Abstract
Adsorption of λ-phage on sensitive bacteria Escherichia coli is a classical problem but not all issues have been resolved. One of the outstanding problems is the rate of adsorption, which in some cases appears to exceed the theoretical limit imposed by the law of random diffusion. We revisit this problem by conducting experiments along with new theoretical analyses. Our measurements show that upon incubating λ-phage with bacteria Ymel, the population of unbound phage in a salt buffer decreases with time and in general obeys a double-exponential function characterized by a fast (τ1) and a slow (τ2) decay time. We found that both the fast and the slow processes are specific to interactions between λ-phage and its receptor LamB. Such specificity motivates a kinetic model that describes the interaction between the phage and the receptor as an on-and-off process followed by an irreversible binding. The latter may be a signature of the initiation of DNA translocation. The kinetic model successfully predicts the double exponential behavior seen in the experiment and allows the corresponding rate constants to be extracted from single measurements. The weak temperature dependence of the reversible and the irreversible binding rate suggests that phage retention by the receptor is entropic in nature and that a molecular key-lock interaction may be an appropriate description of the interaction between the phage tail and the receptor.
INTRODUCTION
Bacteriophages are viruses that infect bacteria and were first identified by Frederick Twort and Felix d'Herelle in the early 20th century. In subsequent years, studies of bacteriophage have played an essential role in our fundamental understanding of the molecular underpinning of live phenomena, such as the hereditary nature of DNA and the central dogma of molecular biology (1–3). One of the key steps in bacterial infection is the binding of the viral particle to the outer membrane of a bacterium. For many strains of bacteria and phage, such a binding event requires specific interactions between the phage tail and the bacterial receptors (4). Such specificity can be demonstrated on the grounds that when the receptors are absent or modified, the phage is unable to bind and the bacterium is immune to infection.
Phage adsorption on host bacteria has been studied for many years, leading one to believe that all interesting problems have been solved. This certainly is not the case. It had been realized in early studies (5,6) that phage binding is highly efficient with the rate of adsorption approaching the theoretical limit determined by diffusion models (7). An influential study with bacteria having a defined surface density of receptors suggests that the adsorption rate may even exceed the theoretical limit given the geometrical parameters of the bacterium and the phage (6,7). The effect is remarkable in that despite their small size (radius s ∼ 4 nm) and being sparsely populated on the bacterial cell wall, receptors appear to capture phage on each collision (6). A similarly high capture efficiency was also found in insect pheromone chemoreception systems (8) and has been interpreted in terms of thermal diffusion of sex attractants (9). Adam and Delbruck (10) attempted to explain this fast adsorption by proposing a two-stage capture process, also known as the reaction rate enhancement by dimensional reduction (RREDR). According to this scenario, a phage particle first encounters a bacterium by diffusion in the three-dimensional (3D) fluid. Once adsorbed to the bacterium, the phage undergoes a “random walk” on the bacterial surface, which was modeled as a two-dimensional (2D) fluid, until it is captured by a receptor. This 3D+2D searching strategy was shown, under certain conditions, to be more efficient than searching in 3D space alone. Adam and Delbruck's work was reanalyzed by Berg and Purcell (7) and quantitative bounds were placed on the model. Specifically, Berg and Purcell found that, for the 2D diffusion to be advantageous for phage binding, the adsorption energy should be strong enough to keep the phage on the bacterial wall but weak enough such that the 2D diffusion is not impeded by the interaction. Their model was also the first to predict the adsorption rate k as a function of number of receptors N on a bacterium, and it was shown that the maximum adsorption rate kmax was noticeably less than that reported in Schwartz (6). To reconcile the apparent discrepancy between theory and experiment, Berg and Purcell (7) suggested that bacterial swimming might be the cause for the anomalously fast adsorption seen in the experiment. Since molecular recognition and binding are critical for all biological functions, there are reasons to believe that RREDR may be ubiquitous in all biological systems. A survey of a variety of binding pairs mediated by intracellular membranes has been carried out by McCloskey and Poo (11). However, this study showed no conclusive evidence supporting the notion of RREDR.
This work is motivated in part by the outstanding issues delineated above and in part by the realization that the problem has became more tractable in recent years due to new information accumulated over the past decades on the structure and function of the maltose system (12,13). The high-resolution 3D structures of LamB, the maltoporin hijacked by λ-phage for infection, is now available (14) and its critical sites for phage binding are genetically mapped (15,16). Herein, we report new measurements and analyses of λ-phage adsorption on the Escherichia coli (E. coli) strain Ymel, which carries the wild-type λ-receptor. Our measurement shows that upon incubating λ-phage with Ymel in a defined salt solution (10 mM of MgSO4 and pH 7.4), the free phage initially decrease in number rapidly followed by a much slower decline. We found that while the fast decay depends strongly on the bacterial concentration, the slower process does not. This observation suggests that there are multiple steps in phage inactivation, and it is enticing to associate these steps with RREDR. However, careful analyses of the data and in particular a control experiment using CR63 that carries dysfunctional λ-receptors ruled out such a possibility. Specifically, we found that CR63 shows no discernible phage adsorption using the same protocol, indicating that depletion of phage particles from the bulk solution cannot be a result of nonspecific binding on bacterial membrane. In other words, if membrane adsorption and subsequent 2D diffusion are significant for the uptake of phage particles, these intermediate steps are not sensitive to our experimental detection. The observed receptor specificity also suggests that the long-term inactivation of λ-phage must be due to interactions between phage receptor LamB and the protein J of the phage tail (17).
A kinetic model for phage adsorption is proposed, consisting of reversible adsorption and desorption, and irreversible binding. These processes are characterized respectively by three rate constants k, k′, and k″. Given the experimental condition of low multiplicity of infection (moi), the model can be solved analytically. It is shown that all the rate constants can be extracted from single measurements under different conditions. Our measured adsorption rate constant k is lower than the theoretical limit imposed by random diffusion and allows us to deduce the average number of receptors per bacterium, N ∼ 300, using Berg and Purcell's model (7). The surprising finding of this study is that the very slow reversible and irreversible binding processes (1/k′ ∼ 1/k″ ∼ 1000 s) are nearly independent of temperature. These observations suggest that translocation of phage DNA to the host cells must be at least as long as 1/k″ for the Ymel strain and that the receptor-bound phage are likely to be trapped by an entropic force. A simple calculation shows that the entropic barrier for the phage particle to be released or permanently bound is ∼15 kBT.
This article is organized in the following fashion. We begin by a discussion of experimental procedures and characterizations of bacterial and phage strains by light scattering and by optical microscopy. The main body of the article, the middle two sections, describes the experimental findings and the interpretations of the results. In particular, the predictions of our theoretical model are also presented there (a more detailed derivation can be found in the Appendix). The final section contains the summary and possible future experiments.
MATERIALS AND PROCEDURES
Bacteriophage λ (cI857, sam7) acquired from the New England BioLabs (Ipswich, MA) was used to infect the bacterial strain Ymel. The λ-phage targets the outer membrane protein LamB (a porin for maltose and maltodextrin transport (18)) for binding and possibly for DNA translocation (13). The λ-phage were purified using cesium-chloride density-gradient centrifugation (38,000 rpm, 16 h, 18°C) and dialyzed three times in the λ-dilution buffer (10 mM MgSO4, 20 mM Tris buffer, and pH 7.4). The phage stock typically contained ∼1011–1012 plaque forming units per milliliter, and was stored at 4°C. The activity of the phage stock was routinely checked, and no appreciable decay (less than a factor of 10) was observed during the period of approximately six months.
Two different bacterial strains, Ymel (F+mel-1 supF58 λ−) and CR63 (F+supD60 lamB63) (19), were used for the adsorption measurements. Both strains were derived from E. coli K12. Standard protocols were used for bacterial culture, which consists of overnight growth of Ymel/CR63 in M9 media supplemented with either 0.4% w/v glucose or maltose. The bacteria were inoculated at 1:100 in the fresh medium and grown for 4 h to the midexponential phase (OD550 ∼ 0.3 and cell density ∼ 3 × 108 cm−3). The liquid cultures were maintained at 37°C and constantly shaken at 200 rpm. Earlier experiments by Schwartz (6) showed that when bacteria were grown in glucose, the LamB receptors were expressed at the basal level (N ∼ 10) whereas in the presence of maltose, an elevated expression of LamB (N ∼ 3000) was found. However, in our experiment, different carbon sources appear to show little difference in phage adsorption, suggesting that maltose alone may not be sufficient in stimulating LamB expression in the Ymel strain we used.
The bacteria were washed and resuspended in the λ-buffer (10 mM MgSO4 adjusted to pH 7.4) before the adsorption measurements. To avoid phage loss due to adsorption to container walls, all reactions were carried out in glass tubes instead of plastic. The adsorption curves were generated by adding phage to samples of a fixed bacterial concentration at specific time intervals, and the reactions were terminated simultaneously for all samples by a 54× dilution using the λ-buffer. This procedure enabled adsorption measurements to be carried out within a time span as short as 1 min. The phage particles were mixed with the bacteria at moi 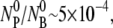 where
where 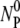 and
and 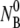 are, respectively, the initial phage and the bacterial concentrations, and
are, respectively, the initial phage and the bacterial concentrations, and 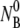 is typically ∼108 cm−3. The reactions were carried out in a constant temperature bath (model No. RTE-110; Neslab Instruments, Portsmouth, NH) with temperature T being controlled to within 1°C during the measurement. The diluted samples were centrifuged at 16,000 RCF for 1 min, and the concentrations of free phage in the supernatants were determined on soft-agar plates using Ymel as indicator bacteria.
is typically ∼108 cm−3. The reactions were carried out in a constant temperature bath (model No. RTE-110; Neslab Instruments, Portsmouth, NH) with temperature T being controlled to within 1°C during the measurement. The diluted samples were centrifuged at 16,000 RCF for 1 min, and the concentrations of free phage in the supernatants were determined on soft-agar plates using Ymel as indicator bacteria.
To ascertain that Ymel carries functional wild-type LamB receptors, we have performed sequence analyses of that stretch of DNA on the bacterial genome. The lamB gene was amplified by colony PCR using pfuUltraII (Cat. No. 600670; Stratagene, La Jolla, CA) and the following two primers:
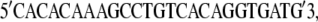 |
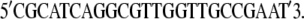 |
These DNA sequences are located 58 bps upstream and 55 bps downstream, respectively, of the lamB gene (20). The PCR product was sequenced at the DNA Sequencing Center of The University of Pittsburgh School of Medicine, Biomedical Research Support Facilities. The primers used for forward sequencing were
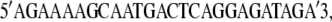 |
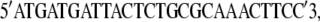 |
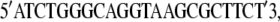 |
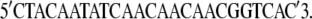 |
The first 400 bps were also sequenced in reverse using the reverse primer 5′GAAGTCGATCATATGAACGTCATG′3 to ensure the accuracy of the first 50 bps. The sequence was then compared to the known sequence of the MG1655 strain as described in the ASAP database (20). The match was 100%, indicating no mutation on the lamB gene of Ymel.
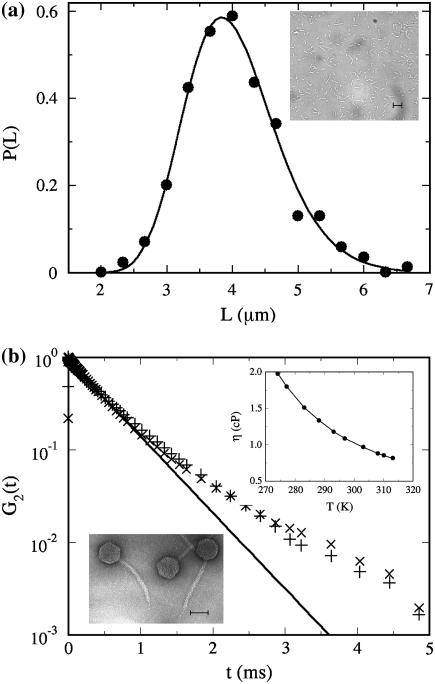
We used bright-field optical microscopy to characterize the bacterial size for Ymel grown to the midexponential phase. Fig. 1 a shows a typical image of the glucose-grown cells along with the size distribution measured with 300 bacteria. The picture indicates that the cell length is varied, which is typical of an actively dividing bacterial culture. The measurements yielded the average length of the bacterium to be 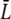 = 2a = 3.8 ± 0.8 μm and the average diameter 2b = 0.8 μm, where a and b are the semimajor and the semiminor axes of the bacterium. For a quantitative comparison with theory, it is also useful to know the diffusion coefficient of λ-phage. We measured this quantity directly using a quasi-elastic light scattering apparatus equipped with an ALV-5000 digital autocorrelator and an HeNe laser. The scattered light intensity from the phage particles was detected by an avalanche diode. Using different scattering angles, we consistently obtained the diffusion coefficient for the λ-phage to be D3 = 6.2 × 10−8 cm2/s at T = 25°C. A typical autocorrelation function measured at the 90° scattering angle is given in Fig. 1 b. We also determined the shear viscosity η of the λ-buffer over a broad range of temperatures, 1 < T < 40°C, using a viscometer (Cannon, State College, PA). From the measured D3 and η, the hydrodynamic radius R of the λ-phage particles was calculated based on the Stokes-Einstein relation D3 = kBT/6πηR. This yielded R = 35 nm, which is comparable to the size of the λ-phage head determined by electron microscopy (21).
= 2a = 3.8 ± 0.8 μm and the average diameter 2b = 0.8 μm, where a and b are the semimajor and the semiminor axes of the bacterium. For a quantitative comparison with theory, it is also useful to know the diffusion coefficient of λ-phage. We measured this quantity directly using a quasi-elastic light scattering apparatus equipped with an ALV-5000 digital autocorrelator and an HeNe laser. The scattered light intensity from the phage particles was detected by an avalanche diode. Using different scattering angles, we consistently obtained the diffusion coefficient for the λ-phage to be D3 = 6.2 × 10−8 cm2/s at T = 25°C. A typical autocorrelation function measured at the 90° scattering angle is given in Fig. 1 b. We also determined the shear viscosity η of the λ-buffer over a broad range of temperatures, 1 < T < 40°C, using a viscometer (Cannon, State College, PA). From the measured D3 and η, the hydrodynamic radius R of the λ-phage particles was calculated based on the Stokes-Einstein relation D3 = kBT/6πηR. This yielded R = 35 nm, which is comparable to the size of the λ-phage head determined by electron microscopy (21).
FIGURE 1.
(a) The size distribution of Ymel grown to the midexponential phase (3.5 h) in the M9 medium supplemented with 0.4% glucose. The inset shows a typical image with the scale bar of 5 μm. The size distribution can be mimicked reasonably well by a log-normal distribution 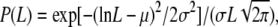 as delineated by the solid line in the figure. Here μ = 1.38 is the mean of ln(L) and σ = 0.2 is its standard deviation. (b) The scattered light intensity-intensity autocorrelation function G2(t) = 〈I(t′)I(t′ + t)〉/〈I(t′) 〉2−1 measured using λ-phage (pluses) and λ-phage heads (crosses) in the λ-buffer. The measurement was carried at room temperature T = 24°C and at the scattering angle of θ = 90°. This corresponds to the scattering wavenumber q(= 4πnsin(θ/2)/Λ) = 1.87 × 105 cm−1, where n is the index of refraction of water and Λ = 633 nm is the wavelength of the HeNe laser. The data shows that the phage tail does not contribute significantly to the decay of the autocorrelation function; i.e., the diffusion constant is essentially determined by the phage head. As can also be seen in the figure, for early times G2(t) decays exponentially and can be compared to the theoretical predictions for spheres diffusing in a medium with viscosity η: G2(t) = exp(−2D3q2t), where D3 = kBT/6πηR is the diffusion coefficient for the sphere of radius R. This measurement yields the hydrodynamic radius R = 35 nm. The inset in the upper corner is the plot of the measured shear viscosity η for the λ-buffer, which is essentially the same as water. The lower inset is an electron microscopy image of intact λ-phage. Here the scale bar is 50 nm (courtesy of R. Hendrix).
as delineated by the solid line in the figure. Here μ = 1.38 is the mean of ln(L) and σ = 0.2 is its standard deviation. (b) The scattered light intensity-intensity autocorrelation function G2(t) = 〈I(t′)I(t′ + t)〉/〈I(t′) 〉2−1 measured using λ-phage (pluses) and λ-phage heads (crosses) in the λ-buffer. The measurement was carried at room temperature T = 24°C and at the scattering angle of θ = 90°. This corresponds to the scattering wavenumber q(= 4πnsin(θ/2)/Λ) = 1.87 × 105 cm−1, where n is the index of refraction of water and Λ = 633 nm is the wavelength of the HeNe laser. The data shows that the phage tail does not contribute significantly to the decay of the autocorrelation function; i.e., the diffusion constant is essentially determined by the phage head. As can also be seen in the figure, for early times G2(t) decays exponentially and can be compared to the theoretical predictions for spheres diffusing in a medium with viscosity η: G2(t) = exp(−2D3q2t), where D3 = kBT/6πηR is the diffusion coefficient for the sphere of radius R. This measurement yields the hydrodynamic radius R = 35 nm. The inset in the upper corner is the plot of the measured shear viscosity η for the λ-buffer, which is essentially the same as water. The lower inset is an electron microscopy image of intact λ-phage. Here the scale bar is 50 nm (courtesy of R. Hendrix).
EXPERIMENTAL FINDINGS
Phage adsorption depends on magnesium sulfate concentrations
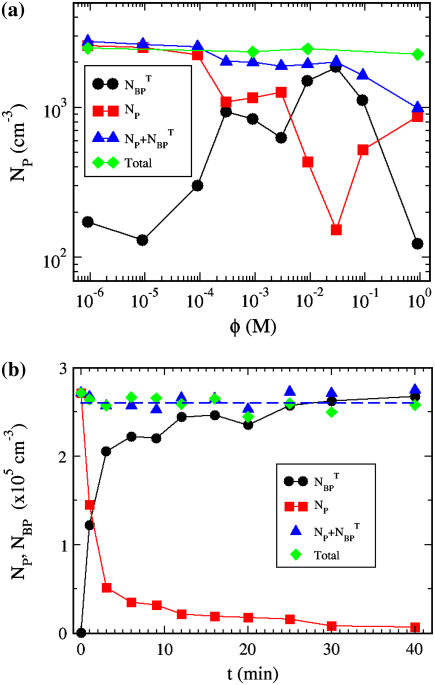
In this experiment, we attempted to establish the optimum magnesium sulfate (MgSO4) concentration for phage adsorption and to evaluate typical errors involved when the titering technique is used to measure the phage concentration. A set of measurements was carried out in the λ-adsorption buffers with different MgSO4 concentrations, in the range 10−6 < φ < 1 M. The reactions were carried out at room temperature and each lasted for 6 min. The phage that were adsorbed onto bacteria, which we call the bacterium-phage complexes 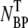 (≡ NBP + NBP*), and the phage that remained in the buffer, which we call the free phage NP, were separated by centrifugation. The pellet resulting from centrifugation includes both transiently bound NBP and permanently bound NBP* complexes. The concentrations of the two populations,
(≡ NBP + NBP*), and the phage that remained in the buffer, which we call the free phage NP, were separated by centrifugation. The pellet resulting from centrifugation includes both transiently bound NBP and permanently bound NBP* complexes. The concentrations of the two populations, 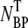 and NP, were measured separately on the indicator bacterial lawns. As shown in Fig. 2 a, the adsorption depends strongly on φ in a highly nonlinear fashion. For low MgSO4 concentrations (φ < 10−4 M), there is almost no adsorption as indicated by the low complex counts
and NP, were measured separately on the indicator bacterial lawns. As shown in Fig. 2 a, the adsorption depends strongly on φ in a highly nonlinear fashion. For low MgSO4 concentrations (φ < 10−4 M), there is almost no adsorption as indicated by the low complex counts 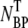 (circles) in the pellets. Phage adsorption increases and reaches a local maximum at φ ∼ 2 × 10−4 M and a global maximum at φ ∼ 3 × 10−2 M. For φ > 0.1 M, the adsorption decreases rapidly. We also noticed in Fig. 2 a that there is a strong correlation between the bacterium/phage complexes
(circles) in the pellets. Phage adsorption increases and reaches a local maximum at φ ∼ 2 × 10−4 M and a global maximum at φ ∼ 3 × 10−2 M. For φ > 0.1 M, the adsorption decreases rapidly. We also noticed in Fig. 2 a that there is a strong correlation between the bacterium/phage complexes 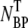 (circles) and the free phage NP (squares), i.e., when one increases the other decreases, or vice versa. Though such a correlation is expected for the low moi, the observation is reassuring, suggesting that the measurement noise is below the systematic change of adsorption as φ is varied. The sum of the two measurements (
(circles) and the free phage NP (squares), i.e., when one increases the other decreases, or vice versa. Though such a correlation is expected for the low moi, the observation is reassuring, suggesting that the measurement noise is below the systematic change of adsorption as φ is varied. The sum of the two measurements (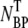 + NP) is delineated by the triangles in Fig. 2 a, which is reasonably constant for φ < 0.1 M and is also consistent with the initial phage concentration
+ NP) is delineated by the triangles in Fig. 2 a, which is reasonably constant for φ < 0.1 M and is also consistent with the initial phage concentration 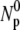 (diamonds) in the reaction. For φ > 0.1 M, the total number of phage in the reaction volume decreases considerably. This may be a result of either the phage being unstable or because the bacteria cannot be infected at high-salt conditions. Based on these observations, the MgSO4 concentration was fixed at 10−2 M for the rest of the measurements. At this concentration, the Debye screening length
(diamonds) in the reaction. For φ > 0.1 M, the total number of phage in the reaction volume decreases considerably. This may be a result of either the phage being unstable or because the bacteria cannot be infected at high-salt conditions. Based on these observations, the MgSO4 concentration was fixed at 10−2 M for the rest of the measurements. At this concentration, the Debye screening length 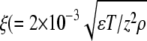 (nm)) is ∼1.5 nm, which is smaller than the receptor radius s = 4 nm, where ɛ = 78 is the dielectric constant of water, T = 300 K is the temperature, z = 2 is the valence of the ion, and ρ is the molar concentration of the ion. The above observation suggests the importance of electrostatic interactions in λ-phage binding to the LamB receptors. Such electrostatic effect in phage adsorption appears to be common in different phage/bacterium systems and has been recognized by early phage workers (22).
(nm)) is ∼1.5 nm, which is smaller than the receptor radius s = 4 nm, where ɛ = 78 is the dielectric constant of water, T = 300 K is the temperature, z = 2 is the valence of the ion, and ρ is the molar concentration of the ion. The above observation suggests the importance of electrostatic interactions in λ-phage binding to the LamB receptors. Such electrostatic effect in phage adsorption appears to be common in different phage/bacterium systems and has been recognized by early phage workers (22).
FIGURE 2.
Phage adsorption is strongly dependent on salt concentrations. (a) The magnesium salt is varied over four decades in concentrations φ and the free λ-phage (squares) are titered after incubating with Ymel bacteria for 6 min in each salt concentration. The different symbols (squares, circles, triangles, and diamonds) correspond to free phage, bacterium/phage complexes, the sum of free phage and bacterium/phage complexes, and initial phage concentration, respectively. (b) The total bacterium/phage complex 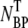 = NBP + NBP* (circles) and free phage NP (squares) concentrations as a function of adsorption time t. The MgSO4 concentration is 10 mM. All measurements in panels a and b were carried out in room temperature.
= NBP + NBP* (circles) and free phage NP (squares) concentrations as a function of adsorption time t. The MgSO4 concentration is 10 mM. All measurements in panels a and b were carried out in room temperature.
To confirm that the adsorption measurements can be carried out over a longer period of time without degradation to phage and/or without phage being depleted by other extraneous sinks (such as the container walls and multiple pipetting), additional control measurements were carried out as a function of time. Both bacterium-phage complexes (circles) and free phage (squares) were titered at different time intervals; the result is displayed in Fig. 2 b. We noted that the free phage concentration NP(t) decreases with time whereas the complexes 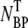 (t) increases with time, demonstrating again a rather strong correlation between the two populations. The sum of NP(t) and
(t) increases with time, demonstrating again a rather strong correlation between the two populations. The sum of NP(t) and 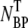 (t) is plotted as triangles and they yield a constant value that is consistent with the initial phage input
(t) is plotted as triangles and they yield a constant value that is consistent with the initial phage input 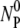 (diamonds) in the reaction. Thus, these control runs demonstrate that the total number of phage particles during the reaction and preparation is conserved NP(t) +
(diamonds) in the reaction. Thus, these control runs demonstrate that the total number of phage particles during the reaction and preparation is conserved NP(t) + 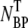 (t) =
(t) = 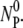 Since NP and
Since NP and 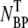 yield essentially similar information, only the free phage population Np(t) was measured in the subsequent experiments. To increase the measurement accuracy,
yield essentially similar information, only the free phage population Np(t) was measured in the subsequent experiments. To increase the measurement accuracy, 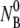 and
and 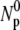 was determined on multiple plates. Exercising these procedures carefully, we found the dominant uncertainty was the run-to-run variations, which amount to ∼30%.
was determined on multiple plates. Exercising these procedures carefully, we found the dominant uncertainty was the run-to-run variations, which amount to ∼30%.
Phage adsorption depends on temperature and the growth medium
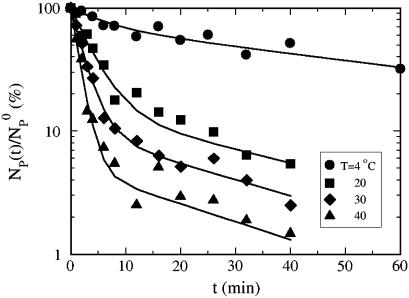
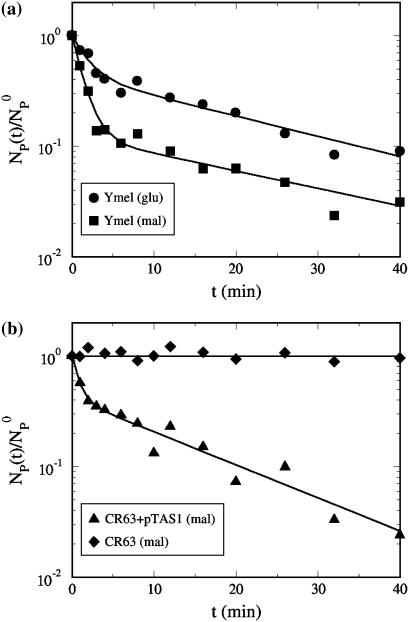
Fig. 3 displays the normalized concentration of the free phage NP(t)/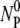 as a function of time for four different temperatures, T = 4, 20, 30, and 40°C. For all temperatures, the phage concentration decreases with time as phage particles are gradually adsorbed onto bacteria. One can observe that the rate of adsorption is strongly affected by T, i.e., the percentage of the free phage being removed from the reaction volume increases significantly (∼30 times) as T increases from 4 to 40°C. The adsorption curves also show interesting features; they all decay rapidly initially and then slowly over long times. Motivated by the earlier findings that the expression level of LamB can be significantly altered by the type of carbon sources present in the growth medium, we grew the Ymel bacteria in M9 supplemented with 0.4% maltose (maximum induction) and 0.4% glucose (minimum induction) (6,13). For comparison, Fig. 4 a displays two sets of data taken at T = 24°C for maltose- and glucose-grown Ymel cells. We noticed that adsorption in maltose-grown cells (squares) is slightly stronger than for glucose-grown cells (circles) as indicated by the faster initial decay. The long-time behavior, however, is essentially the same. This observation is consistent with the expectation that maltose causes cells to produce more receptors, but the extent is not as large as reported for other bacterial strains (Hfr G6, Hfr 3000), where orders-of-magnitude increase in the receptor expression level were observed after induction (6). In our experiment, despite quantitatively different adsorption curves due to varying temperature or growth conditions, a unifying feature is that the phage adsorption kinetics is nonexponential and appears to contain at least two different timescales. It is suggestive that the short- and the long-time decays might correspond to the two-step process proposed by Adam and Delbruck (10). However, the detailed analyses below indicate that this is not the case. Since adsorption curves for the maltose- and the glucose-grown cells are qualitatively the same, in what follows, only the data from the glucose-grown cells are presented. The rate constants for bacteria grown in both media are summarized in Table 1.
as a function of time for four different temperatures, T = 4, 20, 30, and 40°C. For all temperatures, the phage concentration decreases with time as phage particles are gradually adsorbed onto bacteria. One can observe that the rate of adsorption is strongly affected by T, i.e., the percentage of the free phage being removed from the reaction volume increases significantly (∼30 times) as T increases from 4 to 40°C. The adsorption curves also show interesting features; they all decay rapidly initially and then slowly over long times. Motivated by the earlier findings that the expression level of LamB can be significantly altered by the type of carbon sources present in the growth medium, we grew the Ymel bacteria in M9 supplemented with 0.4% maltose (maximum induction) and 0.4% glucose (minimum induction) (6,13). For comparison, Fig. 4 a displays two sets of data taken at T = 24°C for maltose- and glucose-grown Ymel cells. We noticed that adsorption in maltose-grown cells (squares) is slightly stronger than for glucose-grown cells (circles) as indicated by the faster initial decay. The long-time behavior, however, is essentially the same. This observation is consistent with the expectation that maltose causes cells to produce more receptors, but the extent is not as large as reported for other bacterial strains (Hfr G6, Hfr 3000), where orders-of-magnitude increase in the receptor expression level were observed after induction (6). In our experiment, despite quantitatively different adsorption curves due to varying temperature or growth conditions, a unifying feature is that the phage adsorption kinetics is nonexponential and appears to contain at least two different timescales. It is suggestive that the short- and the long-time decays might correspond to the two-step process proposed by Adam and Delbruck (10). However, the detailed analyses below indicate that this is not the case. Since adsorption curves for the maltose- and the glucose-grown cells are qualitatively the same, in what follows, only the data from the glucose-grown cells are presented. The rate constants for bacteria grown in both media are summarized in Table 1.
FIGURE 3.
The temperature-dependent adsorption curves. The bacteria were grown in the minimal M9 medium supplemented with 0.4% glucose. The reactions were carried out in the λ-buffer. The normalized free phage concentration is plotted as a function of time for different temperatures. Here, triangles, diamonds, squares, and circles correspond to T = 40, 30, 20, and 4°C, respectively. The solid lines are fits to the data (see text for details).
FIGURE 4.
(a) Phage adsorption kinetics using Ymel grown in M9 with different carbon supplements. The adsorption measurements were carried out at room temperature and in the λ-buffer. The circles are for the glucose-grown cells and the squares are for the maltose-grown cells. It is clear from the graph that adsorption for maltose-grown cells is faster, indicating a higher level of LamB expression. (b) The same measurement was also carried out using E. coli CR63 (diamonds). In this case, the concentration of free phage remained constant, indicating no discernible adsorption. However, when the CR63 bacteria were transformed by inserting the plasmid pTAS1 that constitutively expresses LamB, the ability for cells to adsorb λ-phage was recovered (triangles).
TABLE 1.
The rate constants
| Glucose-grown cells
|
Maltose-grown cells
|
|||||
|---|---|---|---|---|---|---|
| T (°C) | k (cm3 s−1) | k′ (s−1) | k″ (s−1) | k (cm3 s−1) | k′ (s−1) | k″ (s−1) |
| 1 | 9.6 × 10−13 | 3.8 × 10−4 | 1.7 × 10−3 | 7.4 × 10−13 | 2.5 × 10−3 | 1.0 × 10−3 |
| 3 | 3.0 × 10−12 | 4.0 × 10−3 | 1.5 × 10−3 | 6.0 × 10−12 | 1.7 × 10−3 | 7.2 × 10−4 |
| 4 | 2.3 × 10−12 | 2.1 × 10−3 | 7.9 × 10−4 | 3.6 × 10−12 | 3.6 × 10−3 | 5.2 × 10−4 |
| 10 | 1.6 × 10−11 | 1.2 ×10−3 | 2.0 × 10−4 | 1.3 × 10−11 | 1.1 × 10−3 | 4.7 × 10−4 |
| 15 | 2.0 × 10−11 | 4.2 ×10−3 | 1.4 × 10−3 | 3.5 × 10−11 | 2.0 × 10−3 | 8.4 × 10−4 |
| 20 | 1.2 × 10−11 | 8.5 × 10−4 | 8.0 × 10−4 | 1.4 × 10−11 | 9.7 × 10−4 | 5.7 × 10−4 |
| 24 | 3.3 × 10−11 | 3.0 × 10−3 | 1.2 × 10−3 | 2.2 × 10−11 | 1.8 × 10−3 | 8.4 × 10−4 |
| 30 | 1.5 × 10−11 | 6.2 × 10−4 | 5.7 × 10−4 | 1.6 × 10−11 | 1.9 × 10−3 | 7.8 × 10−4 |
| 35 | 2.0 × 10−11 | 1.4 × 10−3 | 1.2 × 10−3 | 2.7 × 10−11 | 1.8 × 10−3 | 6.0 × 10−4 |
| 37 | 1.6 × 10−11 | 2.2 × 10−3 | 1.5 × 10−3 | 2.6 × 10−11 | 5.4 × 10−3 | 1.5 × 10−3 |
| 40 | 1.6 × 10−11 | 4.3 × 10−4 | 5.1 × 10−4 | 2.9 × 10−11 | 2.3 × 10−3 | 1.2 × 10−3 |
To evaluate the importance of the LamB receptors for λ-phage adsorption and binding, the measurements were also performed using the bacterial strain CR63. The data is displayed as diamonds in Fig. 4 b. This strain of E. coli lacks functional LamB and does not form plaques when plated with wild-type λ-phage (23). Our measurement in Fig. 4 b shows that the bacterium cannot be infected because the critical step of infection, i.e., initial adsorption and subsequent binding, is aborted in CR63. However, this phenotype can be rescued by inserting a high-copy number plasmid pTAS1 (a gift of J. Lawrence) that constitutively expresses LamB and confers ampicillin resistance (24). For this newly created bacterial strain CR63+pTAS1, the phage adsorption is as rapid as maltose-grown Ymel as demonstrated by the triangles in Fig. 4 b. This measurement strongly suggests that LamB receptor is the single determinant of the initial phage adsorption, and it also shows that without functional receptors, the nonspecific binding of phage to other parts of the bacterial membrane is minimal and can be neglected in our measurement.
INTERPRETATIONS OF THE ADSORPTION MEASUREMENTS
The early stage of phage adsorption is a stochastic diffusion process and has been analyzed using diffusion equations by Adam and Delbruck (10), and by Berg and Purcell (7). In our experiment, the adsorption curve does not decay exponentially, suggesting that a single diffusion process is not adequate. To incorporate features of adsorption with multiple stages, we used the first-order rate equations to model the adsorption kinetics. Phenomenologically, the simplest reaction kinetics is a two-stage process given by
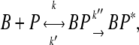 |
where B and P represent bacteria and phage, and BP and BP* represent the transient and the stable bacterium/phage complexes. The constants k, k′, and k″ describe, respectively, the rates of adsorption, desorption, and irreversible binding. This simple reaction scheme is not only convenient for theoretical analyses, as will be shown below, but is also supported by the observation that viable phage can be released by washing the LamB-receptor/phage complexes in the λ-dilution buffer (25). It is useful to point out that this reaction scheme is rather general without referring to detailed physical processes of adsorption. However, the measured (macroscopic) rate constants can be compared with more detailed (microscopic) model calculations. For instance, the adsorption rate k can be compared with Berg and Purcell's model (7), taking into account the finite number of receptors per bacterium. The reversible and irreversible binding rates, k′ and k″, may be similarly calculated if the interactions between the bacterium and the phage are specified. However, in the absence of such information, which clearly is the case here, the measured k′ and k″ can yield insight about the microscopic interactions between the λ-phage and the receptors.
Mathematical predictions of the proposed adsorption kinetics
The number conservation laws for bacteria and phage in the reaction volume allow us to write the following set of ordinary differential equations:
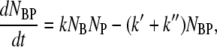 |
(1) |
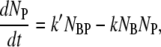 |
(2) |
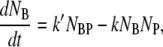 |
(3) |
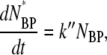 |
(4) |
where NB, NP, NBP, and NBP* are the concentrations of the susceptible bacteria, the free phage, the transient bacterium-phage complexes, and the irreversibly bound (or infected) cells, respectively. Because the reaction is carried out for a short time in the λ-buffer, there are no production terms for either bacteria or phage. Since only the binary collision terms (NBNP) are included, the equations are valid only when the bacterial and phage concentrations are low, i.e., 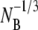 and
and 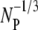 being much greater than the bacterial size. Moreover, Eqs. 1–4 require that no multiple infections occur during the course of the measurements. This latter condition is clearly satisfied in our experiment since moi < 10−3.
being much greater than the bacterial size. Moreover, Eqs. 1–4 require that no multiple infections occur during the course of the measurements. This latter condition is clearly satisfied in our experiment since moi < 10−3.
Physically, k″ specifies the “strength” of the sink for λ-phage. Depending on its value, the kinetics represented by Eqs. 1–4 have the following simple asymptotes:
For k″ = 0, the free phage population and the transient bacterium/phage complexes reach an equilibrium at long times with the equilibrium constant Ka determined by
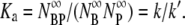
On the other hand, for the intermediate values of k″, the free phage and the complexes diminish at long times,
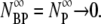
For a very strong sink, k″ ≫
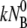 and k″ ≫ k′, the free phage concentration decays exponentially NP(t)/
and k″ ≫ k′, the free phage concentration decays exponentially NP(t)/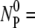 exp(−t/τa), where
exp(−t/τa), where 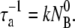 In this case, phage diffusion to the bacterial cell wall is the rate-limiting step of infection, as expected. This diffusion-limited result has been exclusively used for interpreting phage adsorption data in the past (6,26).
In this case, phage diffusion to the bacterial cell wall is the rate-limiting step of infection, as expected. This diffusion-limited result has been exclusively used for interpreting phage adsorption data in the past (6,26).Finally, a single exponential decay can also result if k′ = 0 and k″ ≪
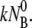 This is the reaction-limited case with the free phage concentration decreasing as NP(t)/
This is the reaction-limited case with the free phage concentration decreasing as NP(t)/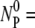 exp(−k″t).
exp(−k″t).
Although the coupled equations, Eqs. 1–4, appear to be complex, they admit analytical solutions for all times t. A detailed derivation is given in the Appendix. The solution that is relevant to the current discussion is the free-phage concentration NP as a function of time t,
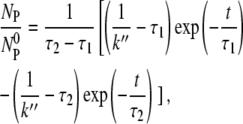 |
(5) |
where τ1 and τ2 are given by
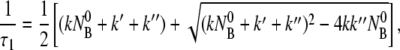 |
(6) |
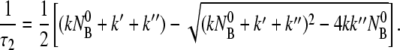 |
(7) |
It is interesting that the general solution to this simple model is a double-exponential function for the free phage population similar to what was observed in the experiment. The equation also shows that the short-time adsorption behavior (t ≪ τ1) is linear in t with Np/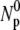 ∼ 1−t/τa, where
∼ 1−t/τa, where 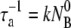 is the reversible adsorption rate that is entirely determined by the bulk phage flux. To fit the experimental data, we reparameterized the equation such that
is the reversible adsorption rate that is entirely determined by the bulk phage flux. To fit the experimental data, we reparameterized the equation such that 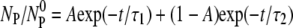 and used A, τ1, and τ2 as the fitting parameters, where
and used A, τ1, and τ2 as the fitting parameters, where 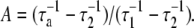 By knowing A, τ1, and τ2, the three rate constants k, k′, and k″ can be extracted from the measurement.
By knowing A, τ1, and τ2, the three rate constants k, k′, and k″ can be extracted from the measurement.
Comparisons between theory and experiment
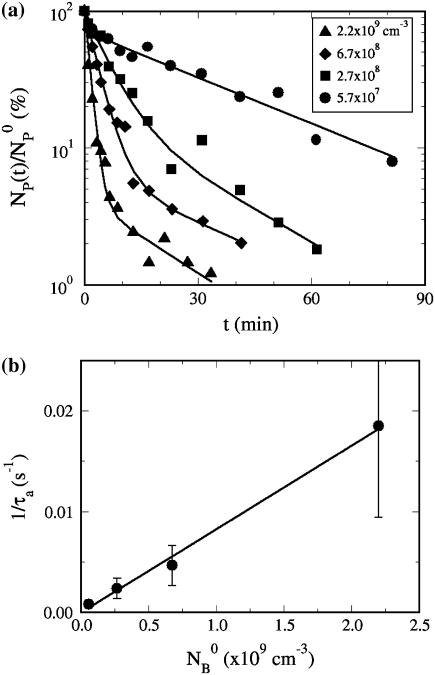
Using the double exponential function derived above, the adsorption curves were fit using a nonlinear regression algorithm (Origin Ver. 7.0). The solid lines in Figs. 3–5a demonstrate that the quality of the fit is adequate, given the noise of the measurements. The validity of the model was further tested by systematically varying the initial bacterial concentration 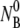 at room temperature. Fig. 5 a shows that when
at room temperature. Fig. 5 a shows that when 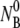 is increased from 5.7 × 107 to 2.2 × 109 cm−3, the concentration of the free phage decays more rapidly. Overall, the model mimics the data well for all concentrations as indicated by the solid lines in the figure. Our model predicts a linear dependence of the adsorption rate 1/τa = ((τ2/τ1−1)A + 1)/τ2 on
is increased from 5.7 × 107 to 2.2 × 109 cm−3, the concentration of the free phage decays more rapidly. Overall, the model mimics the data well for all concentrations as indicated by the solid lines in the figure. Our model predicts a linear dependence of the adsorption rate 1/τa = ((τ2/τ1−1)A + 1)/τ2 on 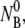 which is seen in Fig. 5 b. The slope of the plot gives the adsorption coefficient k = (0.93 ± 0.4) × 10−11 cm3 s−1, which is reasonably consistent with the individual runs listed in Table 1.
which is seen in Fig. 5 b. The slope of the plot gives the adsorption coefficient k = (0.93 ± 0.4) × 10−11 cm3 s−1, which is reasonably consistent with the individual runs listed in Table 1.
FIGURE 5.
The 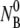 dependence. In panel a, four different measurements (circles, squares, diamonds, and triangles) corresponds respectively to
dependence. In panel a, four different measurements (circles, squares, diamonds, and triangles) corresponds respectively to 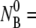 5.7 × 107, 2.7 × 108, 6.7 × 108, and 2.2 × 109 cm−3. The measurements were performed in room temperature. In panel b, the linear dependence of 1/τa vs.
5.7 × 107, 2.7 × 108, 6.7 × 108, and 2.2 × 109 cm−3. The measurements were performed in room temperature. In panel b, the linear dependence of 1/τa vs. 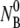 is observed. The slope gives the adsorption constant k = 9 × 10−12 cm3 s−1.
is observed. The slope gives the adsorption constant k = 9 × 10−12 cm3 s−1.
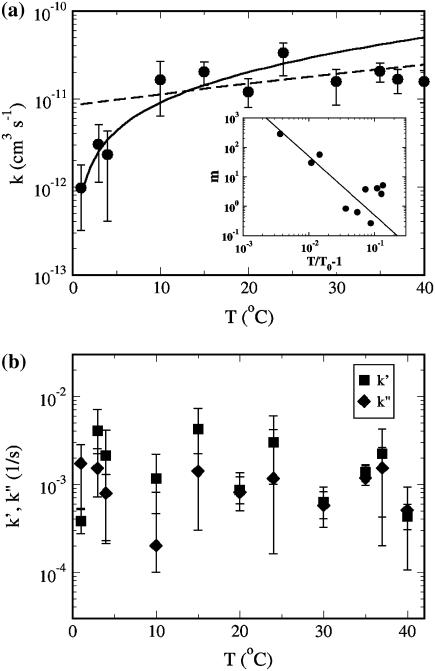
The adsorption data for different temperatures were also analyzed and the results are presented in Fig. 6 a for k and in Fig. 6 b for k′ (squares) and k″ (diamonds). Although the data are somewhat noisy, particularly for k′ and k″, the trend is clear. It shows that while both k′ and k″ are only weakly temperature-dependent, if at all, the reversible adsorption rate constant k is temperature sensitive. For low temperatures, T < 10°C, k increases rapidly with T and for T > 10°C, it levels off with a plateau value 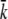 = (2 ± 1) × 10−11 cm3 s−1. We noted that among the three rate constants,
= (2 ± 1) × 10−11 cm3 s−1. We noted that among the three rate constants, 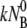 is by far the largest and thus contributes the most to the fast initial decay time τ1, as seen in all of the adsorption curves. In contrast, the dissociation and the irreversible binding rate constants, k′ and k″, are, respectively, 5 and 10 times smaller than
is by far the largest and thus contributes the most to the fast initial decay time τ1, as seen in all of the adsorption curves. In contrast, the dissociation and the irreversible binding rate constants, k′ and k″, are, respectively, 5 and 10 times smaller than 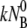 and consequently they contribute marginally to τ1. However, under certain conditions such as low bacterial concentrations, it is feasible that k′ and k″ are comparable to
and consequently they contribute marginally to τ1. However, under certain conditions such as low bacterial concentrations, it is feasible that k′ and k″ are comparable to 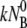 such that initial decay time τ1 of Np is determined by the bulk phage flux
such that initial decay time τ1 of Np is determined by the bulk phage flux 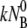 as well as by k′ and k″. If one is unaware of this subtlety and using the initial decay rate (on a semilog plot) to extract the adsorption constant k, its value can be overestimated. It is plausible that some of the early measurements of k could have been influenced by this effect. However, since the initial bacterial concentration
as well as by k′ and k″. If one is unaware of this subtlety and using the initial decay rate (on a semilog plot) to extract the adsorption constant k, its value can be overestimated. It is plausible that some of the early measurements of k could have been influenced by this effect. However, since the initial bacterial concentration 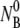 was not given in Schwartz (6) and since there were no estimates for k′ and k″ for the bacterial strain used, it is not possible to estimate the true value of the adsorption constant k in Schwartz's early experiment.
was not given in Schwartz (6) and since there were no estimates for k′ and k″ for the bacterial strain used, it is not possible to estimate the true value of the adsorption constant k in Schwartz's early experiment.
FIGURE 6.
The adsorption rate constants k, k′, and k″. (a) The measured k is nearly independent of T for T > 10°C but it drops sharply for T < 10°C. The dotted line takes into account T-dependent viscosity η of the medium. The solid line takes into account both T-dependent η and the aggregation of receptors (see text for details). The inset depicts the aggregation number m = N/M as a function of the reduced temperature T/T0−1, where T0 = 274 K. The data can be approximated as a power law in reduced temperature as m ∼ (T/T0−1)−α, where the exponent α is ∼2 as delineated by the solid line in the inset. (b) The desorption rate constant k′ and the irreversible binding constants k″. Within noise, these rate constants show no systematic T dependence.
To put our measurements in perspective, we compared the measured adsorption rate constant k with Berg and Purcell's theory (7). Using an electrostatic analogy, these investigators treated the phage receptors as ideal sinks that inactivate the phage upon each collision. An alternative derivation that is easier to follow can be found in Berg (27). For a bacterium with its surface entirely covered with such ideal receptors, the adsorption rate constant is given by kmax = 4πaD3, where a is the radius of the bacterium (assuming it to be a sphere) and D3 is the diffusion coefficient of a phage in the solution. For a finite number of receptors N per bacterium, the adsorption rate constant is reduced by the factor C = k/kmax = sN/(sN + πa), which interestingly depends only on the linear dimensions of the bacterium a and the receptor s. This is a significant result because it shows that 1), the reaction rate with a surface-bound receptor is much higher than the ratio of the receptor area (Ar = πs2N) to total bacterial surface area (At = 4πa2), with the latter corresponding to random target shooting; and 2), the adsorption rate levels-off at a relatively low surface coverage given by NC ∼ πa/s or Ar/At = (π/2)2/NC ≪ 1. This leaves much of the bacterial surface for other receptors of different biological functions. We note that the high binding efficiency for sparsely populated membrane-bound receptors is a general feature of a diffusion process. The underlying physics relies on the fact that a “molecule” that is already in the neighborhood of the cell has a high probability of hitting the surface multiple times, enhancing its capture probability by receptors. It should be emphasized that C given above is exact for a small number of receptors, sN ≪ πa. In this case, receptors are statistically independent with k = k0N, where k0 = 4sD3 is the adsorption rate constant of a single receptor. The result is almost exact for a large number of receptors, sN ≫ πa, where they compete with each other for phage particles. A recent self-consistent calculation by Zwanzig (28) showed that even in the high-coverage limit, the correction to C is very small (approximately a few percent) and is likely not discernible in most measurements. For elongated bacteria with the major- and the minor-semi-axes being a and b, Berg and Purcell (7) found that the maximum rate constant is modified with the result kmax = 4πaD3/ln(2a/b). Correspondingly, the correction factor should be C = k/kmax = sN/(sN + πa/ln(2a/b)) so that the low-coverage limit can be recovered.
Using the experimentally determined a, b, and D3 in the previous section, we found kmax = 7.1 × 10−11 cm3 s−1, which is a factor of 3.6 larger than the measured k at high temperatures. This suggests that the average number of LamB receptors N on bacteria Ymel is below the saturation limit, i.e., N < NC (≡ π(a/s)/ln(2a/b)). The LamB receptor is a homotrimer and its 3D structure has been determined recently by x-ray diffraction (14). Genetic mapping of mutations has further shown that the amino acid residuals (12 of them) responsible for phage adsorption all belong to the outer looping structures of the mature LamB polypeptides (15,16). Using the structural data (14) as well as the genetic map (15,16), the outer radius of the receptor can be estimated to be s ≈ 4 nm. Using the simple assumption that the geometric area is the same as the capture cross-sectional area, we were able to determine the number of functional receptors per bacterium N ≈ πaC/(s(1−C)ln(2a/b)) ∼ 280 ± 70 and the crossover NC ≈ 680 for Ymel. This expression level is considerably higher than the glucose-grown bacteria Hfr G6 and Hfr 3000 used in Schwartz's experiment (6), where N ∼ 10 was reported. Our observation shows that Ymel bacteria constitutively produce LamB receptors at a moderate level and do not respond to the presence of glucose. The lack of glucose effect in the maltose system can often be traced to mutations in malK, which is a part of the maltose transport machinery and is subject to phosphotransferase regulation (12). However, in the case of Ymel, the exact cause is not known. To ascertain that N determined by the adsorption measurements is reasonable, additional experiments were conducted in which fluorescently labeled λ-phage were incubated with Ymel cells and the brightness of the resulting bacteria were measured using fluorescent microscopy (model No. TE300; Nikon, Tokyo, Japan). This experiment yielded an estimate of the average bound phage per bacterium to be a few hundred, in qualitative agreement with the adsorption measurement (29).
Implications of the measured rate constants
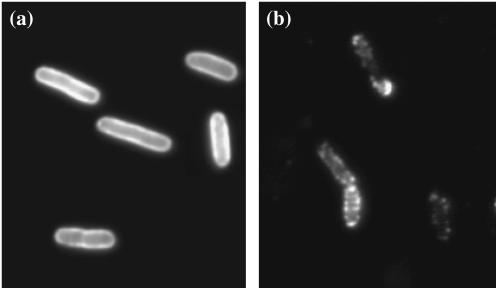
We next turned our attention to the possible biological implications suggested by the temperature dependence of the rate constants k, k′, and k″. The temperature dependence of k may naively be thought to be due to the change in the viscosity of the medium. However, this was shown not to be the case as delineated by the dotted line in Fig. 6 a. Here, the viscosity η of the λ-buffer was measured as a function of T and was found to behave similar to water over the temperature range 1°C < T < 40°C. The significantly reduced adsorption rate at low temperatures suggests that either the LamB receptor experiences a conformational change, reducing its affinity in binding to phage, or its spatial distribution is altered. Nonuniform receptor distributions have been reported previously including a recent study on LamB (30–32). We note that if receptors coagulate at low temperatures, phage adsorption can be reduced based on Berg and Purcell's model (7). A simple calculation shows that if N receptors segregated into M patches of effective radius s*, the new adsorption rate should be characterized by C* = Ms*/(Ms* + πa/ln(2a/b)) = Ns/m1/2/(Ns/m1/2 + πa/ln(2a/b)), where m = N/M is the aggregation number and s* ∼ m1/2s. This yields C*/C = (1 + (πa)/(sNln(2a/b)))/(1 + (πam1/2/(sNln(2a/b))), which reaches a maximum C*/C = 1 when m = 1 and a minimum C*/C = (1 + (πa)/(sNln(2a/b)))/(1 + (πa)/(sN1/2ln(2a/b))) when m = N. For our experiment, assuming that at high temperatures all the receptors are well dispersed and at low temperatures all receptors aggregate into a single patch, the adsorption rate can be reduced by as much as a factor of 12 since (C*/C)min = 0.08. This is close to what we have observed in Fig. 6 a, suggesting that below 10°C, receptors may have segregated into a small number of patches. Heuristically, one can assume a simple power-law dependence of m, m = A0(T/T0−1)−α, where A and α are adjustable parameters and T0 = 274 K is fixed. We found the best values of A and α that fit the measurement in Fig. 6 a are A0 ≈ 5 × 10−3 and α ≈ 2, as delineated by the solid line. The aggregation number m versus the reduced temperature T/T0−1 is plotted in the inset. We also looked for other signs of receptor aggregation using fluorescent microscopy. Here again fluorescently labeled λ-phage were used to react with Ymel at 4°C and at 37°C. After 40 min of incubation with a saturating amount of λ-phage (moi ≈ 2000), the bacterium/phage complexes were washed in λ-dilution buffer, and then examined under the microscope at room temperature. As shown by the images in Fig. 7, the phage population on the bacterial wall is uniformly distributed at the high temperature (Fig. 7 a), but significant granularity was observed at the low temperature (Fig. 7 b). This observation strongly suggests that receptor segregation into patches is a candidate for the decreased phage adsorption at low temperatures. The effect is reminiscent of phase transitions in liquids and in artificial bilayers of mixed lipids (33). However, the biological significance of this effect is unclear and remains to be investigated.
FIGURE 7.
Fluorescent images of bacterium/phage complexes. Fluorescently labeled λ-phage were bound to Ymel at 37°C (a) and at 4°C (b), but the observation was made at room temperature. The labeling was carried out using a saturating amount of λ-phage (moi ≈ 2000) and incubated over 40 min in the λ-buffer. The uniformity of fluorescence indicates how the phage (or LamB receptors) are distributed on the bacterial cell wall. These images indicate that receptor distribution becomes less uniform as T is reduced, and is suggestive that they form large patches.
Among the three rate constants, k′ and k″ are the most intriguing because of their relatively weak temperature dependence and because their underlying biological/physical mechanisms are not known. In the classical two-stage capture model, the mean residence time tr on the bacterial surface may be identified as the inverse of the dissociation rate 1/k′ whereas the mean capture time tc may be identified as the inverse of the irreversible binding rate 1/k″. Since a phage nonspecifically adsorbed on the bacterial surface can dissociate rapidly and it takes many trials before it binds irreversibly, it is generally expected that tr is shorter than tc. Based on 2D diffusion, Berg and Purcell found tc = (1.1 a2/ND2)ln(1.2 a2/Ns2), where D2 is the surface diffusion coefficient, which one expects to be not too different from its 3D counterpart, D3 ∼ 10−8 cm2/s. Since tr < tc ∼ 6 × 10−3 s whereas our measured 1/k′ and 1/k″ are ∼1000 s, it can be concluded that our observations cannot be explained by the classical two-stage capture model. Physically, the above calculation indicates that once the phage touches the bacterial surface by nonspecific binding, it should, statistically speaking, be captured on the timescale of milliseconds, determined by the average squared distance between receptors divided by D2. This implies that the surface process is not rate-limiting (tc < τa) for typical bacterial concentrations encountered in most measurements, i.e., 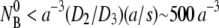 or
or 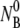 < 1013–1014 cm−3. It may be important to point out that if the nonspecific phage adsorption does indeed take place, it cannot be detected by the current method, which consists of sample dilution plus centrifugation steps. This is demonstrated by the control experiment using CR63. The data in Fig. 4 b leaves little doubt that functional LamB receptors are required for phage capture, and in their absence, NP(t) remains constant over time.
< 1013–1014 cm−3. It may be important to point out that if the nonspecific phage adsorption does indeed take place, it cannot be detected by the current method, which consists of sample dilution plus centrifugation steps. This is demonstrated by the control experiment using CR63. The data in Fig. 4 b leaves little doubt that functional LamB receptors are required for phage capture, and in their absence, NP(t) remains constant over time.
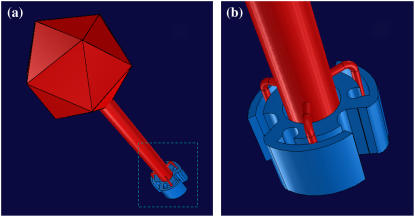
If the adsorbed phage finds the receptor rapidly, k′ and k″ must be related to the specific interactions between the receptor and the phage tail (the gene product of J). Since k′ ∼ 2k″, it may be deduced that the initial binding is transient and that the probability of a captured phage being detached from the receptor is two times greater than it being permanently bound. Moreover, if a receptor can transiently bind to a phage with a typical retention time tb, it is expected that k′−1 = tr + tb. In our experiment, it is clear tb ≫ tr and k′ ≈ 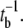 Transient binding of a phage suggests that LamB receptor is not an ideal sink as modeled by theory (7,10). However, because the dissociation time 1/k′ is so long with tb ∼ 1000 s, the short-time adsorption behavior may be adequately mimicked by assuming ideal receptors. The weak temperature-dependence suggests that the phage retention by the receptor is entropic in nature and that a molecular key-lock interaction may be an appropriate description. Fig. 8 displays a hypothetical situation where the phage tip is transiently locked to the receptor. It can be free or permanently bound by rotational diffusion along the axis of the phage tail. Using the Kramer's reaction rate equation and assuming that the entropic energy barrier heights for the reversible and irreversible binding are given respectively by ΔE′ = TΔS′ = n′kBT and ΔE″ = TΔS″ = n″ kBT, we found k′ = Drexp(−n′) and k″ = Drexp(−n″), where we have assumed that once the phage is bound, the kinetics is determined by the rotational diffusion with the rate constant given by Dr (= kBT/8πηR3) ∼ 3.7 × 103 s−1, where R is the radius of the phage head. Using the averaged values for k′ = (1.9 ± 0.4) × 10−3 s−1 and k″ = (1 ± 0.2) × 10−3 s−1, it is found n′ ∼ 14.5 and n″ ∼ 15.1. This corresponds to an entropic barrier of ∼3.6 × 104 J/mole, a value three times smaller than the energy barrier observed in inactivation of bacteriophage φX174 (34). The entropy ΔS′ is related to the logarithm of the probability p′ of a trapped phage making a transition to a free state with ΔS′ = −kBln(p′), and ΔS″ is related to the logarithm of the probability p″ of a trapped phage making a transition to a captured state with ΔS″ = −kBln(p″). In our case, p′ and p″ turn out to be 5.0 × 10−7 and 2.8 × 10−7, respectively. With such a low transitional probability, one may wonder whether some cooperative effects (such as coordinated conformational fluctuations of J and LamB) may play a role. It is also useful to compare the binding rate in bacterium/phage systems with other well known biological reactions such as streptavidin-biotin pairs. The exceptionally tight binding observed in the streptavidin/avidin interaction with biotin (Ka = 1014–1015 M−1) is controlled to a large extent by the slow dissociation kinetics of the protein-ligand complex. This appears to be a common feature of many high-affinity systems in biology. For the wild-type streptavidin, the dissociation rate k′ ≈ 6 × 10−6 s−1 (35). With mutations on conservative sites, k′ increases by two orders of magnitude, making it comparable to what is seen in λ-phage-receptor interactions reported here.
Transient binding of a phage suggests that LamB receptor is not an ideal sink as modeled by theory (7,10). However, because the dissociation time 1/k′ is so long with tb ∼ 1000 s, the short-time adsorption behavior may be adequately mimicked by assuming ideal receptors. The weak temperature-dependence suggests that the phage retention by the receptor is entropic in nature and that a molecular key-lock interaction may be an appropriate description. Fig. 8 displays a hypothetical situation where the phage tip is transiently locked to the receptor. It can be free or permanently bound by rotational diffusion along the axis of the phage tail. Using the Kramer's reaction rate equation and assuming that the entropic energy barrier heights for the reversible and irreversible binding are given respectively by ΔE′ = TΔS′ = n′kBT and ΔE″ = TΔS″ = n″ kBT, we found k′ = Drexp(−n′) and k″ = Drexp(−n″), where we have assumed that once the phage is bound, the kinetics is determined by the rotational diffusion with the rate constant given by Dr (= kBT/8πηR3) ∼ 3.7 × 103 s−1, where R is the radius of the phage head. Using the averaged values for k′ = (1.9 ± 0.4) × 10−3 s−1 and k″ = (1 ± 0.2) × 10−3 s−1, it is found n′ ∼ 14.5 and n″ ∼ 15.1. This corresponds to an entropic barrier of ∼3.6 × 104 J/mole, a value three times smaller than the energy barrier observed in inactivation of bacteriophage φX174 (34). The entropy ΔS′ is related to the logarithm of the probability p′ of a trapped phage making a transition to a free state with ΔS′ = −kBln(p′), and ΔS″ is related to the logarithm of the probability p″ of a trapped phage making a transition to a captured state with ΔS″ = −kBln(p″). In our case, p′ and p″ turn out to be 5.0 × 10−7 and 2.8 × 10−7, respectively. With such a low transitional probability, one may wonder whether some cooperative effects (such as coordinated conformational fluctuations of J and LamB) may play a role. It is also useful to compare the binding rate in bacterium/phage systems with other well known biological reactions such as streptavidin-biotin pairs. The exceptionally tight binding observed in the streptavidin/avidin interaction with biotin (Ka = 1014–1015 M−1) is controlled to a large extent by the slow dissociation kinetics of the protein-ligand complex. This appears to be a common feature of many high-affinity systems in biology. For the wild-type streptavidin, the dissociation rate k′ ≈ 6 × 10−6 s−1 (35). With mutations on conservative sites, k′ increases by two orders of magnitude, making it comparable to what is seen in λ-phage-receptor interactions reported here.
FIGURE 8.
Interactions between a bacteriophage and a receptor. Panel a depicts the hypothetical key-lock interactions between λ-phage and a LamB receptor. The detailed drawing inside the dotted line box is shown in panel b. As the drawing suggests, the rotational diffusion of the phage particle can make it either free or permanently bound to the receptor.
CONCLUSIONS
To summarize, we have presented experimental data demonstrating that phage binding to their hosts in a defined medium proceeds in two distinctive steps characterized by a fast and a slow exponential relaxation. However, this two-step process is distinctively different from the classical two-stage capture model, which postulates that the efficient phage binding results from membrane-assisted diffusion or reaction rate enhancement by dimensional reduction (RREDR). We reach this conclusion based on the observation that E. coli CR63, which lacks functional receptors, aborts the phage-binding ability completely. However, this phenotype can be reversed if a plasmid that expresses LamB is introduced. The observed two-step process thus is an intrinsic property of receptor-phage interaction and is independent of processes mediated by the bacterial membrane. We modeled the receptor and phage interaction as a kinetic process consisting of reversible on/off and irreversible binding events. The analytic solution of this model adequately describes our experimental observations and allows the three rate constants, k, k′, and k″, to be extracted from a single measurement. We found that the reversible binding rate k can be accounted for satisfactorily by the Berg-Purcell model, yielding a LamB receptor number of a few hundred for E. coli Ymel. The surprising finding of our experiment is that the LamB receptor appears to be able to retain the adsorbed phage for a long time, ∼1000 s, before it is inactivated. This finding suggests that the receptor targeting efficiency may not be what optimizes the infection process but, instead, that the binding affinity of a phage to the receptor is what matters. Such a phenotype appears to be more critical in natural habitats where the collision rate between bacteria and phage is low. Therefore, to ensure successful infection, the ability to bind tightly to a receptor becomes a significant attribute.
The present study lays the foundation for future investigations of other outstanding issues related to phage/receptor interactions, such as the relevance of the membrane-mediated processes in the enhancement or impediment of reaction rates, and the potential connection between our observed irreversible binding and phage DNA translocation dynamics. Both of these issues may be addressed by carefully designed fluorescent microscopy experiments that allow dynamics on a microscopic scale to be resolved, a feat which is not feasible in the current experiment. There are also issues related to nonideal aspects of LamB receptors, such as finite mobility of receptors on the cell wall and the reversible nature of phage/receptor binding. The latter issue has been addressed by the work of Axelrod and Wang (36) but a detailed comparison between their theory and experiments has not been made to our knowledge. Finally, the bacterium/phage system can be a useful paradigm for studying evolution (or arms-race) of competing species in a laboratory setting, where molecular recognition and binding can be quantified by the measurement and the mathematical model presented.
Acknowledgments
We thank Q. W. Yip and B. Li for their early involvements in the project. We also thank R. Hendrix, R. Duda, M. Shtrahman, K.W. Lee, and C. Yeung for helpful discussions. In particular, C. Yeung pointed out the possible connection between a weak T-dependent rate constant and the entropic nature of phage-receptor interactions. We are grateful to the gift of pTAS1 given to us by J. Lawrence.
This work was partially funded by the National Science Foundation under the grant No. DMR-0242284.
APPENDIX
In this section, we present mathematical solutions to the rate equations given in Eqs. 1–4. Fortuitously, under the physically relevant conditions, the equations can be solved analytically. The mass conservation laws, Eqs. 2 and 3, imply that ΔNP = ΔNB should hold at all times, yielding the following relationships: NB = 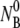 -NBP-NBP* and NP =
-NBP-NBP* and NP = 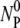 -NBP-NBP*. Substituting these two equations into Eq. 1 and reorganizing we found:
-NBP-NBP*. Substituting these two equations into Eq. 1 and reorganizing we found:
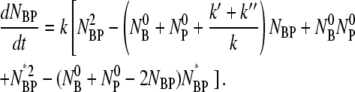 |
(8) |
Equations 8 and 4 complete the description of the problem. In general, this set of differential equations cannot be solved analytically due to the nonlinear terms. To make progress, we used the fact that the moi (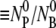 ) is very small in our experiment such that
) is very small in our experiment such that 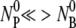 and the nonlinear terms
and the nonlinear terms 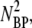
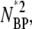 and NBPNBP* are negligible compared to linear terms. This allows Eq. 8 to be linearized with the result
and NBPNBP* are negligible compared to linear terms. This allows Eq. 8 to be linearized with the result
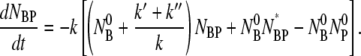 |
(9) |
By taking the time derivative on both sides of this equation and replacing dNBP*/dt with Eq. 4, we arrive at a single differential equation to describe the adsorption process of bacteria and phage:
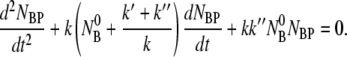 |
(10) |
This equation along with the initial conditions, NBP(0) = 0 and dNBP(0)/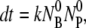 uniquely determine the time evolution of the population of bacteria/phage complexes. It is interesting that Eq. 10 is mathematically identical to a damped harmonic oscillator and allows the solution NBP ∼ exp(iωt). Here the frequency ω obeys the equations
uniquely determine the time evolution of the population of bacteria/phage complexes. It is interesting that Eq. 10 is mathematically identical to a damped harmonic oscillator and allows the solution NBP ∼ exp(iωt). Here the frequency ω obeys the equations
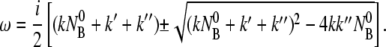 |
(11) |
For the problem at hand, it can be shown that ω is imaginary and the solution to NBP is purely relaxational with two different decay times given by 1/τ1,2 = −iω+,− or
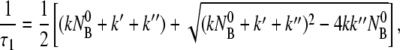 |
(12) |
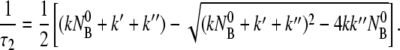 |
(13) |
It is evident from Eqs. 12 and 13 that the fast relaxation time τ1 scales approximately as the inverse sum of the rate constants with τ1 ∼ (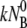 + k′ + k″)−1, and the slow relaxation time τ2 is given approximately by τ2 ∼ max(
+ k′ + k″)−1, and the slow relaxation time τ2 is given approximately by τ2 ∼ max(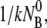 1/k″). The solution to NBP is then
1/k″). The solution to NBP is then
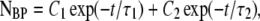 |
(14) |
where the coefficients C1 and C2 are determined by the initial conditions (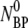 = 0 and dNBP/dt|0 =
= 0 and dNBP/dt|0 = 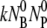 ) with the result:
) with the result:
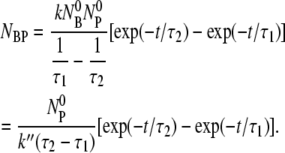 |
(15) |
One can also calculate the population of the stable complexes NBP* as a function of time by integrating Eq. 4:
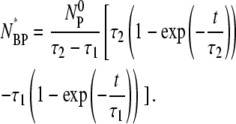 |
(16) |
In our experiment, since the free phage NP are measured, the observable is NP = 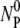 -NBP-NBP*. Using Eqs. 15 and 16, we find
-NBP-NBP*. Using Eqs. 15 and 16, we find
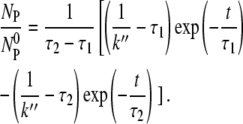 |
(17) |
From Eqs. 12 and 13, a useful relation can be derived as
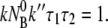 |
(18) |
It is instructive to check the limiting cases of the above equations. For instance, it can be shown that for a very large k″, τ1 ∼ 1/k″ and 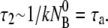 Inserting these relations in Eq. 17 gives the exponential decay
Inserting these relations in Eq. 17 gives the exponential decay 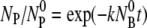 of the free phage in the reaction volume. This corresponds to the diffusion-limited process and is the equation used by all the previous phage adsorption studies. The other useful limits can also be obtained and are discussed in the main text.
of the free phage in the reaction volume. This corresponds to the diffusion-limited process and is the equation used by all the previous phage adsorption studies. The other useful limits can also be obtained and are discussed in the main text.
Editor: Byron Goldstein.
References
- 1.Hershey, A. D., and M. Chase. 1952. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 36:39–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luria, S. E., and M. Delbruck. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 28:491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crick, F. 1970. Central dogma of molecular biology. Nature. 227:561–563. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz, M. 1980. Interaction of phages with their receptor proteins. In Virus Receptors (Receptors and Recognition, Series B). L. L. Randall and L. Philipson, editors. Chapman and Hall, London, UK.
- 5.Schlesinger, M. 1932. Adsorption of bacteriophages to homologous bacteria. II. Quantitative investigation of adsorption velocity and saturation. Estimation of the particle size of the bacteriophage. Z. Hyg. Infektionskr. 114:149–160. [Google Scholar]
- 6.Schwartz, M. 1976. The adsorption of coliphage lambda to its host: effect of variation in the surface density of receptor and in phage-receptor affinity. J. Mol. Biol. 103:521–536. [DOI] [PubMed] [Google Scholar]
- 7.Berg, H. C., and E. M. Purcell. 1977. Physics of chemoreception. Biophys. J. 20:193–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaissling, K. E., and J. Thorson. 1980. Insect olfactory sensilla: structural, chemical and electrical aspects of the functional organization. In Receptors for Neurotransmitters, Hormones and Pheromones in Insets. D. B. Satelle, L. M. Hall, and J. G. Hildebrand, editors. Elsevier/North-Holland Biomedical Press, Amsterdam, The Netherlands.
- 9.Futrelle, R. P. 1984. How molecules get to the detectors, the physics of diffusion of insect pheromones. Trends Neurosci. 7:116–120. [Google Scholar]
- 10.Adam, G., and M. Delbruck. 1968. Reduction of dimensionality in biological diffusion processes, in Structural Chemistry and Molecular Biology. A. Rich and N. Davidson, editors. W.H. Freeman & Company, San Francisco.
- 11.McCloskey, M. A., and M.-M. Poo. 1986. Rates of membrane-associated reactions: reduction of dimensionality revisited. J. Cell Biol. 102:88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartz, M. 1987. The maltose regulon. In Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology. F. C. Neidhardt, editor. American Society for Microbiology, Washington, DC.
- 13.Boos, W., and H. Shuman. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62:204–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schirmer, T., T. A. Keller, Y. F. Wang, and J. P. Rosenbusch. 1995. Structural basis for sugar translocation through maltoporin channel at 3.1 Ångström resolution. Science. 267:512–514. [DOI] [PubMed] [Google Scholar]
- 15.Gehring, K., A. Charbit, E. Brissaud, and M. Hofnung. 1987. Bacteriophage lambda receptor site on the Escherichia coli K-12 LamB protein. J. Bacteriol. 169:2103–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis, G., L. Brennan, S. Stretton, and T. Ferenci. 1991. Genetic mapping of starch- and lambda-receptor sites in maltoporin: identification of substitutions causing direct and indirect effects on binding sites by cysteine mutagenesis. Mol. Microbiol. 5:2293–2301. [DOI] [PubMed] [Google Scholar]
- 17.Wang, J., M. Hofnung, and A. Charbit. 2000. The C-terminal portion of the tail fiber protein of bacteriophage lambda is responsible for binding to LamB, its receptor at the surface of Escherichia coli K-12. J. Bacteriol. 182:508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Randall-Hazelbauer, I., and M. Schwartz. 1973. Isolation of the bacteriophage lambda receptor from Escherichia coli. J. Bacteriol. 116:1436–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bachmann, B. J. 1987. Derivations and genotypes of some mutant derivatives of Escherichia coli K12. In Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology. F. C. Neidhardt, editor. ASM Press, Washington, DC.
- 20.Glasner, J. D., P. Liss, G. Plunkett III, A. Darling, T. Prasad, M. Rusch, A. Byrnes, M. Gilson, B. Biehl, F. R. Blattner, and N. T. Perna. 2003. ASAP, a systematic annotation package for community analysis of genomes. Nucleic Acids Res. 31:147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ptashne, M. 1986. A Genetic Switch, Gene Control and Phage Lambda. Blackwell Scientific and Cell Press, Palo Alto, CA.
- 22.Puck, T. T., A. Garen, and J. Cline. 1950. The mechanism of virus attachment to host cells. J. Exp. Med. 93:65–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roa, M., and D. Scandella. 1976. Multiple steps during the interaction between coliphage lambda and its receptor protein in vitro. Virology. 72:182–194. [DOI] [PubMed] [Google Scholar]
- 24.Kolko, M. M., L. A. Kapetanovich, and J. G. Lawrence. 2001. Alternative pathways for siroheme synthesis in Klebsiella aerogenes. J. Bacteriol. 183:328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz, M. 1975. Reversible interaction between coliphage lambda and its receptor protein. J. Mol. Biol. 99:185–201. [DOI] [PubMed] [Google Scholar]
- 26.Hadas, H., M. Einav, I. Fishov, A. Zaritsky. 1997. Bacteriophage T4 development depends on the physiology of its host Escherichia coli. Microbiology. 143:179–185. [DOI] [PubMed] [Google Scholar]
- 27.Berg, H. C. 1993. Random Walks in Biology. Princeton University Press, Princeton, NJ.
- 28.Zwanzig, R. 1990. Diffusion-controlled ligand binding to spheres partially covered by receptors: an effective medium treatment. Proc. Natl. Acad. Sci. USA. 87:5856–5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moldovan, R. The interaction between lambda phage and its bacterial host. PhD thesis. Department of Physics and Astronomy, University of Pittsburgh, Pittsburgh, PA.
- 30.Bayer, M. E. 1968. Adsorption of bacteriophages to adhesions between wall and membrane of Escherichia coli. J. Virol. 2:346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zgaga, V., M. Medic, E. Salaj-Smic, D. Novak, and M. Wrischer. 1973. Infection of Escherichia coli envelope-membrane complex with lambda phage: adsorption and penetration. 79:697–108. [DOI] [PubMed]
- 32.Gibbs, K. A., D. D. Isaac, J. Xu, R. W. Hendrix, T. J. Silhavy, J. A. Theriot. 2004. Complex spatial distribution and dynamics of an abundant Escherichia coli outer membrane protein, LamB. Mol. Microbiol. 53:1771–1783. [DOI] [PubMed] [Google Scholar]
- 33.Veatch, S. L., and S. L. Keller. 2003. Separation of liquid phase in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys. J. 85:3074–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Incardona, N. L. 1974. Mechanism of adsorption and eclipse of bacteriophage phiX174. III. Comparison of the activation parameters for the in vitro and in vivo eclipse reactions with mutant and wild-type virus. J. Virol. 14:469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chilkoti, A., and P. S. Stayton. 1995. Molecular origins of the slow streptavidin-biotin dissociation kinetics. J. Am. Chem. Soc. 117:10622–10628. [Google Scholar]
- 36.Axelrod, D., and M. D. Wang. 1994. Reduction-of-dimensionality kinetics at reaction-limited cell surface receptors. Biophys. J. 66:588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]