Abstract
The conformation of single laminin molecules adsorbed on synthetic substrates is directly observed making use of the phase magnitude in tapping mode atomic force microscopy (AFM). With AFM, it is not possible to differentiate the proteins on the substrate if use is made of the height signal, since the roughness of the material becomes of the same order of magnitude as the adsorbed protein, typically 10 nm height. This work shows how AFM can be exploited to reveal protein conformation on polymer materials. Different laminin morphologies are observed on a series of different copolymers based on ethyl acrylate and hydroxyethyl acrylate as a function of the surface density of −OH groups: from globular to completely extended morphologies of the protein molecules are obtained, and the onset of laminin network formation on some substrates can be clearly identified. The results stress the importance of the underlying synthetic substrate's surface chemistry for the biofunctional conformation of adsorbed proteins.
INTRODUCTION
When in vivo or in vitro with standard culture media, cells do not interact directly with the surface of a synthetic material, but with extracellular matrix (ECM) proteins somehow immobilized onto it (1,2). Cell adhesion involves different phenomena in which different biological molecules participate: ECM proteins, cell membrane proteins, and cytoskeleton proteins that interact to convey information, transcribe factors, and regulate gene expression (3). Cell adhesion is mediated by a family of transmembrane receptors, the most important being the integrin family, which recognize and bind specific amino-acid sequences along ECM proteins (4) such as the arginine-glycine-aspartic acid tripeptide (RGD), thought to be one of the main adhesion motifs in many ECM proteins, e.g., fibronectin, laminin, vitronectin (5). The concentration, distribution, and mobility of ECM proteins adsorbed on a surface thus play a fundamental role in the biofunctionality of a synthetic material, and are clue factors for the biological response of a substrate. In this work we show that the phase magnitude in tapping mode atomic force microscopy (AFM) is able to reveal single protein molecule conformations on polymer surfaces and we make use of it to study surface-induced changes in laminin adsorbed on copolymers of poly(ethyl acrylate-co-hydroxyethyl acrylate), P(EA-co-HEA). This system provides a family of materials with varying hydrophilicities, in which the surface density of hydroxyl groups can be varied through the ratio of both co-monomers in the material, while keeping the types of chemical functionalities unchanged throughout the series.
Laminins are trimeric molecules of α, β, and γ chains with molecular masses of 140–400 kDa. Several laminin isomorphs are known, with a large number of genetically distinct chains (α1 to α5, β1 to β3, and γ1 to γ3) (6). The laminins are important glycoprotein components of basement membranes, where they provide interaction sites for many other constituents, including cell surface receptors (7–9). Laminin plays an important role in neural cell migration, differentiation, and neurite growth (10–13), and it has been used as a coating for improving nerve cell adhesion and growth on different substrates (14–16).
ECM proteins can adopt different morphologies depending on the substrate onto which they adsorb. Protein adsorption on material surfaces is a process driven both by energy (several noncovalent interactions between the molecular groups of the substrate's surface and of the protein, such as hydrogen-bonding, electrostatic, or van der Waals interactions) and by entropy: the release of bound water molecules of the protein as it unfolds to adsorb on the surface means a significant entropy increase (17,18). Clearly, this second mechanism favors materials with hydrophobic character as better protein adherents, but it is the interplay of both mechanisms that determines the amount and the conformation of the adsorbed proteins. It is known that fibronectin, for example, adsorbs mainly on hydrophobic surfaces, and that its conformation depends on the hydrophilic degree of the surface (19). The von Willebrand factor adsorbs both on hydrophilic and on hydrophobic surfaces, but shows different molecular conformations that affect its function (20). An important factor in cell response is the way in which adhesion motifs are presented to the cell receptors: integrins are able to recognize differences induced by the substrates in the orientation, spacing, and microenvironment of the RGD motifs of the adsorbed proteins (21,22).
There are only few experimental methods available to study the surface-dependent conformations of adsorbed proteins. AFM is one of the most powerful ones, but it is in need of special conditions so as to generate adequate images, i.e., the substrate must possess a very smooth topography so that the height image is able to detect the protein molecule against it, typically some 10 nm height above the surface. This is the reason why, in AFM studies of protein conformation on substrates, only model surfaces have been used so far: α-macroglobulin on graphite (23), fibronectin on silica and mica (24), laminin and collagen on mica (25), and supramolecular assemblies (fibrillin and type VI collagen microfibrils) on silicon wafers and glass coverslips (26,27). Synthetic polymers are employed in many biotechnological processes in which the adsorbed protein layer interfaces to the biological media, and their surfaces differ greatly from those model surfaces. To our knowledge, the direct observation of ECM proteins on these commonly used materials has not been reported, mainly because surface roughness becomes of the same order of magnitude as the adsorbed protein. This work shows that the phase magnitude in tapping mode AFM is the experimental magnitude to be exploited to obtain significant information on protein conformation on polymer substrates.
MATERIALS AND METHODS
Substrates preparation
Copolymer sheets were prepared from a solution of the following monomers: ethyl acrylate (EA) (99% pure; Aldrich, Steinheim, Germany) and hydroxyethyl acrylate (HEA) (96% pure; Aldrich), with the desired proportion, using 0.1 wt % of benzoin (98% pure; Scharlau, Barcelona, Spain) as photoinitiator and 2 wt % of ethylene glycol dimethacrylate (98% pure; Aldrich) as crosslinking agent. The polymerization was carried out up to limiting-conversion. Five monomer feed compositions were chosen, given by the weight fraction of EA in the initial mixture of 1, 0.7, 0.5, 0.3, and 0. After polymerization, low molecular mass substances were extracted from the material by boiling in ethanol for 24 h and then drying in vacuo to constant weight.
Small disks (∼5 mm diameter) were cut from the polymerized sheets to be used in AFM.
Laminin adsorption
Laminin from Engelbreth-Holm-Swarm murine sarcoma basement membrane (L-2020, 1 mg/ml; Sigma, Steinheim, Germany) was adsorbed on the different substrates by immersing the material sheets in a 1:500 physiological solution (NaCl 0.9%) for 10 min. After that, the sample was dried by exposing its surface to a nitrogen flow for a few minutes.
Atomic force microscopy
In the tapping mode AFM, a cantilever oscillates with the probing tip close to its free resonance frequency with given amplitude. The interaction between the sample and the probe gives rise to a shift in the probe vibration respective to that measured in a free oscillation, i.e., with the probe far away from the sample. The vertical displacement (height) needed to keep the set amplitude provides information about the topography of the system. On the other hand, the measured phase shift may be caused by differences in the viscoelastic properties in different parts (or phases) of the sample, and in this sense it can provide some information about the morphology of the system. However, differences in phase lag may be caused by geometric features such as edges, etc., and can be a mere reflection of the topography of the system. There are several strategies for programming the apparatus parameters to obtain both accurate surface topographies (height) and morphologies (phase). Recent studies have shown that only when the amplitude of the vibrating cantilever is programmed to be equal to that of the free cantilever, does the height of the topography represent a true surface topography, and that a much harder tapping is necessary to observe maximum phase shift contrast between stiff and soft regions of the material (28).
AFM was performed in a NanoScope III from Digital Instruments (Santa Barbara, CA) operating in the tapping mode in air; Nanoscope 4.43r8 software version was used. Si-cantilevers from Veeco (Manchester, UK) were used with force constant of 2.8 N/m and resonance frequency of 75 kHz. The phase signal was set to zero at the resonance frequency of the tip. The tapping frequency was 5–10% lower than the resonance one. Drive amplitude was 200 mV and the amplitude setpoint Asp was 1.4 V. The ratio between the amplitude setpoint and the free amplitude Asp/A0 was kept equal to 0.7.
Several AFM images were analyzed to calculate the fraction of the surface area covered by the protein. The experimentally measured intensity images were converted to binary images by using Otsu's method, which chooses the threshold to minimize the interclass variance of the thresholded pixels (29).
RESULTS AND DISCUSSION
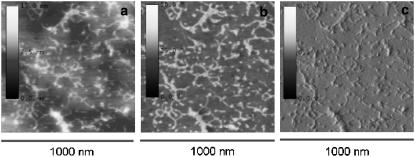
Fig. 1 shows the height, phase, and amplitude magnitudes on a very smooth surface (surface roughness: Rmax = 2.8 nm, root mean-square (RMS) = 0.3 nm) achieved for one of the copolymer samples (50:50). It must be understood as a control experiment in which the structure of the adsorbed laminin, as obtained by the phase mode, can be compared to those depicted on the height and amplitude modes. This experiment supports the idea that the phase magnitude is able to reveal the shape of the protein on the substrate. It is important since we are going to make use of it on normal (not so smooth) polymer surfaces, where the height magnitude cannot be used for revealing the protein morphology on the material.
FIGURE 1.
AFM images for the P(EA-co-HEA) 50:50 copolymer scanned on a very smooth area of the sample (Rmax = 2.8 nm, RMS = 0.3 nm). The three characteristic magnitudes of the tapping mode were taken simultaneously: height (a), phase (b), and amplitude (c). The image shows that the phase mode is able to reveal the conformation of the protein on the substrate as compared both with the height and amplitude magnitudes.
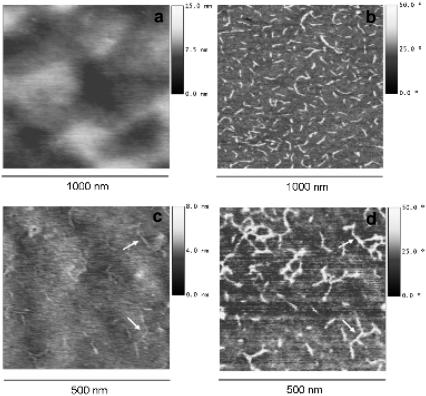
Fig. 2 shows the height and phase magnitudes for two of the copolymer samples. It is observed that while single protein molecules are clearly revealed in the phase signal picture (the dimension of a single laminin molecule is ∼70–90 nm length and <8 nm height), the roughness of the material, even if in the range of 15 nm nanometers (Table 1), masks the laminin molecules which consequently can hardly be distinguished from the substrate. Fig. 2, a and b, do not allow us to distinguish the laminin adsorbed on the substrate in the height magnitude. Nevertheless, in some cases (Fig. 2, c and d), the height magnitude is able to detect some evidence of the protein profile but it is not as sensitive as the phase magnitude. Similar results are obtained for the rest of copolymers. Contrary to what is obtained when studying protein conformations on very smooth surfaces (23–27), for normal polymer substrates with biomedical applications the height magnitude is not appropriate. By contrast, the phase magnitude is the feature capable of revealing the conformation of even single laminin molecules as a function of substrate chemistry.
FIGURE 2.
AFM images for the P(EA-co-HEA) 70:30 (a,b) and the 50:50 copolymers (c,d). The height image (left) shows a uniform surface in the 15 nm scale (a) and some evidence of the adsorbed protein (c), note the small arrows. The phase image (right) shows laminin adsorbed on the substrate. The maximum height scale is 15 nm, the scanned area is 1 × 1 μm.
TABLE 1.
Roughness parameters for the different samples calculated on 1 × 1 μm2 before laminin adsorption
| Sample | Rmax (nm) | RMS (nm) |
|---|---|---|
| PHEA | 24.3 | 3.1 |
| P(HEA-co-EA) 70/30 | 8.5 | 1.5 |
| P(HEA-co-EA) 50/50 | 10.6 | 1.1 |
| P(HEA-co-EA) 30/70 | 17.1 | 1.5 |
| PEA | 6.9 | 1.2 |
Root mean-square (RMS) and the difference in height between the highest and lowest point in the surface (Rmax).
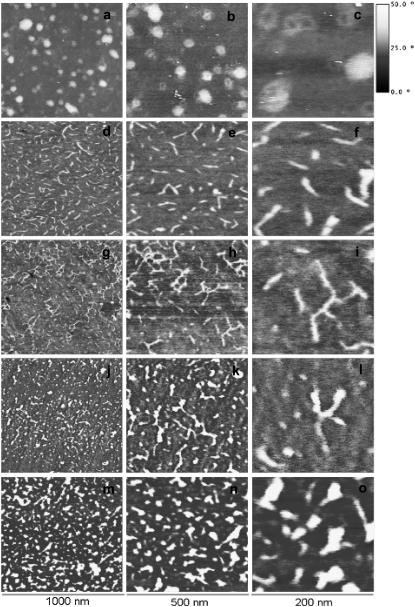
Fig. 3 shows the protein conformation after adsorption on the different copolymers. Protein molecules show globularlike morphology on the hydrophilic PHEA and gradually extend as the amount of −OH groups on the surface diminishes, up to a point in which the protein conformation tends again to a more compact, less extended conformation. Additionally, the formation of a laminin network takes place on the 50:50 copolymer in which the N-terminal domain of all three chains of the protein are linked (Fig. 3 g). This polymerized supramolecular aggregation is the typical form of laminin in the basement membrane and its formation depends on time, temperature, and concentration (30). The formation of protein networks on our surfaces must be conditioned not only by the different conformation of the molecule on the substrate, but also by the surface density of adsorbed protein.
FIGURE 3.
AFM phase images of laminin on the different copolymers at different magnifications. (a–c) PHEA homopolymer; (d–f) P(EA-co-HEA) 30:70; (g–i) P(EA-co-HEA) 50:50; (j–l) P(EA-co-HEA) 30:70; and (m–o) PEA homopolymer. The first column shows 1 × 1 μm, the second column shows 500 × 500 nm and the third one 200 × 200 nm. The vertical scale is the same for all images.
The copolymer substrates employed in this work are based on the random combination of ethyl acrylate and hydroxyethyl acrylate monomers, which have a vinyl backbone chain with the −COOCH2CH3 and −COOCH2CH2OH side groups, respectively, on them. Their copolymerization gives rise to a substrate in which the surface density of −OH groups can be modulated without modifying any other chemical structure in the system. The concentration of −OH groups determines the hydrophilicity of the substrate. The interaction of the protein domains with the chemical functionalities of the substrate and with water determines the molecule's adsorbed conformation. We have scanned the surface of the materials after immersion in the physiological solution (without laminin), and no effect was found due to the salts present in the physiological solution.
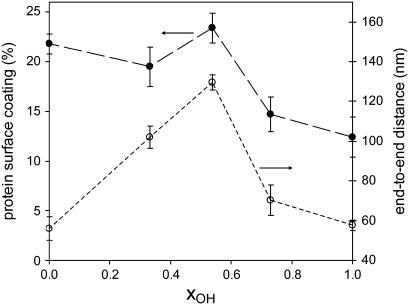
It is remarkable that the protein tends to spread on the substrate until it reaches its extended cross-shaped characteristic profile with three arms for the 50:50 sample (9). From that composition on, increasing or decreasing the −OH density in the material results in a less extended conformation of the protein, that ends in a globular conformation for both the purely hydrophilic (PHEA) and the purely hydrophobic (PEA) substrates. Fig. 4 shows the estimated average end-to end distance, the so-called displacement length, obtained from measurements on the AFM images as a function of the molar fraction of −OH groups in the copolymer, xOH. It ranges from ∼60 nm for the globular conformations of the protein up to ∼115 nm in the extended one, for the 50:50 sample. Even though the quantitative measurements in the phase magnitude might not reflect the accurate dimensions of the protein, the nonmonotonic dependence of the end-to-end distance as a function of the fraction of −OH groups in the surface of the substrate is evident. In explaining this complex behavior, several factors should be taken into account that might be influencing the interaction of specific sites on the laminin molecule with the polymer molecules: the relative amounts of the −CH2 and −CH3 hydrophobic groups and of the polar and more hydrophilic −COO and −OH groups, the number of water molecules in the substrate (which increases with its −OH content), and the spacing between those functionalities, which varies with the amount of absorbed water and thus is a function of the substrate's hydrophilicity.
FIGURE 4.
Fraction of the substrate surface coated with laminin (•) and average end-to-end distance (○) of the protein as a function of the molar fraction of −OH groups in the system as calculated from the AFM phase images. The error bars represent the mean ± SD after measuring at least 20 molecules on three different pictures. The line between points is only a guide to the eye.
The different conformations of laminin influence differently the biological performance of the substrates. The interaction between laminin domains and the cell membrane is regulated by several cell receptors (mainly integrins and nonintegrin binding proteins) (30). Some integrin receptors recognize specific peptide sequences in the protein, and their binding is strongly affected by the quantity, distribution, and spatial orientation on the substrate. Protein density on the substrate, as well as its conformation, modifies the availability of the binding sequences to cell receptors and influences the biological success of the artificial substrate. The copolymer series investigated in this work has already been used for in vitro culture of neural progenitors stemming from rat embryonic brain explants, and the best performance was found around the composition 50:50 (16). However, when Schwann cells were cultured on these substrates, better adhesion results were obtained for the more hydrophobic compositions (EA contents >80%) (31). These results suggest that the global conformation of the protein on the substrate is not the only parameter that influences cell adhesion: laminin shows similar globular conformation of the hydrophilic PHEA and the hydrophobic PEA, but cell adhesion only takes place on the hydrophobic component. Protein folding on these two substrates must be following different patterns that lead to externally similar globulelike protein conformations, but with differently presented binding domains exposed to integrins according to in vitro cell response (18,31).
The amount of adsorbed protein has been also estimated from the AFM phase images. Fig. 4 shows the fraction of the substrate surface covered by the protein as a function of the molar fraction of −OH groups in the material. The trend of the curve can be explained as the superposition of two independent mechanisms. On the one hand, the number of adsorbed proteins (per unit surface area) decreases as the hydrophilicity of the material increases. On the other hand, the specific area associated to each protein has the nonmonotonic dependence shown in Fig. 4 for the end-to-end distance. The curve depicted in Fig. 4 for the fraction of the material surface covered by the protein shows a maximum at approximately the same composition, as does the end-to-end distance curve, but superposed on a monotonically decreasing curve as surface density of −OH increases. A more thorough study of different adsorption times and of the adsorption kinetics as a function of surface chemistry is being performed.
CONCLUSIONS
Surface chemistry-dependent conformations of single laminin molecules were directly observed making use of the phase signal in tapping mode-AFM. This represents a major step in the investigation on surface-induced conformations of proteins since it allows one to observe single protein molecules on normal surfaces, i.e., in those in which the surface roughness is in the same order of magnitude as the protein height.
We made use of this technique to identify different laminin conformations on a set of copolymers in which the −OH surface density was modulated without changing any other chemical group. Protein molecules show globularlike morphology on the hydrophilic PHEA and gradually extend as the amount of −OH groups on the surface diminishes, up to a point in which the protein conformation tends again to a more compact, less extended conformation.
Acknowledgments
AFM was performed under the technical guide of the Microscopy Service at the Universidad Politécnica de Valencia. The specific software for the image analysis was programmed by Dr. David Moratal Pérez. Authors acknowledge the reviewer's comments and suggestions, which improved the quality of the manuscript.
The support of the Universidad Politécnica de Valencia through project No. 5696 and the Spanish Ministry of Science through project No. MAT2006-08120 (including the FEDER financial support) is kindly acknowledged.
Editor: Alberto Diaspro.
References
- 1.Boyan, B. D., T. W. Hummert, D. D. Dean, and Z. Schwartz. 1996. Role of material surfaces in regulating bone and cartilage cell response. Biomaterials. 17:137–146. [DOI] [PubMed] [Google Scholar]
- 2.Anselme, K. 2000. Osteoblast adhesion on biomaterials. Biomaterials. 21:667–681. [DOI] [PubMed] [Google Scholar]
- 3.Alberts, B., A. Johnson, J. Lewis, M. Raff, K. Roberts, and P. Walter. 2004. Molecular Biology of the Cell. Garland Science, NY.
- 4.Hynes, R. O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell. 110:673–687. [DOI] [PubMed] [Google Scholar]
- 5.Yamada, K. M. 1991. Adhesive recognition sequences. J. Biol. Chem. 266:12809–12812. [PubMed] [Google Scholar]
- 6.Burgeson, R. E., M. Chiquet, D. Deutzmann, P. Ekblom, J. Engel, H. Kleinman, G. R. Martin, G. Meneguzzi, M. Paulsson, J. Sanes, R. Timple, K. Tryggvason, Y. Yamada, and P. D. Yurchenco. 1994. A new nomenclature for the laminins. Matrix Biol. 14:209–211. [DOI] [PubMed] [Google Scholar]
- 7.Mercurio, A. M. 1995. Laminin receptors: achieving specificity through cooperation. Trends Cell Biol. 5:419–423. [DOI] [PubMed] [Google Scholar]
- 8.Beck, K., I. Hunter, and J. Engel. 1990. Structure and function of laminin: anatomy of a multidomain protein. FASEB J. 4:148–160. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki, T., R. Fässler, and E. Hohenester. 2004. Laminin: the crux of membrane assembly. J. Cell Biol. 164:959–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleiman, H. K., F. B. Cannon, G. W. Laurie, J. R. Hasell, M. Aumailley, V. P. Terranova, G. R. Martin, and M. DuBois-Dalcq. 1987. Biological activities of laminin. J. Cell. Biochem. 27:317–325. [DOI] [PubMed] [Google Scholar]
- 11.Heiduschka, P., I. Romann, H. Ecken, M. Schöning, W. Schuhmann, and S. Thanos. 2001. Defined adhesion and growth of neurons on artificial structured substrates. Electrochim. Acta. 47:299–307. [Google Scholar]
- 12.Luckenbill-Edds, L. 1997. Laminin and the mechanism of neuronal outgrowth. Brain Res. Rev. 26:2983–2990. [DOI] [PubMed] [Google Scholar]
- 13.He, W., and R. V. Bellamkonda. 2005. Nanoscale neuro-integrative coatings for neural implants. Biomaterials. 26:2983–2990. [DOI] [PubMed] [Google Scholar]
- 14.Rogers, S. L., P. C. Letorneau, S. L. Palm, J. McCarthy, and L. T. Furcht. 1983. Neurite extension by peripheral and central neuron system neurons in response to substratum-bound fibronectin and laminin. Dev. Biol. 98:212–220. [DOI] [PubMed] [Google Scholar]
- 15.Liesi, P., D. Dahl, and A. Vaheri. 1984. Neurons cultured from developing rat brain attach and spread preferentially to laminin. J. Neurosci. Res. 11:241–251. [DOI] [PubMed] [Google Scholar]
- 16.Soria, J. M., C. Martínez Ramos, M. Salmerón Sánchez, B. Benavent, A. Campillo-Férnandez, J. L. Gómez Ribelles, J. M. Garcia Verdugo, M. Monleón Pradas, and J. A. Barcia. 2006. Survival and differentiation of embryonic neural explants on different biomaterials. J. Biomed. Mater. Res. 79A:495. [DOI] [PubMed] [Google Scholar]
- 17.García, A. J. 2006. Interfaces to control cell-biomaterial adhesive interactions. Adv. Polym. Sci. 203:171–190. [Google Scholar]
- 18.Werner, C., T. Pompe, and K. Salchert. 2006. Modulating extracellular matrix at interfaces of polymeric materials. Adv. Polym. Sci. 203:63–93. [Google Scholar]
- 19.García, A. J., M. D. Vega, and D. Boettiger. 1999. Modulation of cell proliferation and differentiation through substrate-dependent changes in fibronectin conformation. Mol. Biol. Cell. 10:785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raghavachari, M., H. M. Tsai, K. Kottke-Marchant, and R. E. Marchant. 2000. Surface dependents structures of von Willebrand factor observed by AFM under aqueous conditions. Colloids Surf. B Biointerf. 19:315–324. [DOI] [PubMed] [Google Scholar]
- 21.Pierschbacher, M. D., and E. Rouslahti. 1987. Influence of stereochemistry of the sequence Arg-Gly-Asp-Xaa on binding specificity in cell adhesion. J. Biol. Chem. 262:17294–17298. [PubMed] [Google Scholar]
- 22.Houseman, B. T., and M. Mrksich. 2001. The microenvironment of immobilized Arg-Gly-Asp peptides is an important determination of cell adhesion. Biomaterials. 22:943–955. [DOI] [PubMed] [Google Scholar]
- 23.Arakawa, H., K. Umemura, and A. Ikai. 1992. Protein images obtained by STM, AFM and TEM. Nature. 358:171. [DOI] [PubMed] [Google Scholar]
- 24.Bergkvist, M., J. Carlsson, and S. Oscarsson. 2003. Surface-dependent conformations of human plasma fibronectin adsorbed to silica, mica and hydrophobic surfaces, studied with use of atomic force microscopy. J. Biomed. Mater. Res. 64A:349–356. [DOI] [PubMed] [Google Scholar]
- 25.Chen, C. H., and H. G. Hansma. 2000. Basement membrane macromolecules: insights from atomic force microscopy. J. Struct. Biol. 131:44–55. [DOI] [PubMed] [Google Scholar]
- 26.Sherrat, M. J., D. F. Holmes, C. A. Shuttleworth, and C. M. Kielty. 2004. Substrate-dependent morphology of supramolecular assemblies: fibrillin and type-VI collagen microfibrils. Biophys. J. 3211–3222. [DOI] [PMC free article] [PubMed]
- 27.Sherrat, M. J., D. V. Bax, S. S. Chaudhry, N. Hodson, J. R. Lu, P. Saravanapavan, and C. M. Kielty. 2005. Substrate chemistry influences the morphology and biological function of adsorbed extracellular matrix assemblies. Biomaterials. 26:7192–7206. [DOI] [PubMed] [Google Scholar]
- 28.Knoll, A., R. Magerle, and G. Krausch. 2001. Tapping mode atomic force microscopy on polymers: where is the true sample surface? Macromolecules. 34:4159–4165. [Google Scholar]
- 29.Otsu, N. 1979. A threshold selection method for gray level histograms. IEEE T. Syst. Man. Cy. 9:62–66. [Google Scholar]
- 30.Luckenbill-Edds, L. 1997. Laminin and the mechanism of neuronal outgrowth. Brain Res. Rev. 23:1–27. [DOI] [PubMed] [Google Scholar]
- 31.Soria, J. M., C. Martínez Ramos, O. Bahamonde, D. M. Garcia Cruz, M. Salmerón Sánchez, M. A. García Ezparza, C. Casas, M. Guzman, X. Navarro, J. L. Gómez Ribelles, J. M. Garcia Verdugo, M. Monleón Pradas, and J. M. Barcia. 2006. Influence of the substrate's hydrophilicity on the in vitro Schwann cells viability. J. Biomed. Mater. Res. A. In press. [DOI] [PubMed]