Abstract
D gene segments with irregular spacers (DIR) are D gene segments that are specific to higher primates. Their use is controversial because of their G+C-rich long sequences. In the human, it has always been tempting to assume that a complementarity-determining region 3 sequence has been added by terminal deoxynucleotidyltransferase (TdT) activity and is not derived from DIR recombination. Herein, we examine the use of human DIR gene segments by cross-breeding the human Ig heavy chain minilocus pHC1 transgenic mice and TdT-deficient mice. In the absence of TdT and with a defined set of human D gene segments, it is relatively easy to demonstrate that DIR2 is used to form human Ig heavy chains, contributing to 7% of the human heavy chain rearrangements. VHDJH rearrangements (where H is heavy chain) in the minilocus TdT−/− mice use small portions of DIR2 located throughout the coding sequence. These results constitute the strongest evidence to date that DIR gene segments are used to form human antibodies. Additionally, we show that direct and inverted DIR2JH and VHDIR2 rearrangements occur in the minilocus transgenic mice. During these rearrangements, DM2 3′ signal sequence and a new DIR2 5′ signal sequence are used. These rearrangements generally follow the 12/23 recombination rule. Our results at the VHDJH, DJH, and VHD levels indicate that DIR2 is used to form human heavy chains in transgenic mice. The rearrangement of this gene segment likely involves, however, other mechanisms in addition to the classical VHDJH recombination.
The variable region of Ig heavy chains (IgH) and T cell receptor β and δ chains derive from the combination of three separate DNA elements termed the variability (V), diversity (D), and junctional (J) gene segments through a cell-specific process termed VHDJH recombination (where H is heavy chain). Typically, the recombination process is performed in two steps with D to JH rearrangement preceding VH to DJH rearrangement (1–3). The rearrangement is directed by recombination signal sequences (RSSs) flanking each coding gene segment. RSSs are formed of a consensus palindromic heptamer related to the sequence CACAGTG and a nonamer related to the sequence ACAAAAACC separated by 12 ± 1 or 23 ± 1 nucleotide spacers. D gene segments are flanked on both sides by 12 ± 1 spacer RSSs, whereas VH and JH gene segments are flanked by 23 ± 1 spacer RSSs at the 3′ and 5′ sides, respectively. Generally, recombination occurs between segments flanked by RSSs with different spacer lengths (12/23 rule) (4–7). During recombination, coding gene segments are joined in an imprecise manner to form the coding joint, whereas signal sequences are generally brought together without deletions or nucleotide additions to form the signal joint. Exceptions to the precise signal junction rule have, however, been recently described (8). The formation of coding joints is imprecise and includes nucleotide deletions as well as non-germ-line-encoded (N) and germ-line-encoded (P) nucleotide additions. N segments are added by the terminal deoxynucleotidyltransferase (TdT), whereas P (palindromic) nucleotides result from hairpin structures in cleavage intermediates (9–13). Hybrid junctions corresponding to recombination products in which a RSS is joined to a coding element have also been described (14).
To study the mechanisms of Ig VHDJH recombination, we engineered mice transgenic for a human Ig heavy chain minilocus, pHC1 (15–22). One of the D gene segments present in the transgenic minilocus belongs to the D gene segments with irregular spacers (DIR) family. These gene segments are specific to higher primates. In the human, DIR gene segments are adjacent to DM gene segments. Segments from the DIR family are characterized by the presence of RSSs composed of several heptamers and nonamers separated by both 12 ± 1 and 23 ± 1 nucleotide spacers, in both direct and inverted orientation. Additionally, several cryptic heptamers have been described within DIR gene segment coding sequences. DIR gene segments are greater than 180 bp long and are G+C-rich (Fig. 1) (21, 23–25).
Figure 1.
DIR2 gene segment sequence. DIR2 179-nt coding region is flanked on both sides by two RSSs composed of heptamers and nonamers separated by 12-nt spacers. The location of the two most external RSSs is indicated. DM2 coding sequence is indicated by plus signs. Double lines and dashes indicate portions of DIR2 observed in VHDJH rearrangements of pHC1 mice in the TdT+/+ and TdT−/− backgrounds, respectively. The portion of DIR2 coding sequence used in the hybrid joint MI2 is indicated by pound signs.
The use of DIR gene segments in human IgH remains controversial. All DIR genes segments may not have been identified. Because their sequences are G+C-rich, it is difficult to determine whether a sequence derives from a DIR gene segment or was added by TdT activity. Moreover, it is unclear how only short portions of DIR can be used to form the heavy-chain complementarity-determining region 3 (CDR3), the coding sequence of DIR gene segments being too long to be used in their entirety.
In sequencing the Ig VH region from more than 200 transcripts and rearranged genes, we previously suggested that DIR2 is used in human IgH produced in the transgenic minilocus mice. The CDR3 of several VHDJH rearrangements studied could be explained only if portions (6–11 bp) of DIR2 were used singly, in either direct or inverted orientation without D–D fusion (18, 19). These portions derive from different locations along the DIR coding sequence. In these mice, involvement of TdT in the formation of the junctions could not be ruled out.
Herein, we further analyze the mechanisms involved in DIR gene segment recombination. (i) To rule out the involvement of TdT, we analyzed DIR gene segment use in human Ig heavy chain minilocus transgenic mice on the TdT−/− background. We show that in these mice, human VHDJH rearrangements are diverse and show little evidence of N nucleotides. Additionally, we show that 7% of the rearrangements use the human DIR2 gene segment. This is unequivocal evidence of DIR gene segment use to form human antibodies. (ii) We analyzed DIR gene segment use in the first steps of VHDJH recombination, namely, DJH and VHD rearrangements. We show that DIR2 is found in direct VHD and DJH and in inverted DJH rearrangements. Our results at the VHDJH, VHD, and DJH levels clearly document that gene segments of the DIR family can be used in human Ig heavy chains. However, the data suggest the existence of additional mechanisms besides the classical VHDJH recombination process. These mechanisms must involve the same recombination machinery as the classical VHDJH recombination because DIR2 is rearranged in transgenic mice despite the absence of a murine DIR counterpart. The existence of DIR gene segments creates an additional level of diversity to the antibody repertoires of higher primates.
MATERIALS AND METHODS
DNA Construct and Mice.
pHC1, the construct used to establish the transgenic founder line 119, is 60 kb long and has been described (16–20). TdT−/− mice have been described (11, 26). pHC1-transgenic TdT−/− mice were obtained by cross-breeding and selection by Southern blot filter hybridization. Briefly, tail DNA was cut with the BglII restriction enzyme and probed with a HindIII fragment covering the two human VH gene segments to detect the pHC1 transgene and with an EcoRI–HindIII fragment covering TdT exons IV–VI (a gift from D. Mathis, Université Louis Pasteur, Strasbourg, France) to analyze the TdT locus. Probing with the TdT probe results in two bands of 6.5 and 1 kb in TdT+/+ mice but results in two bands of 2.1 and 1.5 kb in TdT−/− mice.
VHDJH, VHD, and DJH PCR Amplifications.
Genomic DNA was isolated from spleen, bone marrow, and liver of 2-month-old mice by phenol/chloroform extraction after proteinase K digestion. It was amplified by PCR as described (18–20). The amplification of 1–6 μg of DNA was performed with 40 cycles of 1-min denaturation (94°C), 2-min annealing (50°C to 54°C depending on the primer combination), and 2-min elongation (72°C) and used 1 unit of Taq polymerase. The final cycle was completed by a 7-min elongation at 72°C. VHDJH amplifications were performed by using a VH oligonucleotide complementary to both VH5–251 and ψVH3–105 gene segments (5′-AGGTGCAGCTGGTG(CG)AGTCTG-3′) and a human JH consensus oligonucleotide (5′-ACCTGAGGAGACGGTGACCAGGGT-3′). Direct and inverted DIRJH amplifications were performed by using an oligonucleotide complementary to the DIR2 5′ RSS (DIR2-JH amplification, 5′-GCTGGGGCTCACAGTGC-3′) or to the DIR2 3′ RSS (inverted DIR2-JH amplification, 5′-CGCTGTGACTCGGGGC-3′) and the JH consensus oligonucleotide. Alternatively, DM2 3′ RSS (5′-CAGTTTTTGACGTAGCTATTC-3′) was used to amplify inverted DIRJH rearrangements. Direct and inverted VHDIR amplifications were performed by using an oligonucleotide complementary to the DIR2 3′ RSS (VHDIR amplification) or to the DIR2 5′ RSS (inverted VHDIR amplification) and the VH oligonucleotide. Ten picomoles of each primer was used as appropriate. The PCR products were purified by using Microcon 50 (Amicon) as described by the supplier.
Cloning and Sequencing of PCR Products.
The purified PCR products were blunt-end-ligated into EcoRV-digested pBluescript KS+ plasmids (Stratagene) and the ligation mixture was used to transform Escherichia coli (XL1-Blue) competent cells. The resulting colonies were screened with 32P-labeled oligonucleotides corresponding to the DIR2 (5′-GCCACAGCCTCCGGAGCCCCCG-3′) or VH (5′-TGTATTACTGTG(CT)GAGA-3′) gene segment coding sequences. Double-stranded DNA was prepared from positive colonies and sequenced with a Perkin–Elmer ABI Prism 377 automated DNA sequencer.
Southern Blot Filter Hybridization.
For Southern blot filter hybridization, direct and inverted VHDIR rearrangements were PCR-amplified as described above from 1 μg of splenic or liver DNA or from 10 μg of bone marrow DNA, size-fractionated on 1% agarose gel, and transferred onto Hybond N+ membrane (Amersham). Membranes were probed with an oligonucleotide specific for the DIR2 coding sequence by using Rapid Hyb (Amersham) as described by the supplier.
RESULTS
The Repertoire of Human VHDJH Rearrangements from Minilocus Transgenic TdT−/− Mice Is Diverse and Shows Little Evidence of N Nucleotides.
Eighty and 30 unique human VHDJH rearrangements using the functional VH5–251 and the ΨVH3–105 gene segments, respectively, have been sequenced from five pHC1-transgenic TdT−/− mice (data not shown). The repertoire of human heavy chains produced in these mice is extremely diverse. Every human D gene segment present in the IgH minilocus is used with a preferential use of DHQ52 (28% of the rearrangements). Every human JH gene segment is observed in VHDJH rearrangements. JH4 is preferentially used (34% of the rearrangements). Rearrangements from these mice are characterized by the low frequency of N regions. Only two sequences using conventional D gene segments (2%) show evidence of 1–2 additional nucleotides versus 66% of the rearrangements containing 1–10 additional nucleotides in TdT+/+ minilocus transgenic mice. The frequency of P nucleotides is, however, similar in TdT+/+ and TdT−/− mice. Fifty rearrangements (45% of the rearrangements containing a D gene segment) occurred at sites of short homology between the VH and D gene segments. Thirty-nine rearrangements (35% of the rearrangements containing a D gene segment) occurred at sites of short homology between the D and JH gene segments.
The DIR2 Gene Segment Is Used to Form Human Heavy Chains in Minilocus Transgenic TdT−/− Mice.
Eight of the VHDJH rearrangements (7%) obtained from TdT−/− mice cannot be explained solely by the conventional D gene segments present in the human heavy chain minilocus. These rearrangements can, however, be explained by invoking the use of short portions (5–8 nucleotides) of the DIR2 gene segment (Fig. 2). These short portions derive from various internal areas of the DIR2 coding sequence. As for rearrangements using conventional D gene segments, recombination using DIR2 often occurs at sites of short homology between gene segments. Four rearrangements occurred at sites of short homology between the VH and DIR2 gene segments. Two rearrangements occurred at sites of short homology between the DIR2 and JH gene segments. The frequency of additional nucleotides at the gene segment junctions is higher for DIR2 rearrangement than for conventional D gene segment rearrangement. Two DIR2 rearrangements contain one additional nucleotide at the VHD junction and three DIR2 rearrangements contain one additional nucleotide at the DJH junction.
Figure 2.
CDR3 of eight VHDJH rearrangements using the DIR2 gene segment obtained from minilocus transgenic TdT−/− mice. The VH gene segment 3′ end and part of DIR2 coding sequence are indicated. For each clone, homologies with VH and DIR coding sequences are indicated by dashes. Double lines indicate nucleotides that can derive from either gene segment, suggesting recombination at sites of short homologies between gene segments. The JH gene segment is indicated at the right of the figure. P and N nucleotides are indicated. The CDR3 length is indicated in parenthesis when the rearrangement is productive, whereas nonproductive rearrangements are indicated by a dash.
The DIR2 Gene Segment Is Used in DJH Rearrangements from Human Ig Heavy Chain Transgenic Mice.
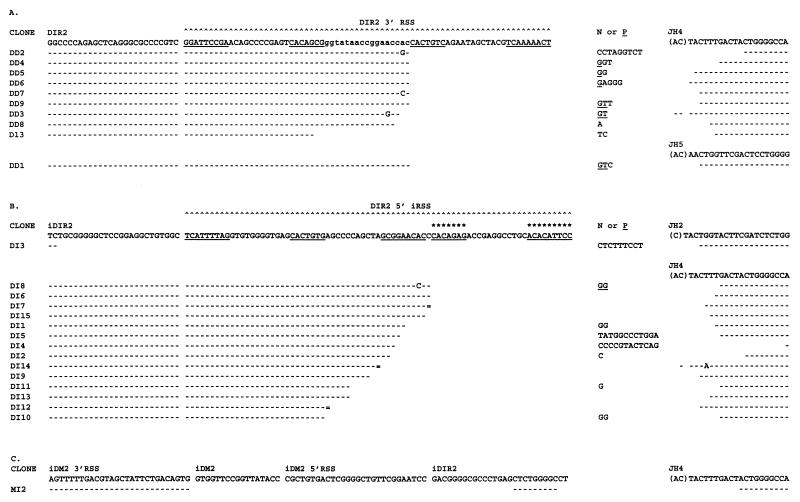
Twenty-five DIR2JH rearrangements have been sequenced from human Ig heavy chain minilocus transgenic TdT+/+ mouse spleens (Fig. 3). Most of the rearrangements (92%) use the JH4 gene segment.
Figure 3.
Junctional sequence of direct and inverted DIR2JH rearrangements obtained from minilocus transgenic mice. Germ-line DIR2 RSS, DIR, and JH coding sequences are indicated in uppercase type at the top. DM2 coding sequence is indicated in lowercase type. Heptamers and nonamers as described (23) are underlined. The heptamer and nonamer of the new 5′ RSS determined from this study are indicated by stars. iDIR, iDM2, and iRSS indicate inverted DIR, DM2, and RSS sequences. For each clone, homologies with VH and DIR coding sequences are indicated by dashes. Double lines indicate nucleotides that can derive from either gene segment. Mutations are indicated in uppercase type in each rearrangement. N and P nucleotides are indicated. (A) Nucleotide sequence of 10 DIRJH rearrangements. (B) Nucleotide sequence of 15 inverted DIRJH rearrangements. (C) Nucleotide sequence of one hybrid joint involving DM2 3′ RSS, DIR2 coding sequence, and JH4.
Ten of the DIR2JH rearrangements result from deletional recombination (Fig. 3A). Most of these rearrangements occur at the JH RSS and at the DM2 3′ RSS. No evidence of rearrangements within the DIR2 coding sequence was found. Because DIR2 and DM2 gene segments are adjacent in the genomic DNA, 9 of the DIR2JH rearrangements cannot be differentiated from DM2JH rearrangements. During these rearrangements, 0–3 nucleotides of the DM2 coding end were removed by exonuclease activity and 0–9 nucleotides were added at the coding joint. Only 1 rearrangement (D13) does not include the DM2 coding sequence. This rearrangement occurred either through DM2 3′ RSS with 19 nucleotides removed by exonuclease activity or through DM2 5′ RSS. If DM2 5′ RSS was used, the D13 rearrangement corresponds to a hybrid joint of DM2 5′ RSS and JH4 coding sequence. In this hybrid joint, two nucleotides were deleted from the signal sequence by exonuclease activity.
Fifteen rearrangements result from inversional recombination (Fig. 3B). Rearrangements by inversion occur at the JH RSS and at the previously described 5′ RSS or at a new DIR2 5′ RSS, located more upstream. Twelve rearrangements occur at the JH RSS and at the new DIR2 5′ RSS. During these rearrangements, 0–16 nucleotides were removed at DIR2 coding end by exonuclease activity and 0–12 nucleotides were added at the coding joint. Two rearrangements (DI12 and DI10) occurred either through the new 5′ RSS with 20 or 21 nucleotides deleted by exonuclease activity or more likely through the previously described 5′ RSS. This RSS is in inverted orientation relative to the DIR2 coding sequence. Therefore, if this RSS was used, the two rearrangements correspond to hybrid joints in which DIR2 5′ inverted RSS is rearranged to JH4 coding sequence. Additionally, one inverted DIR2JH rearrangement occurred within the DIR2 coding sequence (sequence DI3). In rearrangement DI3, 73, 52, or 45 nucleotides were removed if the new 5′ RSS, the previously described inverted 5′ RSS, or the previously described direct 5′ RSS was used, respectively. Alternatively, this rearrangement may have occurred directly within the coding sequence or may have been mediated by DIR2 internal cryptic heptamers in a nonamer independent fashion. However, no clear evidence of a cryptic heptamer near this region can be found. Cryptic heptamers may have, however, been involved in the formation of the rearrangement MI2 (Fig. 3C). This DJH rearrangement was obtained by PCR amplification using an oligonucleotide specific for DM2 3′ RSS. It contains DM2 3′ RSS and JH4 coding sequence separated by 9 nucleotides derived from an internal portion of DIR2. It is noteworthy that several cryptic heptamers can be detected at the 5′ side of the DIR2 portion used. These cryptic heptamers may have mediated the formation of the hybrid joint between the DIR2 coding sequence and the DM2 3′ RSS. In this rearrangement, the DIR2 portion used is located 107 nucleotides 3′ of the DIR2 new 5′ RSS and 61 nucleotides 5′ of DM2 3′ RSS.
The DIR2 Gene Segment Is Used in VHD Rearrangements from Human Ig Heavy Chain Transgenic Mice.
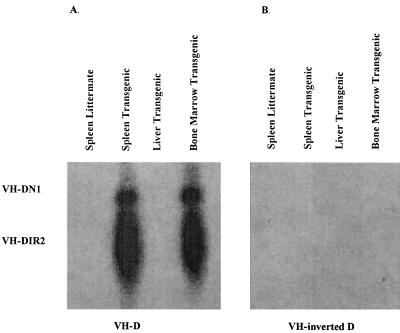
Eleven VHDIR rearrangements have been sequenced from human Ig heavy chain minilocus transgenic mouse spleens (Fig. 4). All of the rearrangements use the functional VH5–251 gene segment and result from deletional recombination. These rearrangements occur at the VH5–251 RSS and at the DIR2 new 5′ RSS. None of the rearrangements occur within the DIR2 coding sequence. No inversional rearrangements were obtained by sequencing. Inverted VHDIR rearrangements are also not detected by PCR followed by Southern blot filter hybridization, whereas deletional VHDIR rearrangements were easily detected in the murine spleen and bone marrow by this method (Fig. 5).
Figure 4.
Junctional sequence of 11 VHDIR2 rearrangements obtained from minilocus transgenic mice. Sequences are indicated as in Fig. 3.
Figure 5.
PCR amplification and Southern blot filter hybridization of direct and inverted VHDIR2 rearrangements. (A) Direct VHDIR rearrangements were PCR-amplified from 1 μg of splenic or liver DNA or from 10 μg of bone marrow DNA of transgenic or littermate TdT+/+ mice and probed with a DIR2-coding-sequence-specific oligonucleotide. The positions of VHDIR2 and VHDN1 rearrangements are indicated. (B) Inverted VHDIR rearrangements were PCR-amplified from 1 μg of splenic or liver DNA or from 10 μg of bone marrow DNA of transgenic or littermate, TdT+/+ mice and probed with a DIR2-coding-sequence-specific oligonucleotide.
DISCUSSION
The use of DIR gene segments in human IgH has long been a subject of controversy. The first point of debate relates to whether or not DIR gene segments are used at all. All D gene segments may not be known. Additionally, because DIR sequences are G+C-rich, it is difficult to determine whether a transcript sequence derives from a DIR gene segment or was added by TdT activity. The second point of contention relates to the mechanisms by which DIR gene segments are used. Indeed, it is unclear how only short portions of DIR are used to form the heavy chain CDR3 because the coding sequence of DIR gene segments is too long to be used in its entirety.
A few human VHDJH sequences have previously been reported that could derive from a DIR gene segment. These rearrangements use up to nine nucleotides of one of the six DIR gene segments previously reported (24, 25, 27–31). Additionally, one sequence was reported that contained 35 nucleotides from DIR (27). It has been suggested that DIR gene segments may contribute to up to 12% of the human IgH repertoire (24). It is, however, intriguing that in the human, CDR3s show large discrepancies compared with DIR germ-line sequences. Sequence alignment often requires the introduction of mismatches and gaps, even in situations where somatic mutation is minimal. These disparities are probably not caused by DIR polymorphism (24). An alternative explanation for these sequences involves TdT activity. An additional possibility is that rearrangements use unknown conventional D gene segments containing DIR-like coding sequences.
Human IgH minilocus transgenic mice constitute an ideal model to study DIR gene segment use in human antibodies. All D gene segments present in the minilocus are well characterized. In the absence of immunization, the human heavy chain repertoire is extremely diverse and contains virtually no evidence of somatic mutation. In the minilocus transgenic mice, endogenous heavy chain gene segment recombination occurs as well. Additionally, in founder line 119, a line we have studied extensively, xenotypic exclusion does not occur. Under these circumstances, selection is more likely to occur on the endogenous murine genes. The nonselected repertoire can also be analyzed by studying the repertoire associated to the ΨVH3–105 gene segment (18–22).
In minilocus transgenic mice, 90% of the human VHDJH rearrangements can be explained by invoking the use of one of the nine conventional D gene segments present in the minilocus D region. The remaining 10% can be explained by the DIR2 gene segment. In minilocus transgenic TdT−/− mice, 93% of the human VHDJH rearrangements can be explained by conventional D gene segments. The remaining 7% can be explained by the DIR2 gene segment. In transgenic TdT−/− mice, N nucleotides are rare in rearrangements using conventional D gene segments, similar to what was shown for murine junctions, as well as in cell lines lacking TdT activity (11, 12, 32, 33). The finding that additional nucleotides are more frequently found in rearrangements using the DIR2 gene segment is in agreement with data indicating that in the human, CDR3 shows large discrepancies compared with DIR germ-line sequences even in situations where somatic mutation is minimal. Our data indicate that these nucleotides are independent of TdT activity and are probably the result of unconventional mechanisms of rearrangement associated with DIR gene segment use. A recent paper suggests that DIR gene segments are not required to explain human IgH rearrangements (34). Herein, we find that 100% of the human heavy chain rearrangements from pHC1 mice can be explained only if the DIR2 gene segment is invoked. The portions of DIR used are located throughout the coding sequence but several hot spots can be observed (Fig. 1). One hot spot corresponds to the previously described DM2 sequence. It is noteworthy that except for the DM2 region flanked by two RSSs, no cryptic heptamer can be detected directly flanking the hot spots.
The portions of the DIR2 coding sequence used in VHDJH rearrangements are located up to 100 nucleotides away from the described RSS. This relates to the second controversial point concerning DIR gene segments: What are the mechanisms involved in DIR gene segment rearrangement? Several nonmutually exclusive hypotheses can be invoked to explain DIR gene segment rearrangement: (i) the TdT is responsible for the sequences, (ii) the sequences derive from unknown gene segments containing DIR-like coding region, (iii) DIR recombination is accompanied by extensive exonuclease activity, (iv) DIR rearrangement occurs through internal cryptic heptamers, (v) DIR gene segment rearrangement occurs at the RSS and is followed by secondary events resulting in the deletion of internal portions of the DIR coding sequence, and (vi) DIR gene segments are used through gene conversion resulting in internal portions of DIR coding sequences under the control of conventional D RSS. Alternatively, this could be the result of the formation of hybrid joints involving the DIR coding sequence and conventional D gene segment RSS. Our results in TdT−/− mice rule out possibility i. Additionally, every gene segment present in the minilocus is known and every D region that does not derive from a conventional D gene segment can be found in the germ-line DIR2 coding sequence, ruling out possibility ii. Exonuclease activity appears to be normal in the DIRJH and VHDIR rearrangements, suggesting that hypothesis iii may not be the only explanation.
Rearrangements at the DIRJH and VHDIR levels occur usually at two main RSSs: DM2 3′ RSS and a new 5′ RSS. It is noteworthy that we have been unable to demonstrate the use of the previously described DIR2 RSS by looking for the signal joints involving these RSSs (data not shown). The use of the two most external RSSs suggests that hypothesis iv in which only internal cryptic heptamers would be used is not the sole mechanism involved in DIR recombination. Only sequences DI3 and MI2 may result from this mechanism. It is noteworthy that DIRJH and VHDIR recombination follows the 12/23 rule. The two RSSs define a 179-nt coding region. This coding region covers the previously described DM2 gene segment. The incorporation of the DM2 gene segment within DIR2 gene segment is also suggested by the fact that we did not find evidence of DM2 3′ RSS use to form inverted DM2 rearrangement. Our results indicate that DIR gene segment recombination requires additional mechanisms besides the classical VHDJH recombination mechanisms. DIR gene segment recombination may be a two-step process in which the first step corresponds to a classical rearrangement through the 5′ or 3′ RSS. This is followed by secondary recombination resulting in the deletion of internal portions of the DIR coding sequence. These secondary recombinations apparently do not require special recombinase machinery because mice rearrange DIR despite the absence of such gene segments in normal mice. Secondary recombination may be directed by the cryptic heptamers located throughout DIR coding sequences. Sequences DI3 and MI2 may be the result of such secondary recombinations. Alternatively, DIR gene segment rearrangement may involve gene conversion mechanisms in which the DIR coding sequence is put under the control of conventional RSSs. However, to date we have not been able to detect evidence for gene conversion in pHC1-transgenic mice. We are currently testing the two possibilities (secondary recombination and gene conversion) with new transgenic miniloci. DIR gene segment recombination may also involve the formation of hybrid joints. Several VHD and DJH rearrangements and more particularly the MI2 rearrangement observed in the minilocus transgenic mice suggest that DIR2 may be involved in such hybrid joints more frequently than conventional D gene segments.
DIR gene segments are specific to higher primates, thus appearing relatively recently in evolution. This study unequivocally shows that these gene segments can be used to form Ig heavy chains. The DIR2 gene segment must be invoked to explain all of the human rearrangements in the transgenic mice even in the absence of TdT. Because of the coding region length, some form of secondary recombination is likely required to explain the use of these gene segments. Several nonmutually exclusive mechanisms that do not require new recombinase machinery can be invoked. The use of DIR gene segments results in proline-, alanine-, and glycine-rich CDR3. Antibodies containing such IgH may have advantages in responses against certain antigens. DIR gene segments add an important level of diversity to the Ig heavy chain repertoires of higher primates.
Acknowledgments
We are most grateful to N. Lonberg for the preparation of the pHC1-transgenic mice and to S. Gilfillan and D. Mathis for the TdT−/− mice. We thank K. Meek and G. Rathbun for their careful review of the manuscript. We thank K. Kamm, S. Hall, and A. Miller for their technical assistance. This work is supported by grants from the National Institutes of Health (AI-31229 and AI-12127).
ABBREVIATIONS
- DIR
D gene segments with irregular spacers
- TdT
terminal deoxynucleotidyltransferase
- H (as subscript)
heavy chain
- IgH
Ig heavy chain
- RSS
recombination signal sequence
- CDR3
complementarity-determining region 3
References
- 1.Max E E. In: Fundamental Immunology. 3rd Ed. Paul W E, editor. New York: Raven; 1993. pp. 315–382. [Google Scholar]
- 2.Hedrick S M, Eidelman F J. In: Fundamental Immunology. 3rd Ed. Paul W E, editor. New York: Raven; 1993. pp. 383–420. [Google Scholar]
- 3.Alt F W, Yancopoulos G D, Blackwell T K, Wood C, Thomas E, Coffman R, Tonegawa S, Baltimore D. EMBO J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Early P, Huang H, Davis M, Calame K, Hood L. Cell. 1980;19:981–992. doi: 10.1016/0092-8674(80)90089-6. [DOI] [PubMed] [Google Scholar]
- 5.Sakano H, Maki R, Kurosawa Y, Roeder W, Tonegawa S. Nature (London) 1980;286:676–683. doi: 10.1038/286676a0. [DOI] [PubMed] [Google Scholar]
- 6.Hesse J E, Lieber M R, Mizuuchi K, Gellert M. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 7.Eastman Q M, Leu T M J, Schatz D G. Nature (London) 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 8.Candeias S, Muegge K, Durum S K. J Exp Med. 1996;184:1919–1926. doi: 10.1084/jem.184.5.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landau R, Schatz D G, Rosa M, Baltimore D. Mol Cell Biol. 1987;7:3237–3243. doi: 10.1128/mcb.7.9.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lafaille J J, DeCloux A, Bonneville M, Takagaki Y, Tonegawa S. Cell. 1989;59:859–870. doi: 10.1016/0092-8674(89)90609-0. [DOI] [PubMed] [Google Scholar]
- 11.Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Science. 1993;261:1175–1178. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- 12.Komori T, Okada A, Stewart V, Alt F W. Science. 1993;261:1171–1174. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- 13.Meier J T, Lewis S M. Mol Cell Biol. 1993;13:1078–1092. doi: 10.1128/mcb.13.2.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis S, Hesse J E, Mizuuchi K, Gellert M. Cell. 1988;55:1099–1107. doi: 10.1016/0092-8674(88)90254-1. [DOI] [PubMed] [Google Scholar]
- 15.Lonberg N, Taylor L D, Harding F A, Troutsine M, Higgins K M, et al. Nature (London) 1994;268:856–859. doi: 10.1038/368856a0. [DOI] [PubMed] [Google Scholar]
- 16.Taylor L D, Carmack C E, Schramm S R, Mashayekh R, Higgins K M, Kuo C C, Woodhouse C, Kay R M, Lonberg N. Nucleic Acids Res. 1992;20:6287–6295. doi: 10.1093/nar/20.23.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor L D, Carmack C E, Huszar D, Higgins K M, Mashayekh R, et al. Int Immunol. 1994;6:579–591. doi: 10.1093/intimm/6.4.579. [DOI] [PubMed] [Google Scholar]
- 18.Tuaillon N, Taylor L D, Lonberg N, Tucker P W, Capra J D. Proc Natl Acad Sci USA. 1993;90:3720–3724. doi: 10.1073/pnas.90.8.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuaillon N, Miller A B, Lonberg N, Tucker P W, Capra J D. J Immunol. 1994;152:2912–2920. [PubMed] [Google Scholar]
- 20.Tuaillon N, Miller A M, Tucker P W, Capra J D. J Immunol. 1995;154:6453–6465. [PubMed] [Google Scholar]
- 21.Tuaillon N, Tucker P W, Capra J D. The Immunologist. 1995;3:269–274. [Google Scholar]
- 22.Tuaillon N, Capra J D. In: The Antibodies. Capra J D, Zanetti M, editors. New York: Gordon & Breach; 1995. pp. 1–39. [Google Scholar]
- 23.Ichihara Y, Matsuoka H, Kurosawa Y. EMBO J. 1988;7:4141–4150. doi: 10.1002/j.1460-2075.1988.tb03309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanz I, Wang S S, Meneses G, Fishbach M. J Immunol. 1994;152:3958–3969. [PubMed] [Google Scholar]
- 25.Moore B B, Meek K. J Immunol. 1995;154:2175–2187. [PubMed] [Google Scholar]
- 26.Gilfillan S, Bachmann M, Trembleau S, Adorini L, Kalinke U, Zinkernagel R, Benoist C, Mathis D. Eur J Immunol. 1995;25:3115–3122. doi: 10.1002/eji.1830251119. [DOI] [PubMed] [Google Scholar]
- 27.Sanz I. J Immunol. 1991;147:1720–1729. [PubMed] [Google Scholar]
- 28.Yamada M, Wasserman R, Reichard B A, Shane S, Caton A J, Rovera G. J Exp Med. 1991;173:395–407. doi: 10.1084/jem.173.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raaphorst F M, Timmers E, Kenter M J H, Van Tol M J D, Vossen J M, Schuurman R K B. Eur J Immunol. 1992;22:247–251. doi: 10.1002/eji.1830220136. [DOI] [PubMed] [Google Scholar]
- 30.Shin E K, Matsuda F, Fujikura J, Akamizu T, Sugawa H, Mori T, Honjo T. Eur J Immunol. 1993;23:2365–2367. doi: 10.1002/eji.1830230947. [DOI] [PubMed] [Google Scholar]
- 31.Minegishi Y, Akagi K, Nishikawa K, Okawa H, Yata J. J Immunol. 1996;156:4666–4671. [PubMed] [Google Scholar]
- 32.Komori T, Pricop L, Hatakeyama A, Bona C A, Alt F W. Int Rev Immunol. 1996;13:317–325. doi: 10.3109/08830189609061755. [DOI] [PubMed] [Google Scholar]
- 33.Gauss H G, Lieber M R. Mol Cell Biol. 1996;16:258–269. doi: 10.1128/mcb.16.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corbett S J, Tomlinson I M, Sonnhammer E, Buck D, Winter G. J Mol Biol. 1997;270:587–597. doi: 10.1006/jmbi.1997.1141. [DOI] [PubMed] [Google Scholar]