Abstract
Prader–Willi syndrome (PWS) and Angelman syndrome are neurogenetic disorders caused by the lack of a paternal or a maternal contribution from human chromosome 15q11-q13, respectively. Deletions in the transcription unit of the imprinted SNRPN gene have been found in patients who have PWS or Angelman syndrome because of a parental imprint switch failure in this chromosomal domain. It has been suggested that the SNRPN exon 1 region, which is deleted in the PWS patients, contains an imprint switch element from which the maternal and paternal epigenotypes of the 15q11-q13 domain originate. Using the model organism Drosophila, we show here that a fragment from this region can function as a silencer in transgenic flies. Repression was detected specifically from this element and could not be observed with control human sequences. Additional experiments allowed the delineation of the silencer to a fragment of 215 bp containing the SNRPN promoter region. These results provide an additional link between genomic imprinting and an evolutionary conserved silencing mechanism. We suggest that the identified element participates in the long range regulation of the imprinted 15q11-q13 domain or locally represses SNRPN expression from the maternal allele.
Prader–Willi syndrome (PWS) and Angelman syndrome (AS) involve oppositely imprinted genes on human chromosome 15q11-q13: the paternally expressed PWS gene(s) and the maternally expressed AS gene (1). A candidate gene for AS has been identified recently (2, 3) whereas the gene(s) for PWS remains elusive. One imprinted gene from the PWS critical region is the gene encoding the small nuclear ribonucleoprotein polypeptide N (SNRPN), which is expressed exclusively from the paternal allele (4–7).
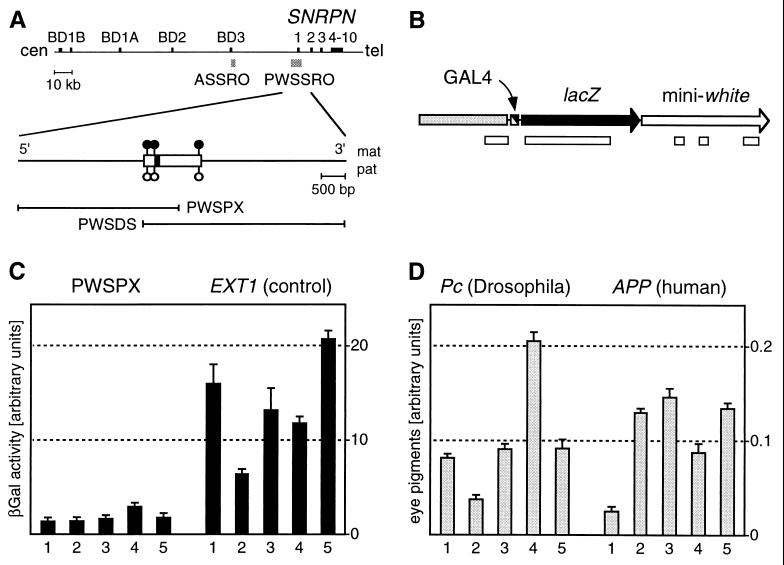
The SNRPN transcription unit has been found to contain small deletions in several PWS and AS families where the syndrome originates from an imprinting defect (8–12). In AS patients of this group, the smallest region of deletion overlap (Fig. 1A, ASSRO) is located several hundred kilobase pairs centromeric to the AS gene. The ASSRO is very close to exon BD3 of the paternally transcribed, noncoding BD transcripts of SNRPN (Fig. 1A; ref. 11). On the other hand, all deletions in PWS-imprinting mutation patients involve a region around exon 1 of SNRPN (Fig. 1A, PWSSRO; ref. 11). It has been suggested that the BD transcripts are required for a switch from the paternal to the maternal epigenotype that would fail in AS patients whereas the region around SNRPN exon 1 might contain a switch initiation site from which both the maternal and paternal epigenotypes originate (11). Alternatively, it has been proposed that the PWSSRO region might be required for erasing maternal and paternal epigenotypes in both germ lines (13).
Figure 1.
(A) Outline of the SNRPN transcription unit indicating regions deleted in AS and PWS patients (ASSRO and PWSSRO, shaded boxes; refs. 10–12). The PWSSRO has been narrowed down recently to ≈3 kb (T. Ohta and R. D. Nicholls, personal communication). Exons are shown as black boxes. Filled and open lollipops in the region of the SNRPN CpG island (open box) represent maternally methylated NotI sites (32). Fragments investigated in this study (PWSPX and PWSDS) are indicated. (B) Schematic outline of the PWSPX transgene. A GAL4 binding cassette (black and white box) immediately 5′ to the promoter mediates binding of the activating protein. The direction of transcription is indicated by arrowheads. Open boxes below the construct denote CpG islands as determined by standard procedures (33) by using grail (34). (C) Quantitative determination of lacZ expression in GAL4-induced transgenic larvae carrying the PWSPX fragment or a control fragment from the nonimprinted EXT1 locus. (D) Quantitative determination of mini-white expression in adult transgenic flies carrying a sequence from the Drosophila Pc locus or a sequence from the human APP locus immediately 5′ to mini-white. Error bars indicate the SD; numbers indicate distinct independent strains.
The fruit fly Drosophila melanogaster has proven to be a valuable genetic model for studying epigenetic mechanisms of gene regulation. Epigenetic phenomena, like position effect variegation and homeotic gene silencing, have been subjected to a genetic dissection. Many of the genes involved were found to encode chromatin-regulating factors (14). Because no covalent modification of DNA has been observed in Drosophila, these results imply that silencing can be sustained by other epigenetic mechanisms. Indeed, it has been shown recently that an imprinting element from the mouse H19 flanking region functions as a silencer in Drosophila (15), suggesting that the fly is a suitable model to investigate certain mechanistic aspects of genomic imprinting.
We show here that a fragment from the region around the SNRPN exon 1 can function as a parent-of-origin-independent silencer in transgenic flies. Repression was specific for this element and could not be observed with nonimprinted human sequences. Additional transgenes resulted in the delineation of the silencer to a 215-bp sequence containing the SNRPN promoter region (16). We propose that the observed silencing is based on an evolutionary conserved mechanism that also is used for allele-specific repression of mammalian imprinted genes.
MATERIALS AND METHODS
Recombinant Plasmids.
The P element vectors were generated by subcloning fragments of interest in the NotI/SpeI linker of pUZ (15). pUZC contains a 2.8-kb XbaI fragment from the human EXT1 locus (chromosome 8; ref. 17) including exon 7 plus flanking sequences. The PWSPX fragment (bp −2651 to +538 relative to bp 1 of SNRPN exon 1) was PCR-amplified by using SpeI cloning primers and was subcloned in pUZ to yield pUZP1 and pUZP2, respectively. pUZP3 contains the PWSDS fragment, a 4.2-kb XbaI fragment (bp −203 to +4094), from the human SNRPN locus. pUZP1Δ contains a 2.4-kb SpeI–XbaI subfragment (bp −2651 to −203) of the PWSPX fragment, and pUZP2Δ contains a 0.7-kb SpeI–XbaI subfragment (bp −203 to +538) of the PWSPX fragment. Base pairs −203 to +12 of a pUZP2Δ clone carrying the human fragment in 5′–3′ orientation were removed by NotI digestion to generate pUZPPΔ. The constructs carrying sequences from the human APP locus (18) and the Drosophila Pc locus (19) each contain a 3-kb cDNA fragment subcloned in the vector pUAST (20).
Transgenic Flies.
Fly stocks were maintained at 25°C on standard medium. Transgenic D. melanogaster were generated (21) using white1118 as host and pUChsΔ2–3 as helper plasmid. Transformed flies were identified by rescue of the white eye phenotype in the F1 generation after backcrossing to the host strain. Only independent founders were used for the generation of stocks. All transgenic strains are homozygous viable and carry the transposon on an autosome. When necessary, transgenes were mobilized by using w; ry506 Sb1 P[ry+ Δ2–3]99B/TM6B as a stable source of transposase (22). Integrity, single copy status, and independence of transgenes were confirmed by genomic Southern blots by using standard procedures (23).
lacZ and mini-white Expression.
Quantitative determination of β-galactosidase (β-gal) activity and red eye pigments was essentially as described (15). All measurements were repeated independently two times. Values from all constructs were compared by using the t test. The difference between lines classified as “silenced” and as “not silenced” was always significant (P < 0.01) as determined by the t test.
Reverse Transcription–PCR Assays.
Transgenic strains were crossed with the 1032.hx GAL4-enhancer trap line (24) or white1118. Total RNA was isolated from wandering third instar larvae by using the S.N.A.P. Total RNA Isolation Kit (Invitrogen) including DNase digestion. cDNA synthesis was performed from 1 μg of total RNA by using the cDNA Cycle Kit (Invitrogen). Control experiments were run in parallel, omitting reverse transcriptase. Ten percent of the cDNA products was amplified by 33 cycles of PCR by using primers specific for SNRPN exon 1/intron 1 (GCGGTCAGTGACGCGATGGAGCG and CCGGATCTGGTTCTCCAGAACAAAGGAC) and lacZ (GAGCCTGCTAAAGCAAAAAAGAAGTCACC and CGTAACCGTGCATCTGCCAGTTTGAGG). PCR was performed separately for SNRPN and lacZ. Assays then were pooled and samples were separated on a 1.8% agarose gel stained with ethidium bromide.
RESULTS
Generation of Transgenic Flies Carrying Fragments from the 15q11-q13 Imprinting Center.
To identify putative silencing elements from the SNRPN exon 1 region (Fig. 1A), we made use of a GAL4 competition system established previously (ref. 24; Fig. 1B). Fragments of interest were subcloned in a vector containing the lacZ gene under the control of an hsp70 minimal promoter. Activation of this promoter can be conferred by GAL4 protein that is supplied in trans from a strain expressing the GAL4 gene under developmental control. The mini-white transformation marker, which is required for the deposition of the fly’s red eye pigment, lies downstream of the lacZ gene (Fig. 1B). Using P element-mediated transformation, we generated several independent transgenic fly lines from the construct containing the PWSPX fragment (Fig. 1A) and a control construct containing a random fragment of nonimprinted human DNA.
Repression of lacZ and mini-white Expression.
To determine the effect of these sequences on lacZ expression, we crossed transgenic strains with flies expressing GAL4 under developmental control. Third instar larvae from these crosses were homogenized, and β-gal activity was quantitatively determined by using chlorophenolred-β-d-galactopyranoside. This revealed a profound silencing effect in all of the strains transgenic for the PWSPX fragment compared with all of the strains transgenic for the control fragment (Fig. 1C). Silencing was found independently of the parental origin of the transgene (data not shown), and the PWSPX fragment was sufficient to mediate an ≈8-fold reduction in β-gal activity (P < 0.001).
From the appearance of the flies transgenic for the PWSPX fragment, it was evident that also the more distantly located mini-white transformation marker (Fig. 1B) was expressed at lower levels. Quantitative determination of red eye pigments revealed that pigment levels are reduced significantly in flies carrying the PWSPX fragment compared with control flies (P = 0.01; data not shown). Thus, the fragment appears also to be capable of long distance repression of the mini-white promoter. However, the effect was not as pronounced as for the lacZ gene (data not shown).
Additional controls were performed to demonstrate that the introduction of foreign DNA into Drosophila has no effect on the expression of neighboring reporter genes. We compared white expression in two groups of five strains each carrying either a transgene with DNA from the human APP locus (ref. 18; chromosome 21q21-q22) or a transgene with DNA from the Drosophila Pc locus (19). In these constructs, the mini-white gene is placed immediately 3′ to the APP and Pc sequences, respectively, as is the lacZ gene in the constructs described above. Eye pigment quantification of adult age-matched flies yielded an average value of 0.105 for the human-related transgene vs. 0.102 for the Drosophila-related transgene (Fig. 1D). Thus, mini-white expression is not affected by the presence of the human sequences. The difference between the two transgenes is not significant as determined by the t test (P = 0.87).
Characterization and Delineation of the Silencing Element.
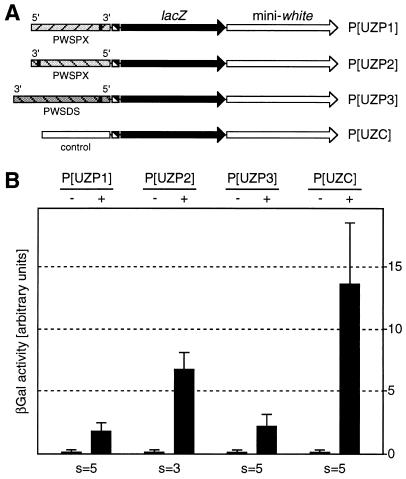
To initiate the characterization of the silencing element, we analyzed lacZ expression in GAL4-induced fly lines transgenic for P[UZP2] (Fig. 2A). This construct contains the PWSPX fragment in 3′–5′ orientation. Chlorophenolred-β-d-galactopyranoside assays performed under the same conditions as previously used revealed a reduced silencing activity compared with P[UZP1] (Fig. 2B). This result could be caused by a directionality effect in the sense that silencing is stronger in the 3′ direction of the PWSPX fragment. Alternatively, reduced silencing could be attributed to a distance effect indicating that the silencing element is close to the repressed hsp70–lacZ promoter in P[UZP1] but farther away in P[UZP2]. If the silencer were located to the immediate vicinity of SNRPN exon 1, it also should be present in the PWSDS fragment that shares with the PWSPX fragment 740 bp around SNRPN exon 1 (Fig. 1A). Therefore, we analyzed lacZ expression in GAL4-induced strains transgenic for P[UZP3], a construct containing the PWSDS fragment in 3′–5′ orientation (Fig. 2A). These strains showed a degree of silencing that was indistinguishable from P[UZP1], suggesting that silencing is bidirectional (Fig. 2B). Reduced silencing of the P[UZP2] lacZ gene could then be explained by a distance effect that would be consistent with the moderate repression observed for the more distant mini-white gene in P[UZP1].
Figure 2.
Quantitative determination of lacZ expression in transgenic larvae carrying fragments from the PWS-imprinting control region. (A) Structure of transgenes. (B) Relative β-gal activity in the presence (+) or absence (−) of GAL4. The depicted values are mean values for all strains analyzed. s, number of strains; error bars indicate the SD.
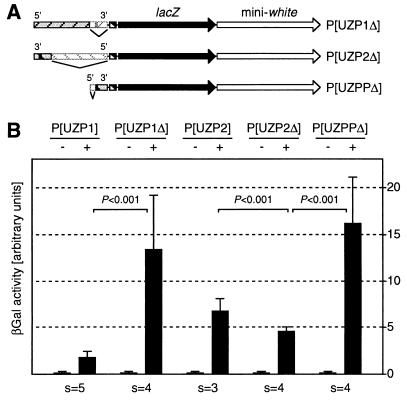
To further characterize and delineate the SNRPN silencing element, we generated strains transgenic for deleted derivatives of the original constructs. In P[UZP1Δ], the 740-bp region of P[UZP1] overlapping with P[UZP3] is deleted whereas in P[UZP2Δ] only the 740-bp region of P[UZP2] shared with P[UZP3] is retained (Fig. 3A). Chlorophenolred-β-d-galactopyranoside assays showed that β-gal activity in larvae transgenic for P[UZP1Δ] was drastically increased compared with P[UZP1] (Fig. 3B). Furthermore, β-gal activity was moderately decreased from P[UZP2Δ] compared with P[UZP2] (Fig. 3B). Expression levels from P[UZP1Δ] were similar to P[UZC] controls confirming that the 740-bp fragment was required for silencing. Expression levels from P[UZP2Δ] are similar to P[UZP1] although repression is not as strong as from the latter transgene. These experiments thus confirmed the previous assumptions that silencing is bidirectional and shows a distance effect. In addition, the deletions uncovered a 740-bp fragment contained largely in the SNRPN CpG island (Fig. 1A) that is necessary and also is partially sufficient for silencing.
Figure 3.
The effect of deletions on PWSPX-dependent silencing. (A) Structure of deleted transgenes. P[UZP1Δ] is derived from P[UZP1] and contains a 740-bp deletion of the region around SNRPN exon 1. P[UZP2Δ] is derived from P[UZP2] and contains a 2.4-kb deletion leaving the 740-bp region around SNRPN exon 1. P[UZPPΔ] is derived from P[UZP2Δ] and contains a 215-bp deletion of the SNRPN promoter region. (B) Quantitative determination of lacZ expression in transgenic larvae carrying deleted constructs. Relative β-gal activity is shown in the presence (+) or absence (−) of GAL4. The depicted values are mean values for all strains analyzed (s, number of strains). Error bars indicate the SD, and P values were determined by the t test.
The only functionally characterized element within the 740-bp fragment is the SNRPN promoter region (16). To determine the effect of this region on silencing, we deleted the region from −203 bp to +12 bp relative to SNRPN exon 1 (Fig. 3A, P[UZPPΔ]). The deletion resulted in a clear loss of silencing with lacZ expression levels being similar to the P[UZC] control construct and the P[UZP1Δ] deletion construct (Fig. 3B). Because the remaining DNA of P[UZPPΔ] is entirely derived from the SNRPN CpG island (9, 16), this result demonstrates also that the presence of CpG-rich DNA per se is not sufficient to induce silencing in the fly.
SNRPN and lacZ Promoter Activity in Transgenic Flies.
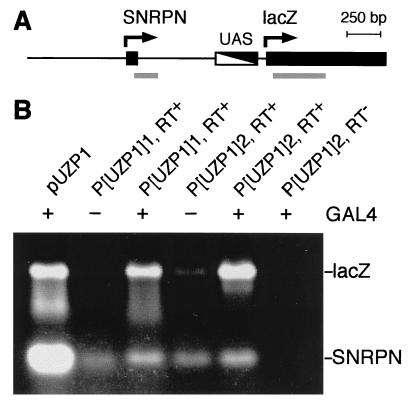
Because the SNRPN promoter is contained in all repressive fragments, silencing could have resulted from a competition between the SNRPN and the lacZ promoters for GAL4-mediated activation. If this were the case, SNRPN transcription should be induced strongly upon GAL4 introduction. To determine the state of activity of the SNRPN and the lacZ promoters, we performed reverse transcription-PCR (Fig. 4A). We isolated total RNA from the same larval stage that was used for the determination of β-gal activity. A control PCR from pUZP1 plasmid yielded a strong signal for lacZ and a very strong signal for SNRPN (Fig. 4B, lane 1). In the absence of GAL4, no signal, or a very faint signal, was detectable for lacZ mRNA. In contrast, a strong signal was observed in the presence of GAL4 indicating a substantial GAL4-mediated induction of lacZ expression (Fig. 4B). The signal for SNRPN mRNA was only fractional compared with the signal amplified from the plasmid template. This signal always could be detected and depended only a little on GAL4 induction (Fig. 4B). Thus, we concluded that promoter competition can hardly account for the observed silencing effect. We also noticed that low levels of SNRPN mRNA were present even in uninduced and silenced larvae. This result may point to the requirement of DNA methylation for vertebrate genomes to achieve a more stringent repression of transcription. Because the gene number of the unmethylated (25, 26) Drosophila genome is relatively small compared with vertebrates, a certain level of “transcriptional noise” may be tolerated and may explain the dispensability of DNA methylation in the fly (27).
Figure 4.
Determination of the transcriptional status of the SNRPN and the lacZ promoters in transgenic larvae carrying the PWSPX fragment. (A) Schematic illustration of the central part of the P[UZP1] transgene. SNRPN exon 1 and the 5′ portion of the lacZ gene are depicted as black boxes, and the GAL4 binding sites (UAS) are depicted as a black and white box. Arrows indicate the direction of transcription. Gray lines designate the sequences amplified by PCR. (B) Result of reverse transcription-PCR from two different strains transgenic for P[UZP1] in the presence (+) or absence (−) of GAL4. PCR products were separated on an agarose gel stained with ethidium bromide. Lane 1 shows control PCR products from a plasmid template, and lane 6 shows the absence of reverse transcription-PCR products when reverse transcriptase was omitted.
DISCUSSION
The recent identification of a comparatively small element encompassing a presumptive imprinting center on human chromosome 15q11-q13 (11) presented the opportunity to use the advantages of Drosophila transgenesis to dissect further and characterize this element. Using an established GAL4 competition system (24), we demonstrated here that an isolated sequence from the human SNRPN locus was able to confer strong repression on a minimal hsp70 promoter. The degree of silencing was comparable with the repression induced by strong endogenous Drosophila silencers (24) and also with the silencing element previously identified in the mouse H19 upstream region (15).
The results obtained with the deleted constructs show that the region from −203 bp to +12 bp relative to SNRPN exon 1 is essential for silencing. Yet, the isolated region from −203 bp to +538 bp (Fig. 3, P[UZP2Δ]) revealed only partial silencing. This result might indicate that additional sequences, e.g., farther upstream and downstream to the SNRPN promoter, are required to achieve the full extent of repression as observed with the large fragments (PWSPX and PWSDS).
Several lines of evidence demonstrate the specificity of the observed silencing effect. (i) Silencing was observed only with fragments from the PWSSRO region and not with a randomly chosen control fragment from a nonimprinted region (chromosome 8q24). In addition, various transgenic strains carrying fragments from the human APP locus (chromosome 21q21-q22) did not reveal any silencing effects. (ii) Small deletions that leave a considerable amount of DNA from the 15q11-q13 region on the transgenes resulted in complete loss of silencing. Because the expression levels of these deletion constructs (P[UZP1Δ] and P[UZPPΔ]) were highly similar to the control construct, this result establishes also the “neutrality” of the control DNA. (iii) A role of CpG islands per se in the observed silencing is highly unlikely because the strongly expressed P[UZPPΔ] construct retains >500 bp of the SNRPN CpG island. In addition, the basic construct used for the generation of all P elements contains several additional CpG islands (Fig. 1B) without any incidence of silencing (24). And, (iv) silencing cannot be explained by promoter competition because the SNRPN promoter appears to be expressed at low levels from our transgenes and not to be induced by GAL4. Therefore, we concluded that specific signals in the SNRPN promoter region mediated the observed silencing effects.
Previous experiments with Drosophila transgenes derived from the mouse H19 locus uncovered a 1.2-kb silencing element in the upstream region of this imprinted gene (15). This fragment is highly overlapping with the sequence required for imprinting of H19 transgenes (28). Because deletion of the element on mouse transgenes resulted in parent-of-origin-independent H19 expression, involvement of silencing in the allele-specific inactivation of the mouse H19 gene was suggested (15). The results presented in this study support the notion that Drosophila is a system in which mammalian imprinting signals can elicit a molecular response. Silencing elements identified in Drosophila and imprinting signals identified in mammals would thus be highly overlapping sequences. In addition, the similarities between silencing and imprinting imply that genomic imprinting makes use of mechanisms that are evolutionarily conserved between fly and mouse. The availability of elaborated genetic tools in Drosophila should allow us to identify trans-acting factors involved in the process.
It has been proposed previously that the region around SNRPN exon 1 contains an imprint switch element required for the establishment of both the paternal and the maternal epigenotypes on human chromosome 15q11-q13 (11). The silencing we observed in Drosophila would reflect one direction of the switch, i.e., the generation of a repressed chromosomal environment on 15q11-q13 (maternal imprint). Erasure of this imprint in primordial germ cells must then require an activating factor that interacts with the silencer and displaces the silencing proteins. A maternally derived deletion of this target sequence, as observed in fathers of PWS imprinting mutation patients, would therefore result in a failure to erase the maternal imprint in the male germ line and, consequently, in the transmission of a paternal chromosome with a (grand)maternal imprint. The failure to observe, in mothers of AS patients, a block of the paternal to maternal imprint switch by a paternally derived deletion of the silencer can be explained by the fact that SNRPN is likely to play a role in PWS (29) and therefore cannot be deleted in normal individuals. Deletions in AS imprinting mutation families never affect SNRPN exon 1 but affect exon BD3 or an element close by (Fig. 1A, ASSRO). This region appears to encode a factor that may interact with the silencer to establish the maternal imprint (11). In this respect, it is also interesting to notice that the ASSRO did not reveal a silencing activity comparable with the PWSSRO in transgenic flies (unpublished results).
A striking feature of imprinting mutation patients is the finding that the deletion of the imprinting control element affects methylation and expression of genes over a domain of ≈2 Mb (8, 9, 12). The silencing effect we describe here, however, functions probably not over distances greater than a few kilobases. This effect could be explained by a limited degree of evolutionary conservation. On the other hand, silencing could be reinforced by additional elements dispersed over the 2-Mb imprinted domain that would not be present in our transgenes. A precedent for this kind of silencing can be found in the 350-kb Drosophila bithorax complex where a few chromosomal elements nucleate silencing complexes of the Polycomb group. The stability of silencing appears to require an interaction of the silencing proteins with additional secondary sites along the entire complex (30, 31). It must be noted, however, that instead of being involved in the long range repression of the 15q11-q13 imprinted domain, the silencer could act also as a local repressor of SNRPN transcription from the maternal allele.
Our results further substantiate the notion that an evolutionary conserved silencing mechanism is involved in the local or long range monoallelic repression of imprinted genes (15). In addition, the precise delineation of the silencing element to a region of a few hundred base pairs should serve as a starting point for the molecular characterization of the imprint switch element (11) deleted in PWS patients.
Acknowledgments
We thank T. Ohta and R. D. Nicholls for the communication of unpublished results, B. Brückner and H. Niemann for transgenic fly strains, and H. J. Lüdecke for the control DNA fragment. We also thank B. Dittrich for helpful discussions and M. A. Surani for valuable comments on the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft ( to B.H. and R.P.), the Human Frontier Science Program Organization (to B.H.), and the Fonds der Chemischen Industrie (to R.P.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: PWS, Prader–Willi syndrome; AS, Angelman syndrome; SNRPN, small nuclear ribonucleoprotein polypeptide N; β-gal, β-galactosidase.
References
- 1.Lalande M. Annu Rev Genet. 1996;30:173–195. doi: 10.1146/annurev.genet.30.1.173. [DOI] [PubMed] [Google Scholar]
- 2.Kishino T, Lalande M, Wagstaff J. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 3.Matsuura T, Sutcliffe J S, Fang P, Galjaard R-J, Jiang Y-h, Benton C S, Rommens J M, Beaudet A L. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 4.Özçelik T, Leff S, Robinson W, Donlon T, Lalande M, Sanjines E, Schinzel A, Francke U. Nat Genet. 1992;2:265–269. doi: 10.1038/ng1292-265. [DOI] [PubMed] [Google Scholar]
- 5.Glenn C C, Porter K A, Jong M T C, Nicholls R D, Driscoll D J. Hum Mol Genet. 1993;2:2001–2005. doi: 10.1093/hmg/2.12.2001. [DOI] [PubMed] [Google Scholar]
- 6.Reed M L, Leff S E. Nat Genet. 1994;6:163–167. doi: 10.1038/ng0294-163. [DOI] [PubMed] [Google Scholar]
- 7.Nakao M, Sutcliffe J S, Durtschi B, Mutirangura A, Ledbetter D H, Beaudet A L. Hum Mol Genet. 1994;3:309–315. doi: 10.1093/hmg/3.2.309. [DOI] [PubMed] [Google Scholar]
- 8.Reis A, Dittrich B, Greger V, Buiting K, Lalande M, Gillesen-Kaesbach G, Anvret M, Horsthemke B. Am J Hum Genet. 1994;54:741–747. [PMC free article] [PubMed] [Google Scholar]
- 9.Sutcliffe J S, Nakao M, Christian S, Örstavik K H, Tommerup N, Ledbetter D H, Beaudet A L. Nat Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- 10.Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, Nicholls R D, Horsthemke B. Nat Genet. 1995;9:395–400. doi: 10.1038/ng0495-395. [DOI] [PubMed] [Google Scholar]
- 11.Dittrich B, Buiting K, Korn B, Rickard S, Buxton J, Saitoh S, Nicholls R D, Poustka A, Winterpacht A, Zabel B, et al. Nat Genet. 1996;14:163–170. doi: 10.1038/ng1096-163. [DOI] [PubMed] [Google Scholar]
- 12.Saitoh S, Buiting K, Rogan P K, Buxton J L, Driscoll D J, Arnemann J, König R, Malcolm S, Horsthemke B, Nicholls R D. Proc Natl Acad Sci USA. 1996;93:7811–7815. doi: 10.1073/pnas.93.15.7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson-Smith A C. Nat Genet. 1996;14:119–121. doi: 10.1038/ng1096-119. [DOI] [PubMed] [Google Scholar]
- 14.Eissenberg J C, Elgin S C R, Paro R. In: Chromatin Structure and Gene Expression. Elgin S C R, editor. Oxford: Oxford Univ. Press; 1995. pp. 147–171. [Google Scholar]
- 15.Lyko F, Brenton J D, Surani M A, Paro R. Nat Genet. 1997;16:171–174. doi: 10.1038/ng0697-171. [DOI] [PubMed] [Google Scholar]
- 16.Huq A H M M, Sutcliffe J S, Nakao M, Shen Y, Gibbs R A, Beaudet A L. Genome Res. 1997;7:642–648. doi: 10.1101/gr.7.6.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ludecke H J, Ahn J, Lin X, Hill A, Wagner M J, Schomburg L, Horsthemke B, Wells D E. Genomics. 1997;40:351–354. doi: 10.1006/geno.1996.4577. [DOI] [PubMed] [Google Scholar]
- 18.Kang J, Lemaire H G, Unterbeck A, Salbaum J M, Masters C L, Grzeschik K H, Multhaup G, Beyreuther K, Müller-Hill B. Nature (London) 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 19.Paro R, Hogness D S. Proc Natl Acad Sci USA. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 21.Spradling A L, Rubin G M. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 22.Robertson H M, Preston C R, Phillis R W, Johnson-Schlitz D M, Benz W K, Engels W R. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.Zink D, Paro R. EMBO J. 1995;14:5660–5671. doi: 10.1002/j.1460-2075.1995.tb00253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urieli-Shoval S, Gruenbaum Y, Sedat J, Razin A. FEBS Lett. 1982;146:148–152. doi: 10.1016/0014-5793(82)80723-0. [DOI] [PubMed] [Google Scholar]
- 26.Patel C V, Gopinathan K P. Anal Biochem. 1987;164:164–169. doi: 10.1016/0003-2697(87)90381-2. [DOI] [PubMed] [Google Scholar]
- 27.Bird A P. Trends Genet. 1995;11:94–100. doi: 10.1016/S0168-9525(00)89009-5. [DOI] [PubMed] [Google Scholar]
- 28.Elson D A, Bartolomei M S. Mol Cell Biol. 1997;17:309–317. doi: 10.1128/mcb.17.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Y, Nicholls R D, Butler M G, Saitoh S, Hainline B E, Palmer C G. Hum Mol Genet. 1996;5:517–524. doi: 10.1093/hmg/5.4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang A, O’Connor M B, Paro R, Simon J, Bender W. Development (Cambridge, UK) 1995;121:1681–1689. doi: 10.1242/dev.121.6.1681. [DOI] [PubMed] [Google Scholar]
- 31.Pirrotta V. Curr Opin Genet Dev. 1997;7:249–258. doi: 10.1016/s0959-437x(97)80135-9. [DOI] [PubMed] [Google Scholar]
- 32.Glenn C C, Saitoh S, Jong M T C, Filbrandt M M, Surti U, Driscoll D J, Nicholls R D. Am J Hum Genet. 1996;58:335–346. [PMC free article] [PubMed] [Google Scholar]
- 33.Gardiner-Garden M, Frommer M. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 34.Uberbacher E C, Xu Y, Mural R J. Methods Enzymol. 1996;266:259–281. doi: 10.1016/s0076-6879(96)66018-2. [DOI] [PubMed] [Google Scholar]