Abstract
A new and severe disease of maize caused by a previously unknown fungal pathogen, Cochliobolus carbonum race 1, was first described in 1938. The molecular events that led to the sudden appearance of this disease are described in this paper. Resistance to C. carbonum race 1 was found to be widespread in maize and is conferred by a pair of unlinked duplicate genes, Hm1 and Hm2. Here, we demonstrate that resistance is the wild-type condition in maize. Two events, a transposon insertion in Hm1 and a deletion in Hm2, led to the loss of resistance, resulting in the origin of a new disease. None of the other plant species tested is susceptible to C. carbonum race 1, and they all possess candidate genes with high homology to Hm1 and Hm2. In sorghum and rice, these homologs map to two chromosomal regions that are syntenic with the maize Hm1 and Hm2 loci, indicating that they are related to the maize genes by vertical descent. These results suggest that the Hm-encoded resistance is of ancient origin and probably is conserved in all grasses.
Keywords: disease resistance genes, plant–fungal interaction, host-specific toxin, Hm1, Hm2
The question of how infectious disease originates and evolves is of great interest to both plant and animal pathologists. The complex and interactive nature of this phenomenon, which may span millions of years of coevolution between the host and its pathogen, makes it challenging to uncover the key event(s) responsible for the genesis of a disease. Not surprisingly, therefore, the molecular events that have resulted in the origin of any disease in plants have remained largely unknown. One possible exception is the southern corn leaf blight of maize, caused by race T of Cochliobolus heterostrophus (Helminthosporium maydis), in which the severity of a rather benign but already existing disease was enhanced dramatically because of the serendipitous appearance of a protein in maize mitochondria (1).
A unique opportunity to address the issue of the molecular origin of a disease is offered by the leaf spot and ear mold disease of maize. In 1938, this disease was first reported in Indiana on an inbred line, Pr (2), which was bred in Iowa from an open-pollinated cultivar, Proudfit Reid (3). Disease symptoms included grayish tan necrotic spots with concentric rings on the foliage, and infection often resulted in severe molding of the ears and premature killing of the plant. The causal agent for the disease was a fungus that resembled H. maydis, and so it was designated race 1 of H. maydis.
Further characterization of the pathogen revealed that it was a new species, and it was renamed Helminthosporium carbonum (referring to the charred appearance of an infected ear; Fig. 1; ref. 4). The sexual stage of H. carbonum was discovered in 1959, identifying the pathogen as an ascomycete belonging to the genus Cochliobolus (5). Genetic studies revealed that pathogenicity of C. carbonum race 1 is determined by a single locus, Tox2, which also confers the ability to produce H. carbonum (HC) toxin (6, 7). HC toxin is a cyclic tetrapeptide of the structure cyclo (d-Pro-l-Ala-d-Ala-l-aminoepoxyoxodecanoic acid) (8, 9). It appears to be the sole determinant of pathogenicity. Genetic variants that do not produce HC toxin are unable to colonize much beyond the site of penetration and, therefore, cause only chlorotic or necrotic flecks on leaves (10, 11). The Tox2 locus has been cloned and found to encode the enzymes required for the biosynthesis of HC toxin (11, 12). It is not clear how HC toxin allows colonization of susceptible maize. However, by virtue of its inhibitory action on histone deacetylases, HC toxin may interfere with the induction of defense genes in maize, thereby leaving the plant vulnerable to colonization by the pathogen (13).
Figure 1.
Typical charred appearance of susceptible maize ears (right two) under natural conditions of infection with C. carbonum race 1.
Three other races of C. carbonum have been reported that are slightly to moderately pathogenic on maize (14). However, their definition as races was based only on the morphology of their cultures and disease symptoms. In fact, except for race 1, none of the other races exhibits a differential reaction toward any of the maize inbreds or cultivars or produces HC toxin or even contains DNA sequences homologous to the Tox2 locus (11, 14), indicating that they must rely on pathogenicity mechanism(s) that involve factor(s) other than HC toxin.
C. carbonum race 1 is one of the most aggressive pathogens of maize. Fortunately, most maize germplasm is resistant. A dominant gene, Hm1 (named after H. maydis), confers complete protection (15), and it maps to the long arm of chromosome 1 (1L). Another gene, Hm2, provides only partial, adult plant resistance. It maps to 9L (16). The cloning of the Hm1 gene has revealed that, by encoding HC toxin reductase (HCTR), Hm1 inactivates HC toxin, and this result is sufficient to prevent infection (17, 18). As discussed in this paper, Hm2 also has been cloned and, as expected, is a duplicate of Hm1.
What was the genesis of this disease? Was it caused by the evolution of a new, toxin-producing fungus, or was it caused by the loss of a resistance mechanism of maize? The cloning of the maize Hm1 and Hm2 genes has allowed us to address this question. Here, we report that it was the breakdown of the natural mechanism of resistance (HCTR production), encoded by the Hm1 and Hm2 genes, that led to this disease. The ubiquitous distribution of Hm-homologous sequences in monocots, which in sorghum and rice also are conserved syntenically, suggests that the Hm-encoded disease resistance mechanism is highly conserved.
MATERIALS AND METHODS
Materials.
The susceptible maize inbreds used in this study were obtained from the following sources: Pr from Iowa State University, Ames, IA; K41, K44, and K61 from Kansas State University, Manhattan, KS; and MO21A from L. D. Dunkle, Purdue University, West Lafayette, IN. The Hm2 tester (br2hm1Hm2) was developed from a brachytic2 mutant (br2Hm1Hm2) stock by crossing it to Pr (Br2hm1hm2) followed by self-pollination of a few resulting progeny (17). The disease reaction of all plants used in this study was assessed by inoculating seedlings at the 3–4 leaf stage with C. carbonum race 1 (18).
DNA Extraction and Southern Blotting.
Total DNA from the seedling tissue of all plant species was isolated by a miniprep method (19). For Southern analysis, digested DNA was transferred to nylon membranes (Fisher) and hybridized in a solution containing 5× standard saline citrate (SSC), 5× Denhardt’s solution, 5% Dextran sulfate, 1 mM EDTA, 2 mM Tris (pH 7.5), 0.1% SDS, and salmon sperm DNA (10 mg⋅ml−1) (17). Blots were washed stringently in 2× SSC and 0.5% SDS at room temperature for 15 min followed by three more washes for 15 min each at 65°C in 1× SSC, 0.2× SSC, and 0.1× SSC, each containing 0.1% SDS.
Genomic and cDNA Cloning.
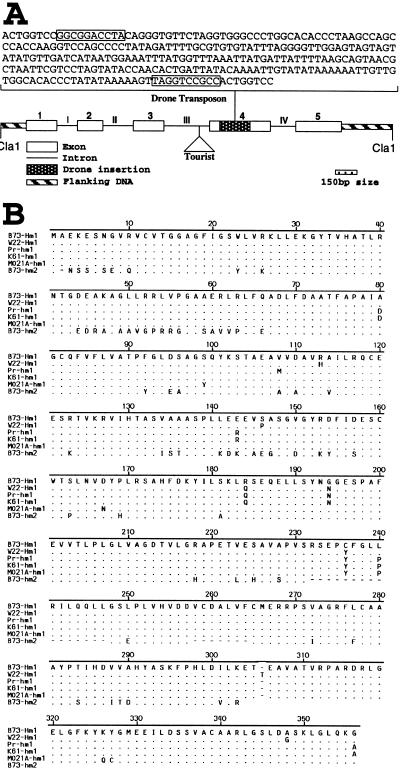
An 11-kb BamHI restriction fragment, which contained the entire hm1 gene, was isolated from a subgenomic library of Pr constructed in Lambda Dash-II vector (Stratagene). The DNA used for library construction was first size-selected for the 10- to 12-kb range by preparative gel electrophoresis. A 5.0-kb ClaI fragment containing hm1 was subcloned in the Bluescript SK+ plasmid vector (Stratagene) and was named pHMC5. The Hm1 cDNA from W22 was cloned from an endosperm-specific library (a gift from Karen Cone, University of Missouri). To characterize the hm2 allele of Pr molecularly, the gene was first cloned (unpublished work) from a B73 genomic library by using a probe derived from the 3′ half of the Hm1-cDNA. The hm2 allele of B73, which is recessive genetically, was subcloned in two parts: pHM216, a 1.6-kb BamHI fragment, which contained part of the hm2 gene that extends from the end of the exon 2 into the 5′ end of the gene (Fig. 2A); and pHM215, a 1.5-kb BamHI/SacI fragment, which contained, in addition to exons 3, 4, and 5, some sequences from the 3′ region of the hm2 gene.
Figure 2.
(A) Structural representation of hm1-Pr showing exons, introns, and two insertions, Tourist and Drone. The entire sequence of Drone (256 bp) is shown with its 10-bp terminal-inverted repeats boxed. The sequences that flank the terminal-inverted repeats represent the 8-bp duplication at the site of insertion. (B) Comparison of the deduced amino acid sequences of cDNA clones of Hm1-B73 and Hm1-W22 (two wild-type alleles of Hm1) with that of the corresponding sequence from the hm1-Pr, hm1-K61, hm1-MO21A, and hm2-B73 genomic clones. Drone insertion and the 8-bp sequence site duplication are not included in the translation products of the hm1-Pr and hm1-K61 sequences. Letters represent amino acids. In the predicted translation products of W22, Pr, K61, MO21A, and hm2-B73, only those amino acids that differ from Hm1-B73 have been shown by letter designations; identical and missing amino acids are shown by dots and dashes, respectively. The 3′ nonmatching sequence of hm2-B73, whose exon and intron boundaries are the same as hm1, is omitted. The complete sequences of W22-Hm1, Pr-hm1, K61-hm1, Mo21-hm1, and B73-hm2 have been deposited in GenBank, accession numbers AF041043, AF041044, AF041045, AF041046, and AF041047, respectively.
PCR Amplification and Cloning.
PCR amplification of exon 4 of hm1 from Pr, K61, and MO21A genomic DNA used the primers 5′-CCGGCGTTCGAGGTGGTGA-3′ and 5′-GATGTCGAGGTGAGGGAAC-3′. The entire coding region of the hm1 alleles of K61 and MO21A was PCR-amplified as follows: The primers 5′-CTGCTCATGACTCATATCAGGCGGTAGC-3′ and 5′-GACCAGCCGACGCAGCAGCCCCGCCTTC-3′ amplified hm1 from 50 nt 5′ of the start codon to the beginning of the exon 2; the primers 5′-CGGATTCGTGTGCTGGTGGGTGTGC-3′ and 5′-TACGGAGGCTGTGTGGATCACTCGC-3′ from exon 2 to the middle of exon 3; the primers 5′-AGTATAAGAGCACGGCGGAAGCTGT-3′ and 5′-CACGTCGTGGATCGTCGG-3′ from the beginning of exon 3 to the middle of exon 4; the primers 5′-ACGCGCTCGTCTTCTGCA-3′ and 5′-GACAGCGAACAATCCAAG-3′ from the middle of exon 4 to the beginning of exon 5; and the primers 5′-ATGGAAGAGATTCTGGATAG-3′ and 5′-CTAAAATTATCGCAAGTCAT-3′ from the beginning of exon 5 to the poly(A) tail site. PCR conditions were as described (17) except that the annealing temperature was 68°C instead of 60°C. All PCR-amplified products were subcloned in a TA cloning vector (Invitrogen), and two separate subclones were sequenced for each PCR product.
DNA Sequencing and Analysis.
Sequencing of both strands of the hm1-Pr subclone in pHMC5 and the Hm1-W22 cDNA clone was accomplished by the chain termination reaction method by using a T7 polymerase sequence kit (United States Biochemical). PCR-derived clones and hm2-B73 subclones pHM216 and pHM215 were sequenced by cycle sequencing with a SequiTherm DNA Polymerase kit (Epicentre Technologies, Madison, WI) using the forward and reverse primers of M13. Amino acid sequence alignment was performed with the dnastar megalign program using a clustal method with a PAM 250 residue weight table, a gap penalty of 10, and a gap length penalty of 10.
Comparative Mapping.
An Hm1-specific probe was mapped by using procedures and populations as described (20, 21), and a comparative genetic map was constructed based on marker orders, recombinational distances, and map positions of flanking markers as described elsewhere (21–23).
RESULTS AND DISCUSSION
The hm1 Gene of Pr Is Interrupted With a Transposon.
From results described in ref. 17, it was apparent that, although an allele of the hm1 gene was present in Pr, the Hm1 transcript was either much diminished or completely missing in leaves of this susceptible inbred. A null mutation in hm1-Pr might be the reason for the lack of the Hm1-specific transcript in Pr. To explore the basis for the recessive phenotype of hm1-Pr, this allele was cloned and sequenced.
Compared with the wild-type Hm1-B73 allele, three types of difference can be observed in the mutant hm1-Pr allele: (i) There is a 256-bp insertion in exon 4 (Fig. 2A). This insertion has characteristics of a transposable element, including 10-bp terminal-inverted repeats and a 8-bp duplication at the site of insertion; (ii) hm1-Pr differs from Hm1-B73 with respect to eight amino acids distributed throughout the length of the encoded protein. All of these changes are conservative, and some occur also in another wild-type allele, Hm1-W22 (Fig. 2B). It seems unlikely that these amino acid polymorphisms contribute to the defect in hm1-Pr; and (iii) intron 3 contains a benign insertion that is related to the Tourist family of transposons (24). This insertion also is present in the wild-type Hm1-B79 allele (data not shown); therefore, the insertion in exon 4, which we have named “Drone,” accounts for the mutant nature of hm1-Pr.
The hm2 Gene Is Deleted Largely in Pr.
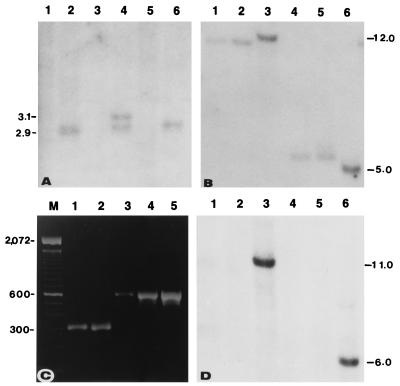
A Southern blot approach was used to examine the structure of the hm2 allele from Pr. DNA from Pr and the Hm2 tester, digested with a number of restriction enzymes, was compared side-by-side for hybridization with two hm2-specific probes, HM215 and HM216. Although no hybridization with HM215 could be detected in any of the lanes carrying the Pr DNA, one or two bands consistently were detected in all lanes containing DNA from the Hm2 tester (Fig. 3A). This result indicated that the region of the hm2 gene corresponding to the DNA present in the 3′ end subclone of hm2 was missing in the hm2 allele of Pr. When the blot was rehybridized with HM216, unique bands were detected in all lanes regardless of whether they carried the DNA from Pr or the Hm2 tester (data not shown), suggesting that the hm2 deletion in Pr did not cover the entire gene.
Figure 3.
(A) Restriction pattern of DNA from Pr (odd-numbered lanes) and the Hm2 tester (even-numbered lanes) showing deletion of parts of the recessive hm2-Pr allele corresponding to probe HM215. DNA of lanes 1 and 2, 3 and 4, and 5 and 6 were digested with SacI, XhoI, and HindIII, respectively. (B) A Southern blot of DNAs from Pr (lanes 1 and 4), K61 (lanes 2 and 5), and MO21A (lanes 3 and 6) inbreds, was digested with EcoRI (lanes 1–3) and SstI (lanes 4–6) and hybridized with a 0.9-kb SstI/XhoI fragment of Hm1. (C) PCR-amplified products from the genomic DNA of inbreds B73 (lane 1), MO21A (lane 2), Pr (lane 3), and K61 (lane 4) and from the pHMC5 subclone of hm1-Pr (lane 5). Lane M contains the 100-bp ladder. The 567-bp amplification product is diagnostic for the presence of the Drone insertion; in the absence of Drone, an amplification product of ≈311 bp is obtained. (D) Southern hybridization of the blot used in B with HM215, revealing the lack of the hm2 deletion, characteristic of Pr and K61, in MO21A.
The Mutant hm1 and hm2 Alleles of Pr Are Conserved in Other Susceptible Inbreds.
Since the discovery of the disease in Pr, additional susceptible inbreds have been found and have been shown to be homozygous recessive at both hm1 and hm2 (4, 15). Inbred K61, which was derived from the open-pollinated cultivar Pride of Saline (3), appeared to have the same mutant alleles of both hm1 and hm2 as Pr. The restriction pattern of the hm1 allele of K61 was identical to that of hm1-Pr (Fig. 3B), and the deletion pattern of the hm2 allele of K61 was identical to the corresponding deletion allele of Pr (Fig. 3D). To confirm that the hm1 allele of K61 contained the Drone insertion, DNA from Pr, K61, and a number of resistant inbreds including B73 was PCR-amplified with two gene-specific primers that flank Drone in hm1-Pr. An amplification product identical in size to that of Pr, suggestive of the presence of Drone, was detected only from the DNA of K61 and not from B73 (Fig. 3C) or any of the other resistant inbreds including Pr1, a near-isogenic resistant version of Pr (4). Subsequent sequence analysis of the entire hm1-K61 confirmed that the susceptible hm1-K61 allele also is the result of Drone insertion. In addition, the hm1-K61 allele also contains seven of the eight amino acid changes present in the hm1-Pr allele (Fig. 2B) and a benign insertion in the intron 3. These results indicate that the sequences of both the hm1 and hm2 alleles of K61 are identical to the corresponding alleles of Pr.
The pedigree relationship between Pr and K61 is untraceable. However, two other susceptible inbreds, K41 and K44, which, like K61, were derived from Pride of Saline, carry the same mutant alleles of hm1 and hm2 as K61 and Pr (data not shown). This suggests that these mutant alleles of hm1 and hm2, which resulted in the appearance of the leaf spot and ear rot disease during the 1930s, may originally have occurred in Pride of Saline (3, 4). The pathogen probably was present in the Midwest before the 1930s; as it has been isolated from old seeds and herbarium specimens (25, 26). Two factors may have hindered the development and, therefore, recognition of the disease before the 1930s. First, maize, being a cross-pollinated species, minimizes the generation of recessive homozygotes in an open-pollinated population, thereby reducing the accumulation of susceptible plants (homozygotes for both hm1 and hm2 in this case). Second, the semiarid climate of Kansas also may have been a factor because the development of disease caused by C. carbonum race 1 is highly dependent on warm and humid conditions. When inbred lines (homozygous for both the mutant alleles) were moved to the humid corn belt, the disease caused by this previously unknown pathogen could be expected to occur.
The Susceptible Inbred MO21A Has a Unique Set of Mutant hm1 and hm2 Alleles.
To address further whether natural mutations in Hm1 and Hm2 have occurred only once in maize, we characterized both hm loci from another susceptible inbred, MO21A, which was derived from Laguna, an open-pollinated cultivar from Mexico (3). Because the genotypic makeup of this inbred in terms of susceptibility to C. carbonum race 1 had never been characterized, a number of genetic crosses were made that revealed that MO21A was homozygous recessive at both the hm1 and hm2 loci. These crosses included an allelism test between MO21A and Pr and outcrosses of MO21A with Pr1 and the Hm2 tester followed by self-pollination of the resulting hybrids.
A Southern blot procedure was used to examine the molecular nature of hm1 and hm2 in MO21A. The hybridization pattern of the hm1 allele of MO21A was distinct from the common pattern of both Pr and K61 (Fig. 3B). The deletion that is characteristic of the recessive hm2 alleles of both Pr and K61 is lacking in MO21A (Fig. 3D). The results from PCR amplification using Drone flanking primers showed that the hm1-MO21A allele lacked Drone (Fig. 3C). Subsequent sequence determination of the entire hm1-MO21A allele has confirmed that it is different from hm1-Pr (Fig. 2B). Although we do not know yet what mutational events have caused defects in the hm1 and hm2 alleles of MO21A, it is clear from the data presented that the defects in the hm1 and hm2 alleles of MO21A are different from the lesions that led to the generation of the common mutant alleles of Pr and K61. Thus, the susceptibility to C. carbonum race 1 has originated naturally at least twice in maize. Given the propensity of genetic instability in maize that has many systems of active transposable elements (27, 28), together with the amount of genetic and breeding research that has gone into characterizing and shaping this crop species, it is not too surprising.
Presence of Hm1-Homologous Sequences in Other Plant Species.
The results suggest that maize normally is not a host of C. carbonum race 1. But, when Hm1 and Hm2 are disrupted, maize can no longer inactivate HC toxin and, as a result, becomes susceptible.
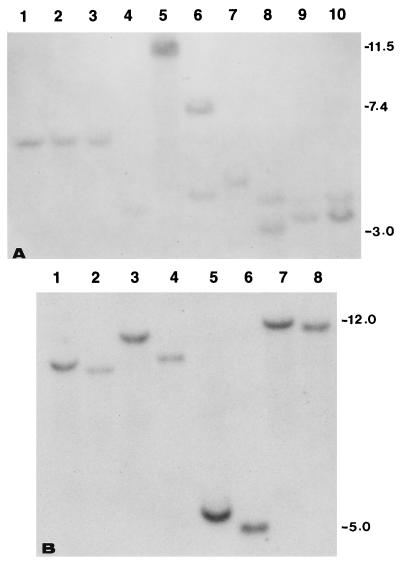
The question then is: Did Hm-encoded resistance evolve exclusively in maize or is it shared by other plant species? To address this question, DNA from monocots was hybridized with the full length Hm1-cDNA under high stringency conditions. Unique bands were detected in all lanes (Fig. 4A), suggesting that all plants tested possess sequences highly homologous to Hm1. In fact, a barley cDNA (GenBank accession no. U77463) (29) and a rice cDNA (GenBank accession no. E10912) have been identified that are more than 70% identical to the maize Hm1 gene at the amino acid level (Fig. 5). An additional homolog of Hm1 has been isolated from barley (sequenced tagged site; GenBank accession no. L43997), which cross-hybridizes specifically to the maize Hm1 gene (Fig. 4B).
Figure 4.
(A) A Southern blot (BglII digestion) showing hm1-homologous restriction fragments in Pr (lane 1), K61 (lane 2), K44 (lane 3), wheat (lane 4), rice (lane 5), sorghum (lane 6), Tripsacum (lane 7), oat (lane 8), and two different accessions of Teosinte (lanes 9 and 10). The blot was probed with the full length Hm1-cDNA and washed under conditions of high stringency. (B) An overlay of autorads of a gel blot that was probed first with Hm1 and then, after stripping, with ABG-459, (the barley STS, GenBank accession no. L43997). DNA of Pr (odd-numbered lanes) and Pr1 (even-numbered lanes) was digested with BamHI (lanes 1 and 2), EcoRI (lanes 3 and 4), SstI (lanes 5 and 6), and EcoRV (lanes 7 and 8).
Figure 5.
Predicted amino acid sequence comparisons of full length cDNAs of maize Hm1-B73 (GenBank accession no. L02540), rice ( DNA Data Bank of Japan accession no. E10912), and barley (GenBank accession no. U77463). Shaded areas represent homologous amino acid residues. A missing amino acid residue is represented by a dash.
There are two obvious explanations for the ubiquitous presence of Hm-hybridizing sequences among monocots. Either the Hm-encoded disease resistance function (HCTR) in maize is recruited, by necessity or serendipity, from a gene(s) that performs an essential but unknown function or it represents a disease resistance mechanism of ancient origin that has evolved in a common ancestor for protection against pathogens that used HC toxin or related cyclic tetrapeptides as part of their pathogenic arsenal. Because there are no obvious pleiotropic effects associated with any of the mutations of Hm1 and Hm2, all of which are null for both the HCTR activity and the resistance function (17, 18), the only nonredundant function of Hm1 and Hm2 is defense against HC toxin. Furthermore, homologs of Hm1 from barley and rice, from which maize diverged at least 70 million years ago (23, 30), are highly conserved in sequence, suggesting that they perform the same function. In fact, all monocots tested possess HCTR activity (31), except the maize inbreds and mutants that are susceptible to C. carbonum race 1 (18).
Syntenic Conservation of Hm1 and Hm2 in Maize, Sorghum, and Rice.
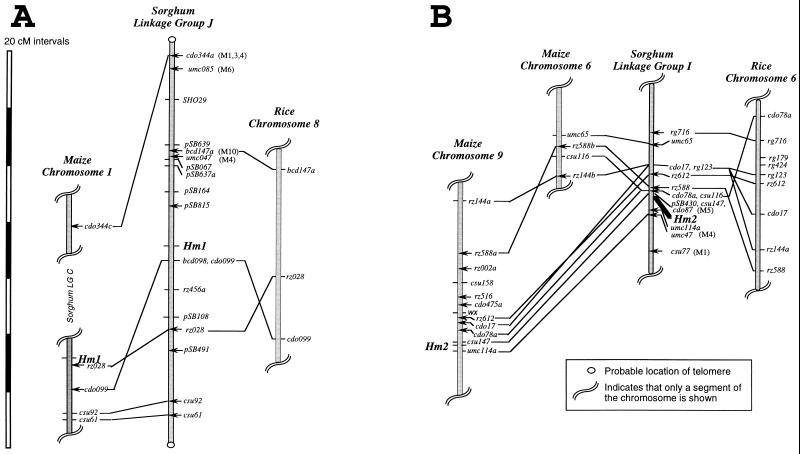
To determine whether any of the homologs in nonmaize plants have ancestral relationships with the maize Hm1 and Hm2 genes, a probe from the 5′ end of the Hm1 gene was mapped in both sorghum and rice. Two restriction fragments, one major and the other minor, were observed in both species. The genomic organization of both Hm1 and Hm2 is conserved among maize, sorghum, and rice. In both sorghum and rice, the major band was mapped to the syntenically conserved region of maize 1L, where Hm1 is located (Fig. 6A). Similarly, the minor band in both sorghum and rice was mapped to the region that is colinear with Hm2 on 9L in maize (Fig. 6B). In agreement with these results, one of the Hm1 homologs of barley appears to map to a location that is related syntenically with the maize Hm2 location (29). Given the conservation in gene composition and colinearity among the genomes of the grass family (30, 32–34), these results clearly indicate that the Hm homologs of both sorghum and rice are orthologs of the maize Hm1 and Hm2 genes. In other words, they are derived from a common ancestor by vertical descent.
Figure 6.
Comparative mapping of an Hm1-specific probe in maize, rice, and sorghum. The genetic maps are based on marker orders, recombinational distances, and populations described elsewhere (21, 22). DNA marker loci indicated by (—) were mapped directly in the cited populations, whereas those indicated by ← were mapped in other populations (ref. 23 and unpublished data), and the appropriate locations were inferred based on map positions of flanking markers. For DNA markers that conflict with the most parsimonious interpretation of chromosomal correspondence between taxa, conflicting map positions are indicated in parentheses (M, maize). Comparative markers were mapped in as few as 56 individuals, and thus reversals in order of closely linked loci (<3 cM) are not conclusive evidence of chromosomal rearrangement.
Conceptual and Evolutionary Implications.
In addition to providing a molecular explanation for the origin of a disease, this study also has a conceptual implication regarding nonhost resistance. A view among plant pathologists is that nonhost resistance in plants is caused by the lack of basic compatibility between host and pathogen. This view is contrasted by the findings of this study, which indicate that nonhost resistance in plants against some pathogens can be caused by the presence of an active, durable resistance function. The susceptibility of maize to C. carbonum race 1 constitutes the first documented case in plants in which the breakdown of a nonhost resistance mechanism has led to the birth of a disease.
The presence of HCTR activity in all monocots tested (31) and the unique absence of this activity in all C. carbonum race 1-susceptible inbreds and mutants of maize, suggest that the function of Hm-homologous sequences in plants specifically is to defend against HC toxin or related molecules. A logical extension of this concept is that HC toxin (or its related cyclic tetrapeptides) may have been important in plant disease before the existence of the Hm-encoded HCTR function. In other words, the recent genesis of the leaf spot and ear mold disease of maize may have uncovered an ancient case of parasitism. Considering the exceptional virulence endowed on C. carbonum race 1 by HC toxin, this factor (or a related metabolite) may have played a significant role in the evolution or geographical distribution of plant species.
Because of the selective toxicity of HC toxin on susceptible maize, it has been categorized as a host-specific toxin capable of inflicting damage only on maize (35, 36). Our results suggest that the host-selective effects of HC toxin and, therefore, the host range of the pathogen are dictated by HCTR activity. The probable antiquity and wide distribution of HCTR function in plants predicts that HC toxin may interfere with some general but fundamental aspect of cell function that renders a plant susceptible to colonization by a fungus. Consistent with this proposal are the recent findings that implicate HC toxin and its structural analogs as potent inhibitors of histone deacetylases (13, 37). Deacetylation of histones may be a prerequisite for the inducible expression of genes (38). Because much of plant defense relies on inducible mechanisms, it has been proposed that suppression of histone deacetylase is the mechanism by which HC toxin permits fungal colonization of maize (13). In fact, a recent study provides some evidence for this (39). Given the ubiquitous ability of HC toxin to inhibit histone deacetylases from all organisms tested (13), one may wonder how dicots, in which HCTR activity has been undetected thus far (18), defend against HC toxin.
Acknowledgments
We thank Yann-Rong Lin and Charlene Chang for help with the comparative mapping of the Hm genes and S. G. Pueppke, A. K. Chatterjee, M. McMullen, N. Yalpani, G. Hu, and P. S. Virk for their helpful comments and critical reading of the manuscript. This work was supported in part by grants from the United States Department of Agriculture Cooperative State Research Service and Pioneer Hi-Bred International Inc.
ABBREVIATIONS
- HC
H. carbonum
- HCTR
HC toxin reductase
- SSC
standard saline citrate
Footnotes
References
- 1.Levings C S, III, Siedow J N. Plant Mol Biol. 1992;19:135–147. doi: 10.1007/BF00015611. [DOI] [PubMed] [Google Scholar]
- 2.Ullstrup A J. Phytopathology. 1941;31:508–521. [Google Scholar]
- 3.Gerdes J T, Behr C F, Coors J G, Tracy W F. Compilation of North American Maize Breeding Germplasm. Madison, WI: Am. Soc. Agron.; 1993. [Google Scholar]
- 4.Ullstrup A J. Phytopathology. 1944;34:214–222. [Google Scholar]
- 5.Nelson R R. Phytopathology. 1959;49:807–810. [Google Scholar]
- 6.Scheffer R P, Ullstrup A J. Phytopathology. 1965;55:1037–1038. [Google Scholar]
- 7.Scheffer R P, Nelson R R, Ullstrup A J. Phytopathology. 1967;57:1288–1289. [Google Scholar]
- 8.Gross M L, McCrery D, Crow F, Tomber K B, Pope M R, Ciufetti L M, Knoche H W, Daly M M, Dunkle L D. Tetrahedron Lett. 1982;23:5381–5384. [Google Scholar]
- 9.Walton J D, Earle E D, Gibson B W. Biochem Biophys Res Commun. 1982;107:785–794. doi: 10.1016/0006-291x(82)90592-7. [DOI] [PubMed] [Google Scholar]
- 10.Comstock J C, Scheffer R P. Phytopathology. 1973;63:24–29. [Google Scholar]
- 11.Panaccione D G, Scott-Craig J S, Pocard J A, Walton J D. Proc Natl Acad Sci USA. 1992;89:6590–6594. doi: 10.1073/pnas.89.14.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott-Craig J S, Panaccione D G, Pocard J A, Walton J D. J Biol Chem. 1992;267:26044–26049. [PubMed] [Google Scholar]
- 13.Brosch G, Ransom R, Lechner T, Walton J D, Loidl P. Plant Cell. 1995;7:1941–1950. doi: 10.1105/tpc.7.11.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones M J, Dunkle L D. Phytopathology. 1993;83:366–370. [Google Scholar]
- 15.Ullstrup A J. J Agric Res. 1941;63:331–334. [Google Scholar]
- 16.Nelson O E, Ullstrup A J. J Hered. 1964;55:195–199. [Google Scholar]
- 17.Johal G S, Briggs S P. Science. 1992;258:985–987. doi: 10.1126/science.1359642. [DOI] [PubMed] [Google Scholar]
- 18.Meeley R B, Johal G S, Briggs S P, Walton J D. Plant Cell. 1992;4:71–77. doi: 10.1105/tpc.4.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dellaporta S L, Wood J, Hicks J B. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- 20.Paterson A H, Lin Y R, Li Z K, Schertz K F, Doebley J F, Pinson S R, Liu S C, Stansel J W, Irvine J E. Science. 1996;269:1714–1718. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- 21.Lin Y R, Schertz K F, Paterson A H. Genetics. 1995;141:391–411. doi: 10.1093/genetics/141.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coe E H, Neuffer M G. In: Genetic Maps. 6th Ed. O’Brien S J, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 157–189. [Google Scholar]
- 23.Ahn S, Tanksley S D. Proc Natl Acad Sci USA. 1993;90:7980–7984. doi: 10.1073/pnas.90.17.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bureau T E, Wessler S R. Proc Natl Acad Sci USA. 1994;91:1411–1415. doi: 10.1073/pnas.91.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheffer R P. In: Phytotoxins and Plant Pathogenesis. Graniti A, editor. H27. Berlin: Springer; 1989. pp. 285–300. [Google Scholar]
- 26.Pringle R B, Scheffer R P. Annu Rev Phytopathol. 1964;2:133–156. [Google Scholar]
- 27.Wessler S R, Bureau T E, White S E. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 28.Osborne B I, Baker B. Curr Opin Cell Biol. 1995;7:406–413. doi: 10.1016/0955-0674(95)80097-2. [DOI] [PubMed] [Google Scholar]
- 29.Han F, Kleinhofs A, Kilian A, Ullrich S E. Mol Plant-Microbe Interact. 1997;10:234–239. doi: 10.1094/MPMI.1997.10.2.234. [DOI] [PubMed] [Google Scholar]
- 30.Bennetzen J L, Freeling M. Trends Genet. 1993;9:259–261. doi: 10.1016/0168-9525(93)90001-x. [DOI] [PubMed] [Google Scholar]
- 31.Meeley R B, Walton J D. In: Advances in Molecular Genetics of Plant-Microbe Interactions. Nester E W, Verma D P S, editors. Vol. 2. Dordrecht, The Netherlands: Kluwer; 1993. pp. 463–475. [Google Scholar]
- 32.Hulbert S H, Richter T E, Axtell J D, Bennetzen J L. Proc Natl Acad Sci USA. 1990;87:4251–4255. doi: 10.1073/pnas.87.11.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitkus R, Doebley J, Lee M. Genetics. 1992;132:1119–1130. doi: 10.1093/genetics/132.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahn S, Anderson J A, Sorrells M E, Tanksley S D. Mol Gen Genet. 1993;241:483–490. doi: 10.1007/BF00279889. [DOI] [PubMed] [Google Scholar]
- 35.Walton J D. Plant Cell. 1996;8:1723–1733. doi: 10.1105/tpc.8.10.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoder O C. Annu Rev Phytopathol. 1980;18:103–129. [Google Scholar]
- 37.Yoshida M, Horinouchi S, Beppu T. BioEssays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 38.Lopez-Rodas G, Brosch G, Georgieva E I, Sendra R, Franco L, Loidl P. FEBS Lett. 1993;317:175–180. doi: 10.1016/0014-5793(93)81271-z. [DOI] [PubMed] [Google Scholar]
- 39.Crane V, Yalpani N, Briggs S. In: Biology of Plant–Microbe Interactions. Stacey Y, Mullin B, Gresshoff P, editors. St. Paul, MN: Int. Soc. Mol. Plant–Microbe Interactions; 1996. pp. 223–226. [Google Scholar]