV) and 84.6% for Mycoplasma hyorhinis; transmissible gastroenteritis virus and porcine respiratory coronavirus (TGEV/PRCV) was not detected. Sequences of the entire genome revealed that these PCV-2 strains belonged to a genotype (named PCV-2b) that has never been reported in Canada. Further sequence analyses on 83 other Canadian PCV-2 positive cases submitted to the provincial diagnostic laboratory during years 2005 and 2006 showed that 79.5% of the viral sequences obtained clustered in the PCV-2b genotype. The appearance of the PCV-2b genotype in Canada may explain the death rate increase related to PMWS, but this relationship has to be confirmed.
Résumé
Émergence du circovirus porcin du génotype 2b chez le porc au Canada. Depuis la fin de l’année 2004, une recrudescence marquée du syndrome de dépérissement en post-sevrage (SDPS) avec une augmentation du taux de mortalité a été observé dans les élevages porcins du Québec. Deux hypothèses furent émises pour expliquer ces observations: 1) présence d’un second pathogène qui exacerbe l’infection primaire au circovirus porcin de type 2 (PCV-2) et 2) présence d’une nouvelle souche de PCV-2 plus virulente. Des échantillons de 13 cas cliniques de SDPS furent soumis au laboratoire de diagnostic provincial du Québec et seulement le virus PCV-2 a pu être détecté dans tous les échantillons. Par contre, d’autres virus ont été détectés par PCR. Entre autres, 61,5 %, 30,8 %, 15,4 %, 69,2 %, 38,5 % et 84,6 % des 13 cas cliniques de SDPS étaient positifs pour le virus du syndrome reproducteur et respiratoire porcin (PRRSV), le virus influenza porcin (SIV), le parvovirus porcin (PPV), le torque teno virus porcin (swTTV), le virus de l’hépatite E porcin (swHEV) et Mycoplasma hyorhinis, respectivement, alors que tous les cas étaient négatifs pour la présence du virus de la gastroentérite transmissible et du coronavirus respiratoire porcin (TGEV/PRCV). Le séquençage complet du génome des 13 virus PCV-2 a révélé que ces virus appartenaient à un génotype (nommé: PCV-2b) qui, jusqu’à présent, n’avait jamais été rapporté au Canada. Le séquençage complet du génome de 83 souches canadiennes du virus PCV-2 soumis à notre laboratoire de diagnostic en 2005 et 2006 a démontré que 79,5 % des séquences virales appartiennent au génotype PCV-2b. L’apparition du génotype PCV-2b au Canada pourrait expliquer l’augmentation du taux de mortalité associé au SDPS mais cette relation de cause à effet reste à être démontrée.
(Traduit par les auteurs)
Introduction
Postweaning multisystemic wasting syndrome (PMWS) is a swine disease initially identified in Canada in 1991 (1). Now, it is known as a worldwide disease, with outbreaks being observed in swine herds of North and South America, Europe, and Asia (1). The disease affects 5- to 12-week-old piglets and is characterized, in part, by weight loss, dyspnea, jaundice, and enlarged lymph nodes, as well as by degeneration and necrosis of hepatocytes, multifocal lymphohistiocytic pneumonia, lymphocytic depletion, and multinucleated giant cell formation (2). The etiological agent responsible for PMWS has been identified as a circovirus particle and named porcine circovirus 2 (PCV-2) (3–5). The PCV-2 is a small nonenveloped virus that possesses a single-stranded ambisense circular DNA genome about 1.76 kb in length (6–9). Viral DNA possesses at least 3 functional open reading frames (ORF): ORF1 encodes the Rep proteins involved in virus replication (10–12), ORF2 encodes the nucleocapsid (NC) protein (13), and ORF3 encodes a protein that induces apoptosis and is also involved in viral pathogenesis in vivo (14,15). Today, it is now recognized that the clinical expression of PCV-2 infection in swine is more complex than previously established, since it can play a pivotal role in several syndromes: porcine dermatitis and nephropathy syndrome (PDNS), porcine respiratory disease complex (PRDC), reproductive failure, granulomatous enteritis, necrotizing lymphadenitis, exudative epidermitis, and congenital tremor (16,17). Consequently, to describe and name all those syndromes in a more convenient terminology, it is now accepted to refer to “porcine circovirus associated disease (PCVAD)”.
At the end of 2004, the swine industry in the province of Quebec experienced a significant increase in death rate related to PCVAD. At that time, no statistical analysis supported this observation and furthermore no data indicating the extent of the increase in death rate was available. Consequently, an epidemiological survey that included producers (for a total of 245 producers) that annually sold on the market 15% (1 000 000 pigs) of the entire Quebec pig production was conducted by Dr. Camille Moore, a private veterinary practitioner in Quebec, to provide valuable information on the severity of the mortality increase (18). This study included all types of production and revealed an increase of 2.39% in the mortality rate in Quebec pig farms in 2005 (7.57%) compared with 2004 (5.18%). More specifically, weaning-finishing production had a mortality rate average of 7.53% in 2005 compared with 5.31% in 2004. Similarly, finishing production had a mortality rate average of 7.66% in 2005 compared with 4.88% in 2004. Interestingly, 56% of the producers indicated that their production had a clinical, pathologic, or laboratory diagnosis of PCVAD at the time of the survey, which was held at the end of 2005. To explain this situation, 2 hypotheses were formulated based on the facts that coinfection with other pathogens is usually necessary to produce the clinical disease and gross lesions typical of PMWS (19–23) and that it is usual in virology to observe pathogenicity variation between different virus isolates (24–26): namely, 1) that the presence of another pathogen that could exacerbate the PCV-2 infection, and 2) that a new and more pathogenic PCV-2 strain was present. Consequently, following the immediate urge to understand what was going on, a PCV-2 genotype, which has never been reported previously in Canada but which has already been identified in Asia and Europe, was identified.
Materials and methods
Clinical cases definition
Thirteen PMWS cases that occurred in 2005 and originated from the province of Quebec were selected because they presented clinical signs related to the PMWS definition and were from affected herds with an increased mortality rate. Those PMWS cases (named: FMV-05-6302, FMV-05-6317, FMV-05-6505, FMV-05-6507, FMV-05-7098, FMV-05-7386, FMV-05-7388, FMV-05-7389, FMV-05-7390, FMV-05-7537, FMV-05-7539, FMV-05-8037, and FMV-05-8574) had been submitted initially to the Quebec provincial animal pathology laboratory (Institut national de santé animal — Ministère de l’agriculture, des pêcheries et de l’alimentation du Québec) for histopathologic evaluation to confirm the clinical diagnosis made by veterinarians. Samples (lung, lymph nodes, liver, spleen, kidneys) from 2 to 4 piglets were submitted for each PMWS-affected herd.
Virus isolation
Four cell lines (PK15A, ST, HRT-G, and MDCK) were used for the isolation of different porcine viruses. The PK15A (porcine kidney) cells were used to isolate PCV-2. The PK15A cells, a subclone of PCV noninfected PK15 cells (27), were maintained in Earle’s minimal essential medium (MEM; Invitrogen Corporation, GibcoBRL, Grand Island, New York, USA), supplemented with 10% fetal bovine serum (FBS), 300 U/mL of penicillin, 300 mg/mL of streptomycin, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 2.5 μg/mL of amphotericin B, and 10 mM HEPES buffer. The ST (swine testis) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen Corporation, GibcoBRL), supplemented with 2% FBS, 300 U/mL of penicillin, 300 mg/mL of streptomycin, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 2.5 μg/mL of amphotericin B, and 10 mM HEPES buffer. The HRT-G (human rectal tumor) cells were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (Invitrogen Corporation, GibcoBRL), supplemented with 300 U/mL of penicillin, 300 mg/mL of streptomycin, 10 U/mL of trypsin, 1 mM sodium pyruvate, 2.5 μg/mL of amphotericin B, and 10 mM HEPES buffer. The MDCK (Madin-Darby canine kidney) cells were maintained in 50:50 Hank’s MEM: Earle’s MEM (Invitrogen Corporation, GibcoBRL), supplemented with 300 U/mL of penicillin, 300 mg/mL of streptomycin, 10 mM HEPES buffer, 2.5 μg/mL of amphotericin B, and 10 U/mL of trypsin. All cells were maintained at 37°C in 5% CO2 atmosphere following their incubation with tissue homogenates. Virus isolation was attempted during 3 consecutive passages with pools of piglet samples (lung, lymph nodes, liver, spleen, kidneys) prepared from the 13 PMWS cases submitted to the Diagnostic Veterinary Virology Laboratory (DVVL) of the Faculté de médecine vétérinaire (FMV) of the University of Montreal.
Polymerase chain reaction diagnostic tests
Viral RNA and DNA were isolated from 140 μL of homogenate suspensions prepared from piglet sample pools for each of the 13 PMWS cases by using commercial kits (QIAamp Viral RNA Mini Kit and QIAamp DNA Mini Kit; Qiagen, Mississauga, Ontario) according to the manufacturer’s instructions. Subsequently, different polymerase chain reaction (PCR) or reverse transcription-polymerase chain reaction (RT-PCR) diagnostic tests (in house or commercially available) were performed to identify porcine pathogens. A commercially available porcine reproductive and respiratory syndrome virus (PRRSV) real time PCR diagnostic test kit (Tetracore, Rockville, Maryland, USA) was used to identify the North American PRRSV genotype according to the manufacturer’s instruction. A PCV-2 nested PCR diagnostic test was performed to identify PCV-2 positive cases, as previously described (28). Alternatively, a PCV-2 real-time PCR diagnostic test was developed by the molecular diagnostic service of the FMV to identify PCV-2 positive cases. Briefly, a set of primers (PCV-Foward: 5′-AGT GAG CGG GAA AAT GCA-3′ and PCV1-AS6: 5′-CAC ACA GTC TCA GTA GAT CAT CC-3′) was used to target the ORF1 gene of PCV-2 viral genome and gave an expected fragment of 226 base pair (bp) in length. A specific fluorogenic PCV-2 DNA probe (5′-(FAM) TGC AGA CCC GGA AAC CAC (BHQ)-3′) was then used to detect the newly synthesized PCR product. The Transmissible gastroenteritis virus and Porcine respiratory corona-virus (TGEV/PRCV) PCR diagnostic test was performed as previously described (29). It is well known that the conserved and variable regions of the 23S ribosomal RNA gene of Mycoplasma permit the identification of the cluster and subsequently the identification of the species (30). This strategy has been used, as previously described (31), to identify by PCR if a Mycoplasma sp. was present in the submitted samples and subsequently to identify which species (hyorhinis or hyopneumoniae) was present in positive cases. The presence of viruses that classified within the Influenza A virus genus, which includes the swine influenza virus (SIV), was determined with a RT-PCR assay targeting the M1 gene, using a specific primer set previously described by others (32). The Swine torque teno virus (swTTV) was detected by using a nested PCR diagnostic test developed by McKeown et al (33). The swine hepatitis E virus (swHEV) was detected according to a nested RT-PCR assay developed by Huang et al (34), by using a commercial kit (QIAGEN OneStep RT-PCR kit; Qiagen) following the manufacturer’s recommendations. The presence of the porcine parvovirus (PPV) genome was evaluated by a nested PCR, using 2 sets of primers (VPS1: 5′-TGG TGG ACC ATT TCT AAC TCC TAT AGT ACC-3′ and VPAS1: 5′-GTT AAT AGT AAA CAC ATG AGA GCT TGT TTC-3′; VPS2: 5′-CAA TAC TGC ACC TGT ATT TCC AAA TGG-3′ and VPAS2: 5′-AAA ATT TTA TTG TTT TTT GGG GAT AAT TGG-3′) that target the VP gene and gave expected fragments of 879 and 526 bp in length for wild type PPV and 1006 and 653 bp in length for the laboratory PPV strain (NADL-2).
Sequencing and phylogenetic analyses
The entire PCV-2 genome was amplified by PCR, using 2 sets of oligonucleotides (SEQ PCV-1NF: 5′-GGA CCC CAA CCC CAT AAA A-3′ and SEQ PCV-1NR: 5′-CCC TCA CCT ATG ACC CCT ATG T-3′; SEQ PCV-2NF: 5′-TGT TTT CGA ACG CAG TGC C-3′ and SEQ PCV-2NR: 5′-CCG TTG TCC CTG AGA TCT AGG A-3′) that produced 2 overlapping PCR products at both ends of 1254 nucleotides (nt) and 1045 nt, respectively. The PCR products were purified by using a commercial kit (QIAquick PCR purification kit; Qiagen) according to the manufacturer’s instruction. Both strands of the purified DNA PCR products were sequenced by using the same primer sets with standard automated sequencing methods (FMV Sequencing Laboratory, Bigdye terminator version 3.1, sequencer: ABI 310; Applied Biosystems, Foster City, California, USA). Resulting sequences were compared with other Canadian PCV-2 strains (8), PCV-2 strains submitted to the DVVL from Quebec, Ontario, Manitoba, and Saskatchewan from early in 2005 until June 2006, as well as other PCV-2 sequences available in GenBank. Software (BioEdit Sequence Alignment Editor version 7.0.5.2; Ibis Therapeutics, Carlsbad, California, USA) using the CLUSTAL W alignment method was utilized and an unrooted phylogenic tree was constructed by using the distance-based neighbor-joining method. Bootstrap values were calculated on 1000 repeats of the alignment. The identification of the PCV-2 sequences used for the phylogenic tree and their respective GenBank accession number are indicated in Table 1.
Table 1.
Identification of porcine circovirus 2 (PCV-2) strains with their geographic origin and GenBank accession number
| Isolate ID | Geographic origin | GenBank accession number | Reference |
|---|---|---|---|
| FMV-05-6302 | Canada/Quebec
↓ |
DQ220739 | Gagnon et al (2006)
↓ |
| FMV-05-6317 | DQ220728 | ||
| FMV-05-6505 | DQ220729 | ||
| FMV-05-6507 | DQ220730 | ||
| FMV-05-7098 | DQ220731 | ||
| FMV-05-7386 | DQ220732 | ||
| FMV-05-7388 | DQ220733 | ||
| FMV-05-7389 | DQ220734 | ||
| FMV-05-7390 | DQ220735 | ||
| FMV-05-7537 | DQ220736 | ||
| FMV-05-7539 | DQ220737 | ||
| FMV-05-8037 | DQ220738 | ||
| FMV-05-8574 | DQ220727 | ||
| 2A | Canada
↓ |
AF027217 | Hamel et al (2000)
↓ |
| 2B | AF112862 | ||
| 2C | AF109398 | ||
| 2D | AF117753 | ||
| 2E | AF109399 | ||
| Imp. 999 | United States | AF055391 | Meehan et al (1998)
↓ |
| Imp. 1010-Stoon | Canada | AF055392 | |
| Imp. 1011-48121 (FRA1) | France
↓ |
AF055393 | |
| Imp. 1011-48285 (FRA2) | AF055394 | ||
| FRA3 | AF201311 | Mankertz et al (2000)
↓ |
|
| GER1 | Germany
↓ |
AF201305 | |
| GER2 | AF201306 | ||
| GER3 | AF201307 | ||
| SPA1 | Spain
↓ |
AF201308 | |
| SPA2 | AF201309 | ||
| SPA3 | AF201310 | ||
| 412 | Canada
↓ |
AF085695 | Wang et al (unpublished)
↓ |
| M226 | AF086836 | ||
| 9741 | AF086835 | ||
| B9 | AF086834 | ||
| ISU-31 | United States/Iowa | AJ223185 | Morozov et al (1998) |
| MLTW98 (TA1) | Taiwan
↓ |
AF154679 | Kuo et al (unpublished) |
| Tainan (TA2) | AF166528 | Yang et al (unpublished) | |
| 26606 | United States/Utah
↓ |
AF264038 | Fenaux et al (2000)
↓ |
| 26607 | AF264039 | ||
| 10489 | United States/Illinois | AF264040 | |
| 40856 | United States/Missouri | AF264041 | |
| 40895 | United States/Iowa | AF264042 | |
| 34464 | Canada | AF264043 | |
| 24657 NL | Netherlands | AF201897 | Wellenberg et al (2000) |
| BF | China
↓ |
AF381175 | Lu et Yang (unpublished)
↓ |
| HR | AF381176 | ||
| BX | AF381177 | ||
| Imp. 1103 | Canada/Alberta | AJ293867 | Meehan et al (2001)
↓ |
| Imp. 1121 | Canada/Saskatchewan | AJ293868 | |
| Imp. 1147 | UK | AJ293869 | |
| IAF2897 | Canada/Québec | AF408635 | Racine et al (2004) |
| SH | China
↓ |
AY291318 | Feng et al (unpublished)
↓ |
| ZJ | AY686764 | ||
| JXII | AY732494 | ||
| JS | AY691679 | ||
| SX04 | AY604430 | Li et al (unpublished) | |
| DG | AY682993 | Wang et al (unpublished)
↓ |
|
| ZC | AY682997 | ||
| ZS | AY596823 | Da et al (unpublished) | |
| NL PMWS 4 | UK
↓ |
AY484416 | Grierson et al (2004)
↓ |
| NL control 6 | AY484412 | ||
| AUT5 | Austria | AY424405 | Exel et al (unpublished) |
| GD | China
↓ |
AY613854 | Song et al (unpublished)
↓ |
| GD-ZJ | DQ017036 | ||
| QD | AY291316 | Xin et al (unpublished) | |
| Henan | AY969004 | Liu et al (unpublished) | |
| 375 | Hungria | AY256460 | Dan et al (2003) |
| HD | China | AY916791 | Jiang et al (unpublished) |
| Fd1 | France
↓ |
AY322000 | de Boisseson et al (2004)
↓ |
| Fd2 | AY321999 | ||
| Fd3 | AY321984 |
Results
Identification of cofactors possibly involved in the appearance of PCVAD
Interestingly, the 13 PMWS cases submitted to the DVVL presented characteristic microscopic lesions of PMWS at various degrees of intensity. The observed lesions were predominantly identified as marked lymphocytic depletion, multinucleated giant cell formation, appearance of inclusion bodies in histiocytes, and multifocal lymphohistiocytic pneumonia. Detection results presented in Table 2 show that these 13 PMWS cases were positive, not only for PCV-2 but also for several other swine viral pathogens. In these PMWS cases, 8 out of 13 (61.5%) were positive for PRRSV, 4 out of 13 (30.8%) were positive for SIV, 2 out of 13 (15.4%) were positive for PPV, 5 out of 13 (38.5%) were positive for swHEV, and 9 out of 13 (69.2%) were positive for swTTV. All PCV-2 positive cases were PCR positive for at least 1 other viral pathogen. The 2 worst cases were simultaneously infected with PCV-2, PRRSV, SIV, swHEV, swTTV, and Mycoplasma hyorhinis (Table 2). Transmissible gastroenteritis virus and porcine respiratory coronavirus was not detected by PCR in any of the 13 PMWS cases studied. No other virus, except for PCV-2, could be isolated from the HRT-G, MDCK, ST, and PK15A cell lines (Table 2). At the 3rd passage, PCR positive results were obtained for a Mycoplasma sp. and subsequently for M. hyorhinis in the cell culture supernatants of 11 samples (Table 2). Cell culture had permitted the growth of M. hyorhinis to a level where it could be identified. To eliminate the possibility of a M. hyorhinis contamination, 8 μg/mL of tylosin was added in the cell culture medium and virus isolation was tried once again. Unfortunately, no beneficial effect on virus isolation was observed, except for a small improvement on PCV-2 isolation (data not shown).
Table 2.
Identification of viral swine pathogens in postweaning multisystemic wasting syndrome (PMWS) cases
| Polymerase chain reaction
|
Virus isolation
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PMWS cases | PCV-2 | PRRSV | TGEV/PRCV | SIV | PPV | swHEV | swTTV | Mycoplasma hyorhinisb | HRT-G/MDCK/ST | PK15Ac |
| FMV05-6302 | + | − | − | − | − | − | + | − | − | + |
| FMV05-6317 | + | − | − | − | − | − | + | + | − | + |
| FMV05-6505 | + | + | − | − | − | − | + | + | − | − |
| FMV05-6507 | + | + | − | − | − | + | + | + | − | + |
| FMV05-7098 | + | + | − | − | − | − | + | − | − | − |
| FMV05-7386 | + | + | − | + | + | − | − | + | − | + |
| FMV05-7388 | + | + | − | − | − | + | − | + | − | + |
| FMV05-7389 | + | + | − | − | − | − | − | + | − | − |
| FMV05-7390 | + | + | − | +a | − | + | + | + | − | + |
| FMV05-7537 | + | − | − | +a | − | + | − | + | − | − |
| FMV05-7539 | + | − | − | − | − | − | + | + | − | − |
| FMV05-8037 | + | − | − | − | + | − | + | + | − | − |
| FMV05-8574 | + | + | − | + | − | + | + | + | − | − |
Swine influenza virus has been confirmed by PCR to be H3N2 virus
Confirmed by PCR on the 3rd passage of cell culture supernatants
Only PCV-2 was isolated in PKISA cells and it was confirmed by PCR at the 3rd passage
PCV-2 = porcine circovirus 2, TGEV/PRCV = transmissible gastroenteritis virus/porcine respiratory coronavirus, PRRSV = porcine reproductive and respiratory syndrome virus, SIV = swine influenza virus, PPV = porcine parvovirus, swHEV = swine hepatitis E virus, swTTV = swine torque teno virus, HRT-G = human rectal tumor cells, MDCK = Madin-Darby canine kidney cells, ST = swine testis cells, PK15A = porcine kidney cells
Sequence analysis of recent PCV-2 Canadian strains
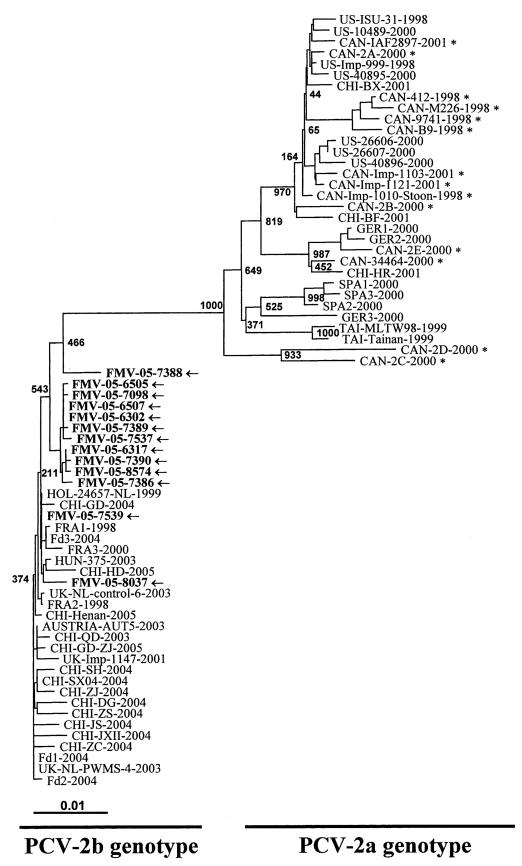
Entire genome sequences obtained from the 13 PCV-2 cases were aligned and compared with PCV-2 reference strains (Table 1). As shown in Figure 1, the PCV-2 strains can be classified in 2 genotypes (PCV-2a and PCV-2b) and the nt sequence identity between both genotypes varies from 94% to 96%. Since the nt sequence identity between strains from PCV-2a genotype varied between 96% to 100% (Figure 1) (8), the overall PCV-2 nt sequences are quite conserved and their classification could be arbitrary. Consequently, it could be more appropriate to look at individual genes or encoded peptide sequences, or both, rather than the entire nt sequences of the viral genome. The most variable protein between the PCV-2a and PCV-2b genotypes was the NC protein, which is encoded by the ORF2 gene, with an amino acid (aa) sequence identity varying from 88% to 94% between genotypes. The 2 other known proteins to be expressed by PCV-2 were less variable between the genotypes than the NC protein, with an aa sequence identity between the genotypes varying from 96% to 99% for Rep protein and from 92% to 98% for ORF3 protein. Results of the sequencing and genotyping of the 83 PCV-2 strains received from the 4 provinces from early in 2005 to June 2006 are shown in Table 3. The highest proportion of PCV-2 strains classified in PCV-2b genotype was found in the province of Quebec with 50 out of 51 (98%). In Ontario, Manitoba, and Saskatchewan, 4 out of 7 (57.1%), 10 out of 23 (43.5%), and 2 out of 2 (100%) were classified in PCV-2b genotype, respectively. Overall, 66 out of 83 (79.5%) PCV-2 entire genome sequences in Canada clustered in the PCV-2b genotype.
Figure 1.
Phylogenetic analysis of the complete genome of porcine circovirus 2 (PCV-2) strains. An unrooted neighbor-joining tree was constructed from aligned nucleic acid sequences of 27 Canadian reference strains (including the newly described 13 sequences identified with an arrow and the older sequences identified with an asterix) and 43 sequences found in GenBank. Original names, country of origin, and GenBank accession number are given in Table 1.
Table 3.
Genotype classification of 2005–2006 porcine circovirus 2 (PCV-2) Canadian strainsa following viral genome sequence analysis
| Genotype classification
|
|||
|---|---|---|---|
| Province | Number of PCV-2 sequenced | PCV-2a | PCV-2b |
| Quebec | 51 | 1 | 50 |
| Ontario | 7 | 3 | 4 |
| Manitoba | 23 | 13 | 10 |
| Saskatchewan | 2 | 0 | 2 |
| Total | 83 | 17 | 66 |
Most of those viruses were obtained from PCVAD affected herds but some were obtained from herds that had shown no clinical sign related to this disease
Discussion
The ST, HRT-G, MDCK, and PK15A cell lines were selected because they are known to be permissive to most of the porcine viruses. As an example, the ST cells are known to permit the replication of porcine enteroviruses, PPV, TGEV/PRCV, and pseudorabies virus (PRV) (35); the HRT-G cells are known to permit the replication of coronavirus like the Porcine hemagglutinating encephalomyelitis virus (36); the MDCK are known to permit the replication of SIV and several other viruses (35,37); and the PK15A cells were used to isolate PCV-2, and they are known to permit the replication of other viruses like classical swine fever virus, African swine fever virus, Vesicular exanthema of swine virus, and vesicular stomatitis virus (35). As mentioned earlier, no virus, except PCV-2, could be isolated in those 4 cell lines from the 13 submitted PMWS cases. Also, no other common viral pathogen could be found by PCR in all 13 PCV-2 positive cases where PMWS disease was observed. Interestingly, even if PCR results from organ samples were negative for the presence of a Mycoplasma sp., following virus isolation assays, most of the cell cultures turned positive for M. hyorhinis. Following these results, we concluded that 84.6% of the PMWS cases were also positive for M. hyorhinis (Table 2), that M. hyorhinis was the most prevalent pathogen found in the 13 PMWS cases but that the Mycoplasma sp. PCR diagnostic test was not sensitive enough to detect the pathogen directly in the submitted samples. In fact, only 2 out of the 13 samples cell culture supernatants were negative for M. hyorhinis (Table 2). Mycoplasma hyorhinis is an extremely common contaminant in cell culture inoculated with swine tissues, and M. hyorhinis has never been considered to be a major problem in the status of swine health. Nevertheless, it was recently implicated in pneumonia, causing lesions similar to those of M. hyopneumoniae (38). Mycoplasma hyopneumoniae has been known to be an important cofactor for the induction of PMWS in PCV-2 infected swine (22). Is it possible that a dual infection with PCV-2 and M. hyorhinis led to the same outcome as an infection with PCV-2 and M. hyopneumoniae? At this time, no data are available to help us answer this question. Nevertheless, it would be a fair assumption to believe that since some M. hyorhinis strains are able to induce pneumonia (38), they may influence the evolution of PCV-2 infection in swine. In the present situation, since the PCR diagnostic test was not able to detect M. hyorhinis in sample homogenates but only in cell culture supernatants, we can assume that the amount of M. hyorhinis was very low, suggesting that the degree of pathogenicity of the M. hyorhinis strains found in the PMWS cases was also very low. The 2 viruses that were found in higher proportion by PCR in PCV-2 positives cases were PRRSV and swTTV with 61.5% and 69.2% positive samples, respectively (Table 2). It is well known that PRRSV is a major pathogen that can lead to PMWS when present in PCV-2 infected swine (20,23). The 61.5% PRRSV prevalence in PMWS cases is quite high but similar to what has already been reported by others (39,40). Until now, swTTV has not been shown to be pathogenic in swine (33). Consequently, the potential role and effect of swTTV during coinfection with PCV-2 is even more obscure and unknown. Nevertheless, as previously reported by others (33), the overall 66.2% prevalence of swTTV infection in swine populations worldwide is similar to our results in regard to the swTTV prevalence in Canada (Table 2). Interestingly, McKeown et al (33) reported that in the province of Quebec, all tested pig sera were positive for swTTV and that the overall swTTV prevalence in Canada was 79.1%. The swTTV prevalence value may vary a lot between countries (33%–100%), but despite this, it is still very high (33). The swHEV virus is more problematic, because it is known to be able to induce a subclinical infection in swine (41,42) and, mostly, because it has to be considered as a zoonotic pathogen (43,44). Similar to swTTV, the prevalence of swHEV infection in the swine population is very high, as shown in Table 2, and it may vary greatly between countries (45,46). Unfortunately, the potential role and effect of swHEV, as well as those of swTTV, during dual infections in swine with PCV-2 remain unknown. Since swTTV and swHEV are recently discovered viruses, many experiments still have to be completed to determine the effect of both viruses on animal health status and to determine their potential synergy during dual infections with PCV-2.
Sequence analysis of the entire genome of recent PCV-2 strains in Canada has helped to identify, for the first time, a new type of circulating PCV-2 strain in North America (Figure 1). In a previous study, Larochelle et al (8) have shown that the PCV-2 strains circulating in Canada were all clustering in the PCV-2a genotype. The PCV-2 nt sequence identity between our 13 PMWS submitted cases was highly conserved, sharing similarities of 99% to 100%. Interestingly, all these 13 new PCV-2 sequences obtained in 2005 clustered in the PCV-2b genotype and, until now, no other older Canadian PCV-2 entire genome sequences have been reported and classified in the PCV-2b cluster, confirming the fact that a new type of strain has appeared in Canada (Figure 1 and Table 1). Although, even if these new Canadian PCV-2 strains clustered in PCV-2b genotype, other older and recent PCV-2b strains have already been reported in Asia and Europe (Figure 1 and Table 1) (47). Except in this report, no Canadian PCV-2 entire genome sequences have been reported between 2002 and 2005, so it is impossible to pinpoint exactly when the new PCV-2b genotype appeared in Canada. Nevertheless, preliminary results of restriction fragment length polymorphism (RFLP) and gene sequence comparisons reported by Carman et al (48,49) suggest that a new PCV-2 genotype, which seems to be related to the PCV-2b genotype, appeared in Ontario in 2004 (48–50). France has experienced severe economical loss associated with PMWS in the past, and commercial exchange (swine importation) with Canada during those years may have favored the introduction of the PCV-2b genotype. Furthermore, the appearance of a new type of circulating PCV-2 strains (PCV-2b strains) seemed to coincide with an increased death rate and PMWS in swine herds across Canada and particularly in Quebec (18,50). It is now obvious that the PCV-2b genotype is more prevalent than the PCV-2a genotype in Quebec and across Canada (Table 3). The same phenomenon, where the appearance of a new type of PCV-2 strain coincided with simultaneous increases in clinical PMWS cases, has also been reported in Hong Kong in 2005 (51). Unfortunately, the relationship between the presence of the PCV-2b genotype strains and the increase of clinical PMWS cases in Quebec has still to be proven without any doubt. Some have argued that PCV-2b genotype strains are not more pathogenic than PCV-2a genotype strains, since they can be found in swine herds both with or without PMWS (47); an observation also made by our research team (data not shown). Nevertheless, the existence of variations in virulence could not be excluded, since others have reported the existence of PCV-2 pathogenicity variations between mutated viruses (15,24). In conclusion, it is obvious that there is a new type of PCV-2 strain circulating in Canadian swine herds. However, experimental infections are needed to prove if this new type of PCV-2 strain is more virulent than previous PCV-2 strains found in Canada during the late 90’s and early 2000’s.
Acknowledgments
We are grateful to Cynthia M. Guilbert for critically reviewing the manuscript. We thank Dr. Réjean Chabot for submitting the 13 2005 PMWS cases to our attention. We also thank Mrs. Danielle Leblanc and Mr. Jozsef Szelei for technical assistance. CVJ
Footnotes
This work was supported by the Ministère de l’agriculture, des pêcheries et de l’alimentation du Québec (MAPAQ), the Fédération des Producteurs de Porc du Québec (FPPQ), the Conseil pour le développement de l’agriculture du Québec (CDAQ), and the Centre d’insémination porcine du Québec Inc. (CIPQ).
References
- 1.Chae C. Postweaning multisystemic wasting syndrome: A review of aetiology, diagnosis and pathology. Vet J. 2004;168:41–49. doi: 10.1016/j.tvjl.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 2.Harding JC, Clark EG. Recognizing and diagnosing postweaning multisystemic wasting syndrome (PMWS) Swine Health Prod. 1997:201–203. [Google Scholar]
- 3.Allan GM, McNeilly F, Kennedy S, et al. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Invest. 1998;10:3–10. doi: 10.1177/104063879801000102. [DOI] [PubMed] [Google Scholar]
- 4.Ellis J, Hassard L, Clark E, et al. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can Vet J. 1998;39:44–51. [PMC free article] [PubMed] [Google Scholar]
- 5.Hamel AL, Lin LL, Nayar GP. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J Virol. 1998;72:5262–5267. doi: 10.1128/jvi.72.6.5262-5267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tischer I, Gelderblom H, Vettermann W, Koch MA. A very small porcine virus with circular single-stranded DNA. Nature. 1982;295:64–66. doi: 10.1038/295064a0. [DOI] [PubMed] [Google Scholar]
- 7.Fenaux M, Halbur PG, Gill M, Toth TE, Meng XJ. Genetic characterization of type 2 porcine circovirus (PCV-2) from pigs with postweaning multisystemic wasting syndrome in different geographic regions of North America and development of a differential PCR-restriction fragment length polymorphism assay to detect and differentiate between infections with PCV-1 and PCV-2. J Clin Microbiol. 2000;38:2494–2503. doi: 10.1128/jcm.38.7.2494-2503.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larochelle R, Magar R, D’Allaire S. Genetic characterization and phylogenetic analysis of Porcine circovirus type 2 (PCV2) strains from cases presenting various clinical conditions. Virus Res. 2002;90:101–112. doi: 10.1016/s0168-1702(02)00141-7. [DOI] [PubMed] [Google Scholar]
- 9.Meehan BM, McNeilly F, Todd D, et al. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J Gen Virol. 1998;79 (Pt 9):2171–2179. doi: 10.1099/0022-1317-79-9-2171. [DOI] [PubMed] [Google Scholar]
- 10.Mankertz A, Hillenbrand B. Replication of porcine circovirus type 1 requires two proteins encoded by the viral rep gene. Virology. 2001;279:429–438. doi: 10.1006/viro.2000.0730. [DOI] [PubMed] [Google Scholar]
- 11.Cheung AK. Transcriptional analysis of porcine circovirus type 2. Virology. 2003;305:168–180. doi: 10.1006/viro.2002.1733. [DOI] [PubMed] [Google Scholar]
- 12.Cheung AK. Comparative analysis of the transcriptional patterns of pathogenic and nonpathogenic porcine circoviruses. Virology. 2003;310:41–49. doi: 10.1016/s0042-6822(03)00096-5. [DOI] [PubMed] [Google Scholar]
- 13.Nawagitgul P, Morozov I, Bolin SR, Harms PA, Sorden SD, Paul PS. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J Gen Virol. 2000;81:2281–2287. doi: 10.1099/0022-1317-81-9-2281. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Chen I, Kwang J. Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J Virol. 2005;79:8262–8274. doi: 10.1128/JVI.79.13.8262-8274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Chen I, Du Q, Chua H, Kwang J. The ORF3 protein of porcine circovirus type 2 is involved in viral pathogenesis in vivo. J Virol. 2006;80:5065–5073. doi: 10.1128/JVI.80.10.5065-5073.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chae C. A review of porcine circovirus 2-associated syndromes and diseases. Vet J. 2005;169:326–336. doi: 10.1016/j.tvjl.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Harding JC. The clinical expression and emergence of porcine circovirus 2. Vet Microbiol. 2004;98:131–135. doi: 10.1016/j.vetmic.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 18.Moore C. Rapport de l’enquête interne sur le syndrome de dépérissement en post-sevrage (SDPS) 2006 Available from Fédération des producteurs de porcs du Québec (FPPQ) “internal inquiry”.
- 19.Allan GM, Kennedy S, McNeilly F, et al. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. J Comp Pathol. 1999;121:1–11. doi: 10.1053/jcpa.1998.0295. [DOI] [PubMed] [Google Scholar]
- 20.Allan GM, McNeilly F, Ellis J, et al. Experimental infection of colostrum deprived piglets with Porcine circovirus 2 (PCV2) and Porcine reproductive and respiratory syndrome virus (PRRSV) potentiates PCV2 replication. Arch Virol. 2000;145:2421–2429. doi: 10.1007/s007050070031. [DOI] [PubMed] [Google Scholar]
- 21.Krakowka S, Ellis JA, Meehan B, Kennedy S, McNeilly F, Allan G. Viral wasting syndrome of swine: Experimental reproduction of post-weaning multisystemic wasting syndrome in gnotobiotic swine by coinfection with porcine circovirus 2 and porcine parvovirus. Vet Pathol. 2000;37:254–263. doi: 10.1354/vp.37-3-254. [DOI] [PubMed] [Google Scholar]
- 22.Opriessnig T, Thacker EL, Yu S, Fenaux M, Meng XJ, Halbur PG. Experimental reproduction of postweaning multisystemic wasting syndrome in pigs by dual infection with Mycoplasma hyopneumoniae and porcine circovirus type 2. Vet Pathol. 2004;41:624–640. doi: 10.1354/vp.41-6-624. [DOI] [PubMed] [Google Scholar]
- 23.Rovira A, Balasch M, Segales J, et al. Experimental inoculation of conventional pigs with porcine reproductive and respiratory syndrome virus and porcine circovirus 2. J Virol. 2002;76:3232–3239. doi: 10.1128/JVI.76.7.3232-3239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fenaux M, Opriessnig T, Halbur PG, Elvinger F, Meng XJ. Two amino acid mutations in the capsid protein of type 2 porcine circovirus (PCV2) enhanced PCV2 replication in vitro and attenuated the virus in vivo. J Virol. 2004;78:13440–13446. doi: 10.1128/JVI.78.24.13440-13446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng XJ, Paul PS, Halbur PG, Morozov I. Sequence comparison of open reading frames 2 to 5 of low and high virulence United States isolates of Porcine reproductive and respiratory syndrome virus. J Gen Virol. 1995;76:3181–3188. doi: 10.1099/0022-1317-76-12-3181. [DOI] [PubMed] [Google Scholar]
- 26.Halbur PG, Paul PS, Meng XJ, Lum MA, Andrews JJ, Rathje JA. Comparative pathogenicity of nine US Porcine reproductive and respiratory syndrome virus (PRRSV) isolates in a five-week-old cesarean-derived, colostrum-deprived pig model. J Vet Diagn Invest. 1996;8:11–20. doi: 10.1177/104063879600800103. [DOI] [PubMed] [Google Scholar]
- 27.Racine S, Kheyar A, Gagnon CA, Charbonneau B, Dea S. Eucaryotic expression of the nucleocapsid protein gene of porcine circovirus type 2 and use of the protein in an indirect immunofluorescence assay for serological diagnosis of postweaning multisystemic wasting syndrome in pigs. Clin Diagn Lab Immunol. 2004;11:736–741. doi: 10.1128/CDLI.11.4.736-741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ouardani M, Wilson L, Jette R, Montpetit C, Dea S. Multiplex PCR for detection and typing of Porcine circoviruses. J Clin Microbiol. 1999;37:3917–3924. doi: 10.1128/jcm.37.12.3917-3924.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim L, Chang KO, Sestak K, Parwani A, Saif LJ. Development of a reverse transcription-nested polymerase chain reaction assay for differential diagnosis of Transmissible gastroenteritis virus and Porcine respiratory coronavirus from feces and nasal swabs of infected pigs. J Vet Diagn Invest. 2000;12:385–388. doi: 10.1177/104063870001200418. [DOI] [PubMed] [Google Scholar]
- 30.Hotzel H, Sachse K, Pfutzner H. A PCR scheme for differentiation of organisms belonging to the Mycoplasma mycoides cluster. Vet Microbiol. 1996;49:31–43. doi: 10.1016/0378-1135(95)00176-x. [DOI] [PubMed] [Google Scholar]
- 31.Kobisch M, Frey J. Detection of Mycoplasma hyponeumoniae from clinical samples and air. Methods Mol Biol. 2003;216:247–256. doi: 10.1385/1-59259-344-5:247. [DOI] [PubMed] [Google Scholar]
- 32.Spackman E, Senne DA, Myers TJ, et al. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J Clin Microbiol. 2002;40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKeown NE, Fenaux M, Halbur PG, Meng XJ. Molecular characterization of porcine TT virus, an orphan virus, in pigs from six different countries. Vet Microbiol. 2004;104:113–117. doi: 10.1016/j.vetmic.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Huang FF, Haqshenas G, Guenette DK, et al. Detection by reverse transcription-PCR and genetic characterization of field isolates of swine Hepatitis E virus from pigs in different geographic regions of the United States. J Clin Microbiol. 2002;40:1326–1332. doi: 10.1128/JCM.40.4.1326-1332.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.American Type Culture Collection [homepage on the internet] [Last accessed March 2, 2007];ATCC cultures, cell lines and hybridomas, cell biology collection search-CRL-1746 (ST cells), CCL-33 (PK15A cells), CCL-34 (MDCK cells) Available at http://www.atcc.org/common/catalog/cellBiology/cellBiologyIndex.cfm.
- 36.Sasseville AM, Boutin M, Gelinas AM, Dea S. Sequence of the 3′-terminal end (8.1 kb) of the genome of porcine haemagglutinating encephalomyelitis virus: Comparison with other haemagglutinating coronaviruses. J Gen Virol. 2002;83:2411–2416. doi: 10.1099/0022-1317-83-10-2411. [DOI] [PubMed] [Google Scholar]
- 37.Clavijo A, Tresnan DB, Jolie R, Zhou EM. Comparison of embryonated chicken eggs with MDCK cell culture for the isolation of Swine influenza virus. Can J Vet Res. 2002;66:117–121. [PMC free article] [PubMed] [Google Scholar]
- 38.Lin JH, Chen SP, Yeh KS, Weng CN. Mycoplasma hyorhinis in Taiwan: Diagnosis and isolation of swine pneumonia pathogen. Vet Microbiol. 2006;115:111–116. doi: 10.1016/j.vetmic.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Wellenberg GJ, Stockhofe-Zurwieden N, Boersma WJ, De Jong MF, Elbers AR. The presence of co-infections in pigs with clinical signs of PMWS in The Netherlands: A case-control study. Res Vet Sci. 2004;77:177–184. doi: 10.1016/j.rvsc.2004.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pogranichniy RM, Yoon KJ, Harms PA, Sorden SD, Daniels M. Case-control study on the association of porcine circovirus type 2 and other swine viral pathogens with postweaning multisystemic wasting syndrome. J Vet Diagn Invest. 2002;14:449–456. doi: 10.1177/104063870201400601. [DOI] [PubMed] [Google Scholar]
- 41.Kasorndorkbua C, Thacker BJ, Halbur PG, et al. Experimental infection of pregnant gilts with swine Hepatitis E virus. Can J Vet Res. 2003;67:303–306. [PMC free article] [PubMed] [Google Scholar]
- 42.Meng XJ, Halbur PG, Haynes JS, et al. Experimental infection of pigs with the newly identified swine Hepatitis E virus (swine HEV), but not with human strains of HEV. Arch Virol. 1998;143:1405–1415. doi: 10.1007/s007050050384. [DOI] [PubMed] [Google Scholar]
- 43.Matsuda H, Okada K, Takahashi K, Mishiro S. Severe hepatitis E virus infection after ingestion of uncooked liver from a wild boar. J Infect Dis. 2003;188:944. doi: 10.1086/378074. [DOI] [PubMed] [Google Scholar]
- 44.Yazaki Y, Mizuo H, Takahashi M, et al. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J Gen Virol. 2003;84:2351–2357. doi: 10.1099/vir.0.19242-0. [DOI] [PubMed] [Google Scholar]
- 45.Meng XJ, Purcell RH, Halbur PG, et al. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci USA. 1997;94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meng XJ, Dea S, Engle RE, et al. Prevalence of antibodies to the hepatitis E virus in pigs from countries where hepatitis E is common or is rare in the human population. J Med Virol. 1999;59:297–302. [PubMed] [Google Scholar]
- 47.de Boisseson C, Beven V, Bigarre L, et al. Molecular characterization of porcine circovirus type 2 isolates from post-weaning multi-systemic wasting syndrome-affected and non-affected pigs. J Gen Virol. 2004;85:293–304. doi: 10.1099/vir.0.19536-0. [DOI] [PubMed] [Google Scholar]
- 48.Carman S, McEwen B, DeLay J, et al. Porcine circovirus-2 associated disease in swine in Ontario (2004 to 2005) Can Vet J. 2006;47:761–762. [PMC free article] [PubMed] [Google Scholar]
- 49.Carman S, McEwen B, Delay J, Cai H, Fairles J. Porcine circovirus type 2-associated disease continued in fall of 2005. Animal Health Laboratory [page on the internet] [Last accessed 7 October 2006];AHL Newsletter. 2006 10(1) Available from http://www.labservices.uoguelph.ca/units/ahl/documents/ANwsl10-1.pdf.
- 50.Delay J, McEwen B, Carman S, van Dreumel T, Fairles J. Porcine circovirus type 2-associated disease is increasing. Animal Health Laboratory [page on the internet] [Last accessed 7 October 2006];AHL Newsletter. 2005 9(3) Available from http://www.labservices.uoguelph.ca/units/ahl/files/ANwsl9-3.pdf.
- 51.Ma ICM, Leung FCC. Genetic characterization and phylogenic analysis of porcine circovirus type 2 (PCV2) in Hong Kong (156) Proc 86th Annu Meet Conf Res Workers Anim Dis. 2005:35. [Google Scholar]