Abstract
Conjugative transfer of plasmid DNA via close cell–cell junctions is the main route by which antibiotic resistance genes spread between bacterial strains. Relaxases are essential for conjugative transfer and act by cleaving DNA strands and forming covalent phosphotyrosine linkages. Based on data indicating that multityrosine relaxase enzymes can accommodate two phosphotyrosine intermediates within their divalent metal-containing active sites, we hypothesized that bisphosphonates would inhibit relaxase activity and conjugative DNA transfer. We identified bisphosphonates that are nanomolar inhibitors of the F plasmid conjugative relaxase in vitro. Furthermore, we used cell-based assays to demonstrate that these compounds are highly effective at preventing DNA transfer and at selectively killing cells harboring conjugative plasmids. Two potent inhibitors, clodronate and etidronate, are already clinically approved to treat bone loss. Thus, the inhibition of conjugative relaxases is a potentially novel antimicrobial approach, one that selectively targets bacteria capable of transferring antibiotic resistance and generating multidrug resistant strains.
Keywords: antimicrobial, bacterial conjugation, bisphosphonates, F plasmid TraI, relaxase inhibition
Conjugative elements are responsible for the majority of horizontal gene transfers within and between bacterial strains (reviewed in ref. 1), as first described for the Escherichia coli F plasmid by Lederberg and Tatum in 1946 (2). Conjugative DNA transfer is also the central mechanism by which antibiotic resistance and virulence factors are propagated in bacterial populations (reviewed in ref. 3). Indeed, it is well established that antibiotic resistance can be rapidly acquired in clinical settings and that such acquisition is critically dependent on conjugative DNA transfer (reviewed in ref. 4). Small-molecule inhibition of conjugation could prove to be a powerful method for curbing the generation and spread of multidrug-resistant strains. Past studies suggested that various antibiotics, polycyclic chemicals, and crude extracts inhibit conjugation at concentrations less than the antibacterial minimum inhibitory concentration (5–11); however, most of these effects have been attributed to nonconjugation-specific inhibition of bacterial growth or DNA synthesis (12–15). This study describes a bottom-up approach used to identify the first small molecule inhibitors of conjugative DNA transfer that target an enzyme of the conjugative system.
The DNA relaxase is a central enzyme in each conjugative system (16–18) and thus is a prime target for inhibition. The conjugative relaxase initiates DNA transfer with a site- and strand-specific ssDNA nick in the transferred strand (T-strand) at the origin of transfer (oriT), forming a covalent 5′-phosphotyrosine intermediate (16, 19–23). The nicked T-strand moves from the donor cell (plasmid+) to the recipient cell (plasmid−) via an intercellular junction mediated by a type IV secretion system (reviewed in refs. 19, 24, and 25). The relaxase completes DNA transfer by reversing the covalent phosphotyrosine linkage and releasing the T-strand. In the F plasmid, this relaxase is located in the N-terminal domain of a large multifunctional protein, TraI (DNA helicase I) (22, 23, 26–28). Some conjugative relaxases use one active-site tyrosine [e.g., IncQ RSF1010 MobA (29), IncP RP4 TraI (30, 31), IncI R64 NikA (32), Agrobacterium Ti VirD2 (33), and Tn5252 MocA/BmgA (34), where R indicates plasmids that propagate antibiotic resistance]. F-like relaxases [e.g., IncF R1 and R100 plasmid TraIs (28), IncN R46 and pCU1 TraIs (35), IncW R388 TrwC (36), and Pseudomonas IncP9 pWW0 TraC (37)] maintain a conserved, bifurcated constellation of two to five tyrosines near their N termini. The most common arrangement is four tyrosines (Y1–Y4; tyrosines 16, 17, 23, and 24 in F TraI) with pairs Y1/2 and Y3/4 separated by a variable linker region. Crystal structures show that all four tyrosines are proximal to a bound metal ion [this study and others (38–41)]. Optimal relaxase cleavage, ligation, and transfer of ssDNA require the metal ion and two catalytic tyrosines, one from each pair (42). F TraI relaxase shares significant sequence identity with relaxases of many R plasmids (e.g., 98% with R100 TraI); thus, the F plasmid serves as a model system for examining conjugative plasmids and the inhibition of conjugative transfer.
In this study we first sought to understand the role that the relaxase enzyme plays in the initiation and termination of DNA conjugation and then sought to use that information to identify potent relaxase-specific inhibitors. Our results establish that the conjugative DNA transfer process can be selectively disrupted by relaxase-targeted compounds, including some clinically approved drugs. This is a potentially novel antimicrobial approach, one that could be used to purge from microbial populations the bacteria capable of propagating antibiotic resistance genes.
Results
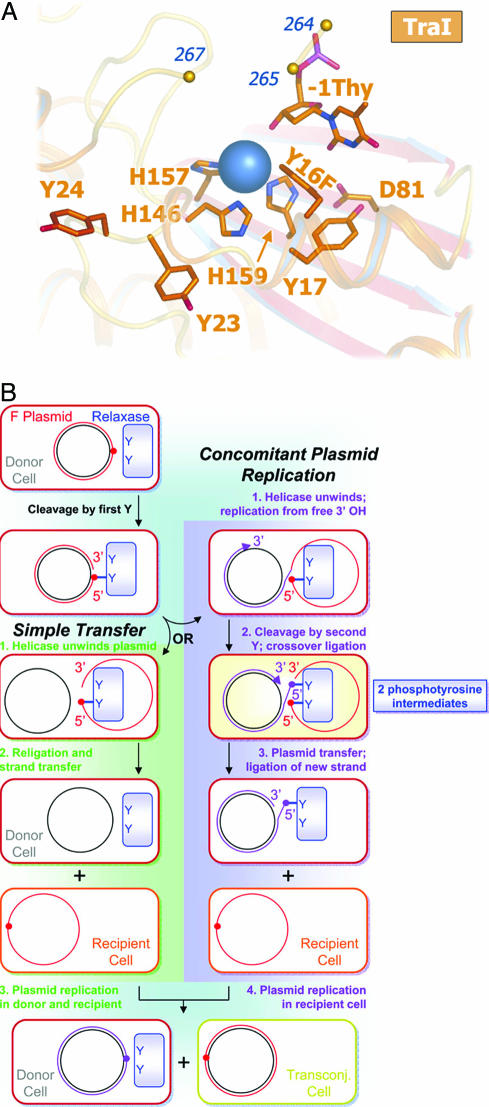
We determined the 2.4-Å crystal structure of the 300-residue N-terminal relaxase domain of F plasmid TraI (N300) with a tyrosine-16 to phenylalanine mutation (Y1 of F TraI; Y16F) [Fig. 1A and supporting information (SI) Table 1]. The structure of N300 is similar to those of a 330-residue F TraI fragment (39, 41) (N330) and the relaxase domain of R388 TrwC (38, 40) (SI Fig. 5). Crystallization required a 9-base ssDNA oligonucleotide consisting of the F oriT nick site sequence. Despite the Y16F mutation, which reduces N300 DNA cleavage 600-fold, we observed electron density for just one DNA base in the active site (Fig. 1A; SI Fig. 6). We interpreted this as the oriT thymidine immediately upstream of the scissile phosphate (−1 Thy). We also observed density for a bound metal ion, chelated by three conserved histidine side chains (soft ligands, i.e., uncharged and high polarizability). Despite unusual soft chelation, we interpret the bound metal as a divalent magnesium ion (a hard center, i.e., low polarizability and a preference for charged ligands) based on bond lengths, electron density, and octahedral coordination. A survey of magnesium-binding proteins in the Protein Data Bank revealed that the chelation of Mg2+ by neutral residues is diagnostic of a site that simultaneously binds to multiple phosphate groups (SI Table 2). Mutation of the metal-chelating residue histidine-159 to glutamic acid (thus reducing the effective charge of the metal site) eliminated relaxase activity (SI Fig. 7). These data indicate that the 2+ charge on the bound metal ion is critical to relaxase function.
Fig. 1.
F TraI N300 Y16F bound to the scissile thymidine and a two-path model of F-like bacterial conjugation. (A) N300 Y16F active site with a metal ion (blue sphere) chelated by three histidines and the −1 Thy 3′-hydroxyl. Y16F occludes a fifth octahedral coordination site. (B) Cleavage by the first tyrosine forms a covalent phosphotyrosine intermediate (red circle) on the T (red) strand. Transfer with CPR diverges from simple transfer when the 3′-hydroxyl left by initial cleavage becomes a substrate for replication (blue strand). The newly created oriT requires a second cleavage event and second phosphotyrosine (purple circle).
We then considered models for conjugative DNA transfer that would require the observed 2+ metal ion and two catalytically competent tyrosine residues (Fig. 1B). In the case of simple transfer (Fig. 1B, green), a single catalytic tyrosine is sufficient for successful intercellular DNA transfer. However, if the free 3′-hydroxyl product of relaxase-mediated DNA cleavage becomes a substrate for concomitant plasmid replication (CPR; Fig. 1B, purple), analogous to rolling circle replication (43), then two tyrosines would be required to resolve the replicative oriT intermediate and release the T-strand to complete transfer. CPR events explain the reported low-frequency generation of greater than unit-length conjugative plasmids (44). Although CPR may not be the primary conjugative pathway, plasmids with relaxases capable of resolving CPR intermediates would be expected to have a selective advantage. Single-tyrosine relaxases could achieve resolution of CPR intermediates through relaxase multimerization (45) or cooperation with a second nonrelaxase protein (46). The ability to resolve undesirable replication products would also confer oriT specific recombinase activity between plasmids (47, 48) or between tandem oriT repeats on the same plasmid (46, 49). However, for our purposes, a key prediction that arose from this model is that multityrosine relaxases are capable of accommodating two phosphotyrosine intermediates simultaneously within their active sites. The need to handle dual phosphotyrosine intermediates would explain the key features of the TraI relaxase outlined above: the presence of two catalytically competent tyrosines and the requirement for a metal ion with an obligate 2+ charge.
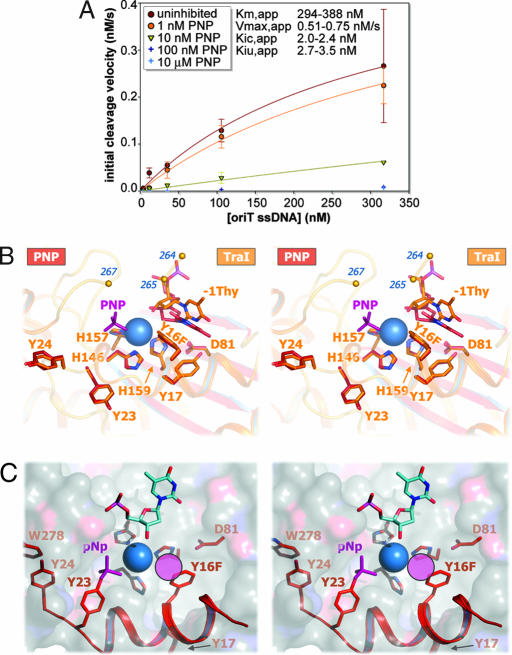
Based on the prediction that multityrosine relaxases employ two simultaneous phosphotyrosine intermediates coordinated to one magnesium ion, we hypothesized that simple bisphosphonates would bind the magnesium center and thus inhibit F TraI relaxase activity and conjugative transfer. To test this hypothesis, a kinetic assay using fluorophore-labeled oriT ssDNA for cleavage by the F TraI relaxase domain (TraI N300) was developed to complement existing radiolabel-based techniques (50, 51). Imidobisphosphate (PNP), a simple and relatively stable bisphosphonate, was the first compound examined in this assay (Fig. 2A). We found that PNP is a nanomolar inhibitor of TraI relaxase activity in vitro. Analysis of cleavage velocity curves revealed that PNP is a mixed-type (specifically, noncompetitive) inhibitor of TraI, with apparent competitive (Kic,app) and uncompetitive (Kiu,app) inhibition constants of 2.0–2.4 nM and 2.7–3.5 nM, respectively. Thus, a simple bisphosphonate serves as a potent inhibitor of a multityrosine relaxase activity in vitro.
Fig. 2.
Relaxase inhibition by PNP. (A) N300 oriT ssDNA cleavage velocity (v0) inhibited by PNP (error bars represent compounded standard errors from time course parameter estimates; n ≥ 3). Competitive/uncompetitive inhibition constants (Kic/Kiu) and uninhibited Michaelis constant/maximum velocity (Km/Vmax) are from nonlinear regression and Cornish-Bowden/Eisenthal direct linear plot analyses. Velocities for 100 and 10,000 nM PNP, estimated from low signal, were excluded from calculations. (B) N300 Y16F active sites with PNP (red) and without (orange). Only one PNP moiety was observed (purple). Y16F occludes the sixth coordination site. Residues 236–263 and 266 were disordered (small orange spheres). (C) Dual phosphotyrosine intermediate conformation model (red) constructed from the N300+PNP structure. Helix αA was rotated ≈120° about the helical axis to match that of TrwC structure 1OMH, and helicity was extended through αA′ and kinked about histidine-146. Important side chains (red sticks), PNP (purple sticks), a hypothetical second phosphate (purple circle), the bound metal (blue sphere), and the scissile thymidine (from 1OMH; blue) are shown over the TraI active-site cleft molecular surface.
To determine whether PNP derives its inhibitory power by binding to the TraI relaxase active site, TraI N300 Y16F crystals were soaked with PNP and the x-ray structure was determined to 3.0-Å resolution (N300+PNP) (SI Table 1). As in the N300 structure, three of the six magnesium octahedral coordination positions are filled by histidine side chains, the fourth is filled by the 3′-hydroxyl of the scissile thymidine, and the fifth is occluded by the Y16F side chain (Fig. 2B). Unlike the N300 structure, a 5σ simulated-annealing omit electron density peak appears in the sixth coordination position (SI Fig. 8), indicating the binding of a single PNP phosphate group within 3.7 Å of the magnesium ion. The second PNP phosphate was not observed, because of either disorder or, more likely, hydrolysis by a water molecule activated by the adjacent 2+ metal (52–55). The N300+PNP structure supports the conclusion that PNP inhibits TraI by binding to the relaxase catalytic site.
The N300+PNP structure revealed a novel phosphate binding site, which allowed us to generate a potential model for the second phosphotyrosine intermediate in the relaxase active site. By rotating the first α-helix (αA; lower right corner of Fig. 1A) to match the orientation observed in the R388 TraI homologue, TrwC (40), and extending helicity through tyrosine-24 (Y4 of F TraI), tyrosine-23 (Y3 of F TraI) reorients such that its side chain hydroxyl overlaps with the N300+PNP phosphate position (Fig. 2C). In this orientation tyrosine-24 also makes an aromatic stacking interaction with the side chain of tryptophan 278 (W278), a residue that is conserved in relaxases that have Y3/4 pairs. This model explains how two phosphotyrosines, one at Y16 and one at Y23, can be accommodated within the active site and in the process fulfill the octahedral coordination geometry of a bound magnesium ion.
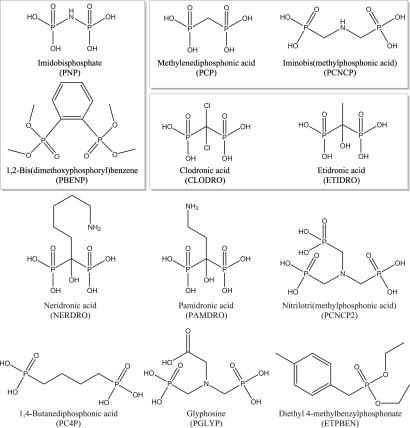
A variety of compounds were then examined for their ability to inhibit the F TraI relaxase in vitro. A coarse screen (200 nM concentrations and pH 7.4) of 11 bisphosphonates and three negative controls (sodium chloride, dibasic potassium phosphate, and ampicillin, none of which exhibited relaxase inhibition) yielded five additional inhibitors: methylenediphosphonic acid (PCP), iminobis(methylphosphonic acid) (PCNCP), etidronic acid (ETIDRO), clodronic acid (CLODRO), and 1,2-bis(dimethoxyphosphoryl)benzene (PBENP) (Fig. 3). Results from this screen reveal that effective inhibitors have two phosphonate moieties separated by three or fewer atoms and have no additional negative charge at pH 7.4. Four clinically approved bisphosphonates were tested; these drugs are used to treat bone loss by inhibiting farnesyl diphosphate synthase (reviewed in ref. 56). The simplest, ETIDRO and CLODRO, inhibited the TraI relaxase, whereas pamidronic acid and neridronic acid, which have an alkyl-amine side chain, did not. Two other inhibitors identified, PCP and PNP, have been used as radioisotope carriers in humans (57, 58). Pyrophosphate was not examined because of its rapid hydrolysis in aqueous solution. The simplest inhibitors, PCP, ETIDRO, and CLODRO, were then characterized further by using a kinetic assay and exhibited purely competitive inhibition, with Kic,app values ranging from 3 to 145 nM (SI Fig. 9). Taken together with the PNP results, these data validate the prediction that F-like conjugative relaxases can accommodate two phosphotyrosine intermediates simultaneously within their active sites. Significantly, these data also establish that bisphosphonates (including clinically approved compounds) potently inhibit the in vitro relaxase activity of F TraI with Ki values in the nanomolar range.
Fig. 3.
Bisphosphonates examined for relaxase inhibition. Chemicals examined for inhibition of TraI activity and F conjugation and for toxicity versus E. coli strains. Boxed chemicals were potent in vitro TraI inhibitors. In cell testing showed that PNP and PBENP were most effective at decreasing F+ population, CLODRO and ETIDRO were most effective at decreasing transconjugant population, and PCP and PCNCP were effective at both.
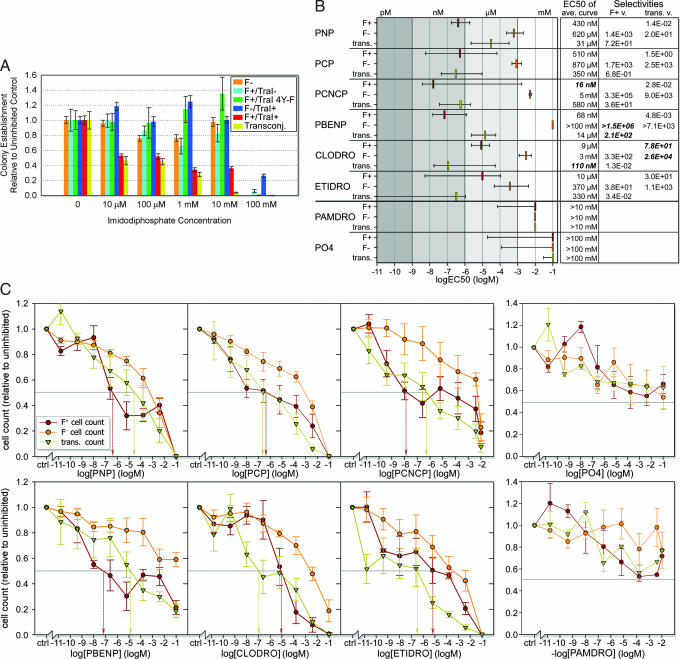
We next asked whether PNP could impact conjugative DNA transfer between living bacterial cells. F+ E. coli were mated with F− E. coli in dilute media and in the presence of increasing concentrations of PNP. The resulting mixture was applied to agar plates with antibiotic selection for transconjugants (newly formed F+ cells). Colony counts revealed that PNP inhibited DNA transfer with an EC50 of ≈10 μM, the lowest concentration tested in this assay (Fig. 4A). We also found that PNP selectively kills F+ donor cells with an EC50 of <10 μM (compared with low millimolar EC50 against F− recipient cells). This suggests that the TraI relaxase sensitizes F+ cells to a toxic effect of PNP. Minimal cell growth was observed in control mating mixtures, so decreases in donor cell count relative to controls are attributed primarily to cell death rather than to a lack of cell growth. Thus, inhibitor-dependant decreases represent bactericidal rather than bacteriastatic effects. Strains containing only the TraI gene (F−/TraI+), an F plasmid lacking the TraI gene (F+/TraI−), or an F plasmid with the four relaxase active-site tyrosines mutated to phenylalanine (F+/TraI 4Y-F) behaved like F− cells in this assay, in that they were resistant to the lethal effects of PNP. The impact on DNA donor cell survival depends on the presence of an F plasmid and a catalytically active relaxase. Thus, the simple bisphosphonate PNP enters living bacteria, inhibits conjugative DNA transfer, and selectively kills cells in an active relaxase-dependent manner. As hypothesized, small bisphosphonates appear most effective against dual tyrosine relaxases. Studies with the single tyrosine relaxase-encoding R27 plasmid reveal plasmid-dependent cell lethality only above 1 mM PNP (data not shown).
Fig. 4.
Effects of F TraI inhibitors on E. coli survival and conjugation. Color coding for lines, bars, and symbols: orange, F−; red, F+/TraI+; cyan, F+/TraI−; green, F+/TraI 4Y-F (inactive relaxase); blue, F−/TraI+; yellow, transconjugants (DNA transfer). (A) Colony counts after PNP incubation (normalized versus uninhibited controls; averaged). Error bars represent standard errors (n ≥ 5). (B) EC50 values from relative cell counts. Colored bars indicate averaged curve EC50 values. Error bars represent ± 1 SD envelope EC50 values (SI Fig. 10). Potent in vitro inhibitors are located above a strong black line, with negative controls below. Selectivities are EC50 ratios for given inhibitors and strains. (C) Relative cell counts with median EC50 values (drop arrows; colored by strain). Error bars represent the standard error (n ≥ 3).
A fluorescence-based 96-well assay was then used to examine further these effects and to screen additional compounds for the ability to impact cell survival and DNA transfer in living bacterial cells. This higher-throughput cell enumeration assay used an oxygen-quenched fluorophore imbedded within a hydrophobic gel; the concentration of live (oxygen-consuming) cells is proportional to fluorescence (59). The six compounds effective at inhibiting TraI relaxase activity in vitro (PNP, PCP, PCNCP, PBENP, CLODRO, and ETIDRO) (Fig. 3), along with two controls (pamidronic acid and K2HPO4) were examined for their impacts on F+ and F− cell survival and on DNA transfer (Fig. 4 B and C). Pamidronic acid and K2HPO4, which had no effect on TraI relaxase activity in vitro, showed little effect on cell survival and DNA transfer in these cell-based assays, exhibiting EC50 values greater than the highest concentrations tested (10 mM and 100 mM, respectively) and no selectivity for F+ over F− cells. In contrast, all six potent in vitro TraI relaxase inhibitors were also effective in living E. coli cells. EC50 values for inhibiting F+ donor cell survival ranged from 10 μM (ETIDRO) to 16 nM (PCNCP), and for inhibiting conjugative DNA transfer from 31 μM (PNP) to 110 nM (CLODRO). These compounds have little effect on F− recipient cells, with EC50 values ranging from 0.34 mM (ETIDRO) to >100 mM (PBENP), which represents 30- to 106-fold selectivity for F+ cells, respectively. In general, PNP and PBENP were more effective at inhibiting F+ cell survival, ETIDRO and CLODRO were more effective at inhibiting DNA transfer, and PCP and PCNCP were effective against both F+ cell survival and DNA transfer (Figs. 3 and 4 B and C). The ranges for EC50 values (Fig. 4B) were derived considering the standard error of the survival curves (Fig. 4C). Thus, for the most extreme case of ETIDRO, the median EC50 for transfer inhibition is 330 nM with a range of 1.1 μM to <10 pM (SI Fig. 10). Taken together, however, these data establish that relaxases can be inhibited with nanomolar affinity within living bacterial cells and that this inhibition both limits DNA transfer and selectively kills microbes harboring conjugative plasmids.
Discussion
This study outlines a potentially novel antimicrobial paradigm that specifically targets the DNA relaxase enzyme required to initiate and terminate the process of bacterial conjugation. The compounds identified could be used alone or in combination with existing antibiotics to treat recalcitrant bacterial infections. Other antibiotics and natural extracts have been reported to disrupt conjugative DNA transfer and the presence of plasmids within actively dividing bacterial cells (5–15). In each case, however, the macromolecular target of those compounds was not understood and their mechanism of action has not been determined. We took a bottom-up approach to targeted DNA conjugation by considering first the mechanism and role of a single enzyme (the DNA relaxase) in this process and then identifying inhibitors to test a specific mechanistic hypothesis (that two phosphotyrosine intermediates can be accommodated in the relaxase active site). Although our data were collected on the relaxase from the well established F plasmid that was first identified in 1946 (2), the F conjugative machinery shares up to 99% sequence identify with R plasmids containing dual tyrosine relaxases that transfer antibiotic resistance in the wild. Thus, our approach may be effective against plasmids involved in propagating a range of resistance genes and virulence factors, many of which play an important role in clinical infections (60–62).
In addition to inhibiting DNA transfer, we show that simple bisphosphonates selectively purge populations of bacteria containing a conjugative plasmid with an active relaxase enzyme. However, because TraI is F plasmid-encoded, not an essential E. coli enzyme, and was not expected to play a significant role in isolated F+ cells, this relaxase-dependent cell lethality was a surprise. Several mechanisms could be envisioned to explain this observation, and future studies will be required to distinguish between them. One possibility is that relaxases engage in cycles of plasmid DNA cleavage and religation that are uncoupled from mating and conjugative DNA transfer and that the disruption of that process results in a competitive disadvantage relative to cells without conjugative plasmids. Indeed, inspection of Fig. 4C reveals that a direct competition appears to exist between plasmid propagation and donor cell survival (note particularly PNP, PCNCP, PBENP, and CLODRO). F+ donor cell survival is enhanced at higher bisphosphonate concentrations to the detriment of plasmid propagation via conjugative transfer. These observations suggest a classic “zero-sum game” in which toxic relaxase-specific bisphosphonate inhibitors pit the interests of endosymbiont plasmids against those of their bacterial hosts.
We show that the clinically approved bisphosphonates etidronate (Didronel) and clodronate (Bonefos), but not other bisphosphonate therapeutics, are potently effective at killing F+ cells and preventing conjugative DNA transfer. These particular compounds could also be combined with existing antibiotics to create potent antimicrobial cocktails. Etidronate and clodronate exhibit low absorption (63, 64) and can be administered at high oral doses (SI Table 3). Extrapolating from our results, approved doses of etidronate and clodronate would be expected kill >90% of plasmid+ cells and to stop >80% of conjugative transfer within the gastrointestinal tract. Such results are relatively mild, given the large bacterial populations present in the gastrointestinal tract or at wound sites, but may be enough shift the balance toward success in a variety of recalcitrant clinical infections, especially given the prevalence of conjugative plasmids within multidrug-resistant bacterial strains. The treatment of skin infections, primary sites of nosocomial antibiotic resistance transfer, using the topical applications of bisphosphonates may also be effective. In summary, this study establishes conjugative relaxases as a unique antimicrobial target. Our results suggest that approved therapeutics could have an immediate impact, alone or in combination with existing antibiotics, in the prevention of resistance propagation during clinical treatment of bacterial infections and in extending the lifetime of our antibiotic arsenal.
Methods
Protein Expression, Purification, and Crystallization.
F plasmid TraI residues 1–300 bearing a phenylalanine substitution for tyrosine-16 (N300 Y16F) were subcloned into the pTYB2 vector (NEB, Ipswich, MA) and overexpressed in E. coli at 30°C. The fusion with intein-chitin-binding-domain (CBD) was purified on chitin resin (NEB) and the CBD tag cleaved with DTT. N300 Y16F crystals were grown at room temperature by the hanging-drop vapor diffusion method in the presence of ssDNA (5′-GGTGTGGTG-3′). The crystals were cryoprotected and frozen in liquid nitrogen for data collection at 77 K. PNP was added to the cryoprotectant for the N300+PNP structure.
Data Collection, Structure Determination, and Refinement.
X-ray diffraction data were collected at the Argonne National Laboratory Advanced Photon Source on the SER-CAT and GM-CAT beamlines. Diffraction data were indexed and scaled with HKL2000 and MOSFLM (CCP4) (65). Initial phases were determined by molecular replacement in Molrep (CCP4) (65) with apo-TraI [Protein Data Bank ID code 1P4D (41)] as a search model (SI Table 1). The Protein Data Bank ID codes for the structures without and with PNP are 2Q7T and 2Q7U, respectively. Structural figures were constructed in PyMol (66).
OriT Cleavage Assays.
Gel-based cleavage assays were performed as previously described (50). Fluorescent cleavage reactions were performed with biotinylated oriT ssDNA, either fluorescein end-labeled or postreaction hybridized to an end-labeled probe. Twelve to 96 parallel reactions were run on 96-well plates (37°C), stopped with SDS and EDTA (80°C), and read on streptavidin-coated 96- or 384-well plates.
Conjugative Mating Assays.
Liquid mating assays were performed as previously described (26) except that HMS174 cells replaced HMS174 (DE3) cells. Where applicable, inhibitor candidates were added at reaction initiation and removed before strain selection/detection. Strains were selected, and cell concentrations were quantified either on agar plates or in antibiotic-laden media on Oxygen Biosensor 96-well round-bottom plates (BD Biosciences) (59). In toxicity assays on agar plates, donor and recipients cells remained separate. In mating assays, donor and recipient cells were mixed at a 1:9 ratio. All inhibitor exposures were 100 min.
Additional Details.
Detailed descriptions of all methods and data analyses are included in SI Methods.
Supplementary Material
Acknowledgments
We thank D.-T. Lesher, J. K. Sampson, D. Haisch, M. J. Miley, S. A. Kennedy, and S. Bencharit for experimental assistance, and we thank J. Lomino, D. G. Teotico, C. D. Fleming, and E. Ortlund for helpful advice. This work was supported by National Institutes of Health Grants CA90604 (to M.R.R.) and GM61020 (to S.W.M.) and a Kirschstein National Institutes of Health Postdoctoral Fellowship (L.M.G.).
Abbreviations
- PCP
methylenediphosphonic acid
- PCNCP
iminobis(methylphosphonic acid)
- ETIDRO
etidronic acid
- CLODRO
clodronic acid
- PBENP
1,2-bis(dimethoxyphosphoryl)benzene
- PNP
imidobisphosphate
- CPR
concomitant plasmid replication.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates for the structures without and with PNP have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2Q7T and 2Q7U, respectively).
This article contains supporting information online at www.pnas.org/cgi/content/full/0702760104/DC1.
References
- 1.Burrus V, Waldor MK. Res Microbiol. 2004;155:376–386. doi: 10.1016/j.resmic.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 2.Lederberg J, Tatum EL. Nature. 1946;158:558. doi: 10.1038/158558a0. [DOI] [PubMed] [Google Scholar]
- 3.de la Cruz F, Davies J. Trends Microbiol. 2000;8:128–133. doi: 10.1016/s0966-842x(00)01703-0. [DOI] [PubMed] [Google Scholar]
- 4.Mazel D, Davies J. Cell Mol Life Sci. 1999;56:742–754. doi: 10.1007/s000180050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura S, Inoue S, Shimizu M, Iyobe S, Mitsuhashi S. Antimicrob Agents Chemother. 1976;10:779–785. doi: 10.1128/aac.10.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George BA, Fagerberg DJ. Am J Vet Res. 1984;45:2336–2341. [PubMed] [Google Scholar]
- 7.Oliva B, Selan L, Ravagnan G, Renzini G. Chemioterapia. 1985;4:199–201. [PubMed] [Google Scholar]
- 8.Michel-Briand Y, Laporte JM. J Gen Microbiol. 1985;131:2281–2284. doi: 10.1099/00221287-131-9-2281. [DOI] [PubMed] [Google Scholar]
- 9.Palomares JC, Prados R, Perea EJ. Chemioterapia. 1987;6:256–260. [PubMed] [Google Scholar]
- 10.Molnar J, Csiszar K, Nishioka I, Shoyama Y. Acta Microbiol Hung. 1986;33:221–231. [PubMed] [Google Scholar]
- 11.Leite AA, Nardi RM, Nicoli JR, Chartone-Souza E, Nascimento AM. J Gen Appl Microbiol. 2005;51:21–26. doi: 10.2323/jgam.51.21. [DOI] [PubMed] [Google Scholar]
- 12.Weisser J, Wiedemann B. Antimicrob Agents Chemother. 1987;31:531–534. doi: 10.1128/aac.31.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Debbia EA, Massaro S, Campora U, Schito GC. New Microbiol. 1994;17:65–68. [PubMed] [Google Scholar]
- 14.Spengler G, Molnar A, Schelz Z, Amaral L, Sharples D, Molnar J. Curr Drug Targets. 2006;7:823–841. doi: 10.2174/138945006777709601. [DOI] [PubMed] [Google Scholar]
- 15.Jung YD, Ellis LM. Int J Exp Pathol. 2001;82:309–316. doi: 10.1046/j.1365-2613.2001.00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byrd DR, Matson SW. Mol Microbiol. 1997;25:1011–1022. doi: 10.1046/j.1365-2958.1997.5241885.x. [DOI] [PubMed] [Google Scholar]
- 17.Pansegrau W, Lanka E. Prog Nucleic Acid Res Mol Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- 18.Lanka E, Wilkins BM. Annu Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 19.Llosa M, Gomis-Ruth FX, Coll M, de la Cruz Fd F. Mol Microbiol. 2002;45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 20.Reygers U, Wessel R, Muller H, Hoffmann-Berling H. EMBO J. 1991;10:2689–2694. doi: 10.1002/j.1460-2075.1991.tb07812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matson SW, Morton BS. J Biol Chem. 1991;266:16232–16237. [PubMed] [Google Scholar]
- 22.Traxler BA, Minkley EG., Jr J Mol Biol. 1988;204:205–209. doi: 10.1016/0022-2836(88)90609-2. [DOI] [PubMed] [Google Scholar]
- 23.Lahue EE, Matson SW. J Biol Chem. 1988;263:3208–3215. [PubMed] [Google Scholar]
- 24.Schroder G, Lanka E. Plasmid. 2005;54:1–25. doi: 10.1016/j.plasmid.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. FEMS Microbiol Lett. 2003;224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 26.Matson SW, Sampson JK, Byrd DR. J Biol Chem. 2001;276:2372–2379. doi: 10.1074/jbc.M008728200. [DOI] [PubMed] [Google Scholar]
- 27.Byrd DR, Sampson JK, Ragonese HM, Matson SW. J Biol Chem. 2002;277:42645–42653. doi: 10.1074/jbc.M205984200. [DOI] [PubMed] [Google Scholar]
- 28.Traxler BA, Minkley EG., Jr J Bacteriol. 1987;169:3251–3259. doi: 10.1128/jb.169.7.3251-3259.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scherzinger E, Lurz R, Otto S, Dobrinski B. Nucleic Acids Res. 1992;20:41–48. doi: 10.1093/nar/20.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pansegrau W, Ziegelin G, Lanka E. J Biol Chem. 1990;265:10637–10644. [PubMed] [Google Scholar]
- 31.Pansegrau W, Schroder W, Lanka E. J Biol Chem. 1994;269:2782–2789. [PubMed] [Google Scholar]
- 32.Furuya N, Nisioka T, Komano T. J Bacteriol. 1991;173:2231–2237. doi: 10.1128/jb.173.7.2231-2237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young C, Nester EW. J Bacteriol. 1988;170:3367–3374. doi: 10.1128/jb.170.8.3367-3374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srinivas P, Kilic AO, Vijayakumar MN. Plasmid. 1997;37:42–50. doi: 10.1006/plas.1996.1268. [DOI] [PubMed] [Google Scholar]
- 35.Paterson ES, More MI, Pillay G, Cellini C, Woodgate R, Walker GC, Iyer VN, Winans SC. J Bacteriol. 1999;181:2572–2583. doi: 10.1128/jb.181.8.2572-2583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Llosa M, Bolland S, de la Cruz F. J Mol Biol. 1994;235:448–464. doi: 10.1006/jmbi.1994.1005. [DOI] [PubMed] [Google Scholar]
- 37.Greated A, Lambertsen L, Williams PA, Thomas CM. Environ Microbiol. 2002;4:856–871. doi: 10.1046/j.1462-2920.2002.00305.x. [DOI] [PubMed] [Google Scholar]
- 38.Boer R, Russi S, Guasch A, Lucas M, Blanco AG, Perez-Luque R, Coll M, de la Cruz F. J Mol Biol. 2006;358:857–869. doi: 10.1016/j.jmb.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 39.Larkin C, Datta S, Harley MJ, Anderson BJ, Ebie A, Hargreaves V, Schildbach JF. Structure (London) 2005;13:1533–1544. doi: 10.1016/j.str.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 40.Guasch A, Lucas M, Moncalian G, Cabezas M, Perez-Luque R, Gomis-Ruth FX, de la Cruz F, Coll M. Nat Struct Biol. 2003;10:1002–1010. doi: 10.1038/nsb1017. [DOI] [PubMed] [Google Scholar]
- 41.Datta S, Larkin C, Schildbach JF. Structure (London) 2003;11:1369–1379. doi: 10.1016/j.str.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Grandoso G, Avila P, Cayon A, Hernando MA, Llosa M, de la Cruz F. J Mol Biol. 2000;295:1163–1172. doi: 10.1006/jmbi.1999.3425. [DOI] [PubMed] [Google Scholar]
- 43.Dressler D. Proc Natl Acad Sci USA. 1970;67:1934–1942. doi: 10.1073/pnas.67.4.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erickson MJ, Meyer RJ. Mol Microbiol. 1993;7:289–298. doi: 10.1111/j.1365-2958.1993.tb01120.x. [DOI] [PubMed] [Google Scholar]
- 45.Pansegrau W, Lanka E. J Biol Chem. 1996;271:13068–13076. doi: 10.1074/jbc.271.22.13068. [DOI] [PubMed] [Google Scholar]
- 46.Furuya N, Komano T. J Bacteriol. 2003;185:3871–3877. doi: 10.1128/JB.185.13.3871-3877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Draper O, Cesar CE, Machon C, de la Cruz F, Llosa M. Proc Natl Acad Sci USA. 2005;102:16385–16390. doi: 10.1073/pnas.0506081102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Llosa M, Bolland S, Grandoso G, de la Cruz F. J Bacteriol. 1994;176:3210–3217. doi: 10.1128/jb.176.11.3210-3217.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker C, Zhang XL, Henderson D, Becker E, Meyer R. Plasmid. 2002;48:186–192. doi: 10.1016/s0147-619x(02)00105-1. [DOI] [PubMed] [Google Scholar]
- 50.Matson SW, Ragonese H. J Bacteriol. 2005;187:697–706. doi: 10.1128/JB.187.2.697-706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ragonese H, Haisch D, Villareal E, Choi JH, Matson SW. Mol Microbiol. 2007;63:1173–1184. doi: 10.1111/j.1365-2958.2006.05576.x. [DOI] [PubMed] [Google Scholar]
- 52.Smirnova IN, Baykov AA, Avaeva SM. FEBS Lett. 1986;206:121–124. doi: 10.1016/0014-5793(86)81352-7. [DOI] [PubMed] [Google Scholar]
- 53.Taylor JS. J Biol Chem. 1981;256:9793–9795. [PubMed] [Google Scholar]
- 54.Tomaszek TA, Jr, Schuster SM. J Biol Chem. 1986;261:2264–2269. [PubMed] [Google Scholar]
- 55.Yount RG, Babcock D, Ballantyne W, Ojala D. Biochemistry. 1971;10:2484–2489. doi: 10.1021/bi00789a009. [DOI] [PubMed] [Google Scholar]
- 56.Russell RG. Ann NY Acad Sci. 2006;1068:367–401. doi: 10.1196/annals.1346.041. [DOI] [PubMed] [Google Scholar]
- 57.Brody KR, Hosain P, Spencer RP, Hosain F, Wagner HN. Br J Radiol. 1976;49:267–269. doi: 10.1259/0007-1285-49-579-267. [DOI] [PubMed] [Google Scholar]
- 58.Huckell VF, Lyster DM, Morrison RT, Cooper JA. Clin Nucl Med. 1985;10:455–462. doi: 10.1097/00003072-198507000-00001. [DOI] [PubMed] [Google Scholar]
- 59.Wodnicka M, Guarino RD, Hemperly JJ, Timmins MR, Stitt D, Pitner JB. J Biomol Screen. 2000;5:141–152. doi: 10.1177/108705710000500306. [DOI] [PubMed] [Google Scholar]
- 60.Ahmed AM, Kawamoto H, Inouye K, Hashiwata Y, Sakaki M, Seno M, Shimamoto T. J Med Microbiol. 2005;54:867–872. doi: 10.1099/jmm.0.46055-0. [DOI] [PubMed] [Google Scholar]
- 61.Tosini F, Visca P, Luzzi I, Dionisi AM, Pezzella C, Petrucca A, Carattoli A. Antimicrob Agents Chemother. 1998;42:3053–3058. doi: 10.1128/aac.42.12.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei ZQ, Chen YG, Yu YS, Lu WX, Li LJ. J Med Microbiol. 2005;54:885–888. doi: 10.1099/jmm.0.46151-0. [DOI] [PubMed] [Google Scholar]
- 63.Villikka K, Perttunen K, Rosnell J, Ikavalko H, Vaho H, Pylkkanen L. Bone. 2002;31:418–421. doi: 10.1016/s8756-3282(02)00841-4. [DOI] [PubMed] [Google Scholar]
- 64.Recker RR, Saville PD. Toxicol Appl Pharmacol. 1973;24:580–589. doi: 10.1016/0041-008x(73)90219-6. [DOI] [PubMed] [Google Scholar]
- 65.Collaborative Computing Project N. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 66.DeLano WL, editor. PyMol. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.