Abstract
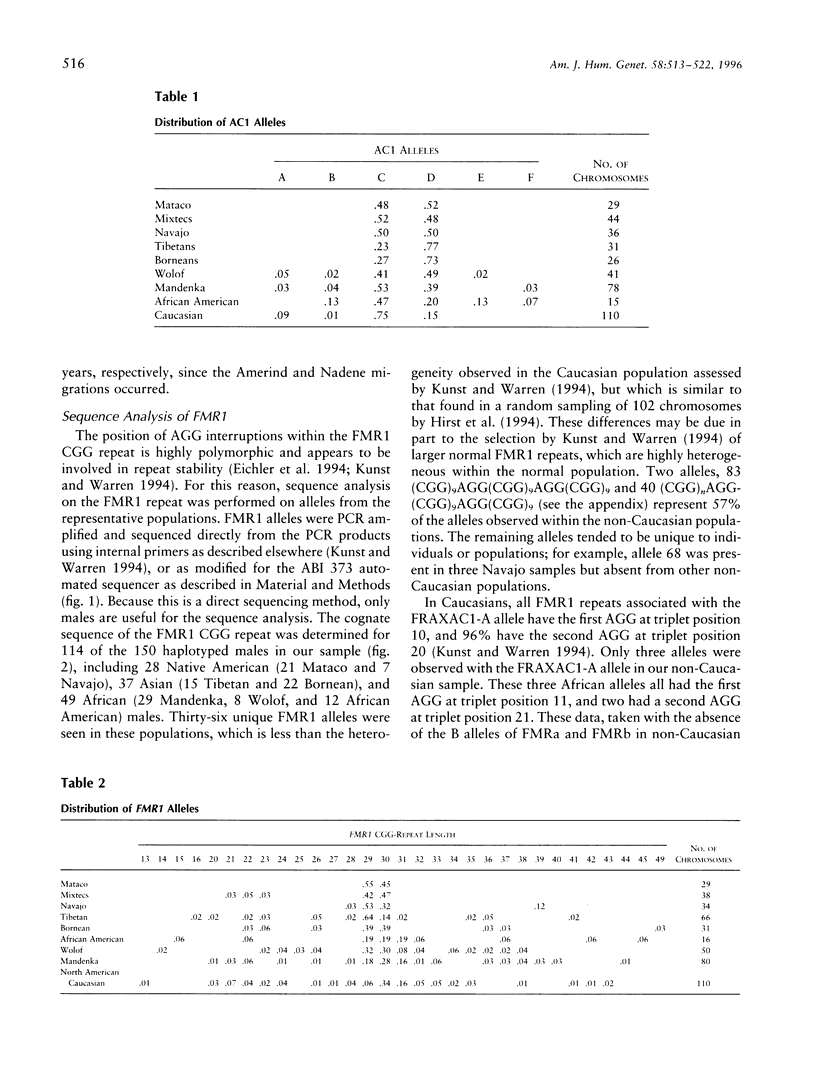
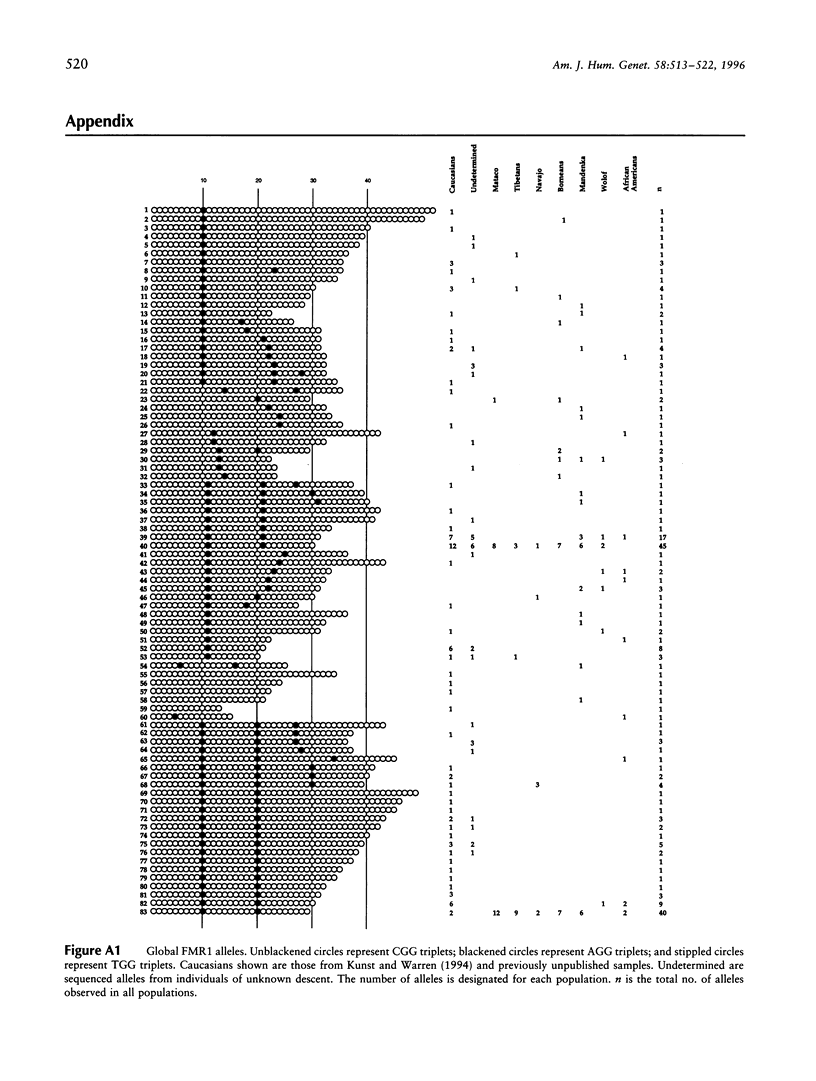
Fragile X syndrome, a frequent form of inherited mental retardation, results from the unstable expansion of a cryptic CGG repeat within the 5' UTR region of the FMR1 gene. The CGG repeat is normally polymorphic in length, and the content is frequently interrupted by AGG triplets. These interruptions are believed to stabilize the repeat, and their absence, leading to long tracts of perfect CGG repeats, may give rise to predisposed alleles. In order to examine the stability of normal FMR1 alleles, the repeat length of 345 chromosomes from nine global populations was examined with the content also determined from 114 chromosomes as assessed by automated DNA sequencing. The FMR1 alleles, defined by the CGG repeat, as well as by the haplotypes of nearby polymorphic loci, were very heterogeneous, although the level of variation correlated with the age and/or genetic history of a particular population. Native American alleles, interrupted by three AGG repeats, exhibited marked stability over 7,000 years. However, in older African populations, parsimony analysis predicts the occasional loss of an AGG, leading to more perfect CGG repeats. These data therefore support the suggestion that AGG interruptions enhance the stability of the FMR1 repeat and indicate that the rare loss of these interruptions leads to alleles with longer perfect CGG-repeat tracts.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arinami T., Asano M., Kobayashi K., Yanagi H., Hamaguchi H. Data on the CGG repeat at the fragile X site in the non-retarded Japanese population and family suggest the presence of a subgroup of normal alleles predisposing to mutate. Hum Genet. 1993 Nov;92(5):431–436. doi: 10.1007/BF00216445. [DOI] [PubMed] [Google Scholar]
- Ashley C. T., Jr, Warren S. T. Trinucleotide repeat expansion and human disease. Annu Rev Genet. 1995;29:703–728. doi: 10.1146/annurev.ge.29.120195.003415. [DOI] [PubMed] [Google Scholar]
- Ashley C. T., Jr, Wilkinson K. D., Reines D., Warren S. T. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993 Oct 22;262(5133):563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- Ballinger S. W., Schurr T. G., Torroni A., Gan Y. Y., Hodge J. A., Hassan K., Chen K. H., Wallace D. C. Southeast Asian mitochondrial DNA analysis reveals genetic continuity of ancient mongoloid migrations. Genetics. 1992 Jan;130(1):139–152. doi: 10.1093/genetics/130.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. T., Houck G. E., Jr, Jeziorowska A., Levinson F. N., Ding X., Dobkin C., Zhong N., Henderson J., Brooks S. S., Jenkins E. C. Rapid fragile X carrier screening and prenatal diagnosis using a nonradioactive PCR test. JAMA. 1993 Oct 6;270(13):1569–1575. [PubMed] [Google Scholar]
- Burke J. R., Ikeuchi T., Koide R., Tsuji S., Yamada M., Pericak-Vance M. A., Vance J. M. Dentatorubral-pallidoluysian atrophy and Haw River syndrome. Lancet. 1994 Dec 17;344(8938):1711–1712. doi: 10.1016/s0140-6736(94)90497-9. [DOI] [PubMed] [Google Scholar]
- Buyle S., Reyniers E., Vits L., De Boulle K., Handig I., Wuyts F. L., Deelen W., Halley D. J., Oostra B. A., Willems P. J. Founder effect in a Belgian-Dutch fragile X population. Hum Genet. 1993 Oct 1;92(3):269–272. doi: 10.1007/BF00244471. [DOI] [PubMed] [Google Scholar]
- Chakravarti A. Fragile X founder effect? Nat Genet. 1992 Jul;1(4):237–238. doi: 10.1038/ng0792-237. [DOI] [PubMed] [Google Scholar]
- Chen Y. S., Torroni A., Excoffier L., Santachiara-Benerecetti A. S., Wallace D. C. Analysis of mtDNA variation in African populations reveals the most ancient of all human continent-specific haplogroups. Am J Hum Genet. 1995 Jul;57(1):133–149. [PMC free article] [PubMed] [Google Scholar]
- Chong S. S., Eichler E. E., Nelson D. L., Hughes M. R. Robust amplification and ethidium-visible detection of the fragile X syndrome CGG repeat using Pfu polymerase. Am J Med Genet. 1994 Jul 15;51(4):522–526. doi: 10.1002/ajmg.1320510447. [DOI] [PubMed] [Google Scholar]
- Edwards A., Hammond H. A., Jin L., Caskey C. T., Chakraborty R. Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genomics. 1992 Feb;12(2):241–253. doi: 10.1016/0888-7543(92)90371-x. [DOI] [PubMed] [Google Scholar]
- Eichler E. E., Holden J. J., Popovich B. W., Reiss A. L., Snow K., Thibodeau S. N., Richards C. S., Ward P. A., Nelson D. L. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet. 1994 Sep;8(1):88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- Fu Y. H., Kuhl D. P., Pizzuti A., Pieretti M., Sutcliffe J. S., Richards S., Verkerk A. J., Holden J. J., Fenwick R. G., Jr, Warren S. T. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991 Dec 20;67(6):1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Hirst M. C., Grewal P. K., Davies K. E. Precursor arrays for triplet repeat expansion at the fragile X locus. Hum Mol Genet. 1994 Sep;3(9):1553–1560. doi: 10.1093/hmg/3.9.1553. [DOI] [PubMed] [Google Scholar]
- Jacobs P. A., Bullman H., Macpherson J., Youings S., Rooney V., Watson A., Dennis N. R. Population studies of the fragile X: a molecular approach. J Med Genet. 1993 Jun;30(6):454–459. doi: 10.1136/jmg.30.6.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer E. J., Pritchard M., Lynch M., Yu S., Holman K., Baker E., Warren S. T., Schlessinger D., Sutherland G. R., Richards R. I. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991 Jun 21;252(5013):1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- Kunst C. B., Warren S. T. Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell. 1994 Jun 17;77(6):853–861. doi: 10.1016/0092-8674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Macpherson J. N., Bullman H., Youings S. A., Jacobs P. A. Insert size and flanking haplotype in fragile X and normal populations: possible multiple origins for the fragile X mutation. Hum Mol Genet. 1994 Mar;3(3):399–405. doi: 10.1093/hmg/3.3.399. [DOI] [PubMed] [Google Scholar]
- Morton N. E., Macpherson J. N. Population genetics of the fragile-X syndrome: multiallelic model for the FMR1 locus. Proc Natl Acad Sci U S A. 1992 May 1;89(9):4215–4217. doi: 10.1073/pnas.89.9.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlé I., Rousseau F., Heitz D., Kretz C., Devys D., Hanauer A., Boué J., Bertheas M. F., Mandel J. L. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991 May 24;252(5009):1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- Oudet C., Mornet E., Serre J. L., Thomas F., Lentes-Zengerling S., Kretz C., Deluchat C., Tejada I., Boué J., Boué A. Linkage disequilibrium between the fragile X mutation and two closely linked CA repeats suggests that fragile X chromosomes are derived from a small number of founder chromosomes. Am J Hum Genet. 1993 Feb;52(2):297–304. [PMC free article] [PubMed] [Google Scholar]
- Oudet C., von Koskull H., Nordström A. M., Peippo M., Mandel J. L. Striking founder effect for the fragile X syndrome in Finland. Eur J Hum Genet. 1993;1(3):181–189. doi: 10.1159/000472412. [DOI] [PubMed] [Google Scholar]
- Pieretti M., Zhang F. P., Fu Y. H., Warren S. T., Oostra B. A., Caskey C. T., Nelson D. L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991 Aug 23;66(4):817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Richards R. I., Holman K., Friend K., Kremer E., Hillen D., Staples A., Brown W. T., Goonewardena P., Tarleton J., Schwartz C. Evidence of founder chromosomes in fragile X syndrome. Nat Genet. 1992 Jul;1(4):257–260. doi: 10.1038/ng0792-257. [DOI] [PubMed] [Google Scholar]
- Richards R. I., Holman K., Kozman H., Kremer E., Lynch M., Pritchard M., Yu S., Mulley J., Sutherland G. R. Fragile X syndrome: genetic localisation by linkage mapping of two microsatellite repeats FRAXAC1 and FRAXAC2 which immediately flank the fragile site. J Med Genet. 1991 Dec;28(12):818–823. doi: 10.1136/jmg.28.12.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scozzari R., Torroni A., Semino O., Sirugo G., Brega A., Santachiara-Benerecetti A. S. Genetic studies on the Senegal population. I. Mitochondrial DNA polymorphisms. Am J Hum Genet. 1988 Oct;43(4):534–544. [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Siomi M. C., Nussbaum R. L., Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993 Jul 30;74(2):291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Smits A. P., Dreesen J. C., Post J. G., Smeets D. F., de Die-Smulders C., Spaans-van der Bijl T., Govaerts L. C., Warren S. T., Oostra B. A., van Oost B. A. The fragile X syndrome: no evidence for any recent mutations. J Med Genet. 1993 Feb;30(2):94–96. doi: 10.1136/jmg.30.2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow K., Doud L. K., Hagerman R., Pergolizzi R. G., Erster S. H., Thibodeau S. N. Analysis of a CGG sequence at the FMR-1 locus in fragile X families and in the general population. Am J Hum Genet. 1993 Dec;53(6):1217–1228. [PMC free article] [PubMed] [Google Scholar]
- Snow K., Tester D. J., Kruckeberg K. E., Schaid D. J., Thibodeau S. N. Sequence analysis of the fragile X trinucleotide repeat: implications for the origin of the fragile X mutation. Hum Mol Genet. 1994 Sep;3(9):1543–1551. doi: 10.1093/hmg/3.9.1543. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. S., Nelson D. L., Zhang F., Pieretti M., Caskey C. T., Saxe D., Warren S. T. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992 Sep;1(6):397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- Torroni A., Chen Y. S., Semino O., Santachiara-Beneceretti A. S., Scott C. R., Lott M. T., Winter M., Wallace D. C. mtDNA and Y-chromosome polymorphisms in four Native American populations from southern Mexico. Am J Hum Genet. 1994 Feb;54(2):303–318. [PMC free article] [PubMed] [Google Scholar]
- Torroni A., Miller J. A., Moore L. G., Zamudio S., Zhuang J., Droma T., Wallace D. C. Mitochondrial DNA analysis in Tibet: implications for the origin of the Tibetan population and its adaptation to high altitude. Am J Phys Anthropol. 1994 Feb;93(2):189–199. doi: 10.1002/ajpa.1330930204. [DOI] [PubMed] [Google Scholar]
- Torroni A., Schurr T. G., Cabell M. F., Brown M. D., Neel J. V., Larsen M., Smith D. G., Vullo C. M., Wallace D. C. Asian affinities and continental radiation of the four founding Native American mtDNAs. Am J Hum Genet. 1993 Sep;53(3):563–590. [PMC free article] [PubMed] [Google Scholar]
- Torroni A., Schurr T. G., Yang C. C., Szathmary E. J., Williams R. C., Schanfield M. S., Troup G. A., Knowler W. C., Lawrence D. N., Weiss K. M. Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics. 1992 Jan;130(1):153–162. doi: 10.1093/genetics/130.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A., Wallace D. C. Mitochondrial DNA variation in human populations and implications for detection of mitochondrial DNA mutations of pathological significance. J Bioenerg Biomembr. 1994 Jun;26(3):261–271. doi: 10.1007/BF00763098. [DOI] [PubMed] [Google Scholar]
- Verkerk A. J., Pieretti M., Sutcliffe J. S., Fu Y. H., Kuhl D. P., Pizzuti A., Reiner O., Richards S., Victoria M. F., Zhang F. P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991 May 31;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Wallace D. C., Torroni A. American Indian prehistory as written in the mitochondrial DNA: a review. Hum Biol. 1992 Jun;64(3):403–416. [PubMed] [Google Scholar]
- Warren S. T., Ashley C. T., Jr Triplet repeat expansion mutations: the example of fragile X syndrome. Annu Rev Neurosci. 1995;18:77–99. doi: 10.1146/annurev.ne.18.030195.000453. [DOI] [PubMed] [Google Scholar]
- Warren S. T., Nelson D. L. Advances in molecular analysis of fragile X syndrome. JAMA. 1994 Feb 16;271(7):536–542. [PubMed] [Google Scholar]
- Zerylnick C., Torroni A., Sherman S. L., Warren S. T. Normal variation at the myotonic dystrophy locus in global human populations. Am J Hum Genet. 1995 Jan;56(1):123–130. [PMC free article] [PubMed] [Google Scholar]


