Abstract
The identification of cell surface antigens is critical to the development of new diagnostic and therapeutic modalities for the management of prostate cancer. Prostate stem cell antigen (PSCA) is a prostate-specific gene with 30% homology to stem cell antigen 2, a member of the Thy-1/Ly-6 family of glycosylphosphatidylinositol (GPI)-anchored cell surface antigens. PSCA encodes a 123-aa protein with an amino-terminal signal sequence, a carboxyl-terminal GPI-anchoring sequence, and multiple N-glycosylation sites. PSCA mRNA expression is prostate-specific in normal male tissues and is highly up-regulated in both androgen-dependent and -independent prostate cancer xenografts. In situ mRNA analysis localizes PSCA expression in normal prostate to the basal cell epithelium, the putative stem cell compartment of the prostate. There is moderate to strong PSCA expression in 111 of 126 (88%) prostate cancer specimens examined by in situ analysis, including high-grade prostatic intraepithelial neoplasia and androgen-dependent and androgen-independent tumors. Flow cytometric analysis demonstrates that PSCA is expressed predominantly on the cell surface and is anchored by a GPI linkage. Fluorescent in situ hybridization analysis localizes the PSCA gene to chromosome 8q24.2, a region of allelic gain in more than 80% of prostate cancers. A mouse homologue with 70% amino acid identity and similar genomic organization to human PSCA has also been identified. These results support PSCA as a target for prostate cancer diagnosis and therapy.
Prostate cancer is the most common cancer diagnosis and the second leading cause of cancer-related death in American men. Despite recent advances in the detection and treatment of localized disease, significant challenges remain in the management of this disease. Current diagnostic modalities are limited by a lack of specificity and an inability to predict which patients are at risk to develop metastatic disease. Prostate-specific antigen (PSA) is effective at identifying men who may have prostate cancer but is often elevated in men with benign prostatic hyperplasia, prostatitis, and other nonmalignant disorders (1). PSA and other current markers fail to discriminate accurately between indolent and aggressive cancers. There is no effective treatment for the 20–40% of patients who develop recurrent disease after surgery or radiation therapy or for those who have metastatic disease at the time of diagnosis. Although hormone ablation therapy can palliate these patients, the majority inevitably progress to develop incurable androgen-independent disease (1).
In an effort to identify potential markers for the diagnosis and treatment of prostate cancer, we have searched for genes up-regulated during prostate cancer progression by using the recently developed LAPC-4 xenograft model of human prostate cancer (2). The LAPC-4 system accurately recapitulates many of the features of advanced human prostate cancer, including progression to androgen independence and metastasis. We have sought to identify genes encoding secreted or cell surface proteins, because they have potential utility as serum markers of prostate cancer, similar to PSA and glandular kallikrein 2, and may be useful for detecting or targeting prostate cancer cells (3). Prostate specific membrane antigen (PSMA) is a recently described cell surface marker of prostate cancer currently being evaluated for the detection of metastatic cells and as a target for mAb and other immunological therapies (4, 5). Another potential advantage to searching for membrane-bound tumor antigens is that they may provide insights into the biology of prostate cancer progression. Her-2/neu, one of a number of growth factor receptors associated with prostate cancer, is being evaluated both as a target for therapy and as an important signaling molecule in androgen-independent prostate cancer (6–8). Other receptors, such as c-met and urokinase plasminogen activator receptor, may mediate prostate cancer metastasis (9–11).
We have identified a number of candidate molecules up-regulated in the LAPC-4 xenograft model by using representational difference analysis (RDA), a PCR-based subtractive hybridization strategy (12). One promising candidate, prostate stem cell antigen (PSCA), is a prostate-specific cell surface antigen expressed strongly by both androgen-dependent and -independent LAPC-4 tumors. PSCA is homologous to a group of cell surface proteins that mark the earliest phases of hematopoietic development. We hypothesize that PSCA may play a role in prostate cancer progression and may serve as a target for prostate cancer diagnosis and treatment.
MATERIALS AND METHODS
Molecular Studies.
RDA of androgen-dependent and -independent LAPC-4 tumors was performed as described (13). Total RNA was isolated by using Ultraspec RNA isolation systems (Biotecx, Houston, TX) according to the manufacturer’s instructions. Northern blot filters were probed with a 660-bp RDA fragment corresponding to the coding sequence and part of the 3′ untranslated sequence of PSCA or an ∼400-bp fragment of PSA. The human multiple tissue blot was obtained from CLONTECH and probed as specified. For reverse transcriptase-coupled PCR (RT-PCR) analysis, first-strand cDNA was synthesized from total RNA by using the GeneAmp RNA PCR core kit (Perkin–Elmer–Roche). For RT-PCR of human PSCA transcripts, primers 5′-TGCTTGCCCTGTTGATGGCAG and 3′-CCAGAGCAGCAGGCCGAGTGCA were used to amplify an ∼320-bp fragment. Thermal cycling was performed for 25 cycles at 95°C for 30 sec, 60°C for 30 sec, and 72°C for 1 min, followed by extension at 72°C for 10 min. Primers for GAPDH (CLONTECH) were used as controls. For mouse PSCA, the primers used were 5′-TTCTCCTGCTGGCCACCTAC and 3′-GCAGCTCATCCCTTCACAAT.
For mRNA in situ hybridization, recombinant plasmid pCR II (1 μg, Invitrogen) containing the full-length PSCA gene was linearized to generate sense and antisense digoxigenin-labeled RNA probes. In situ hybridization was performed on an automated instrument (Ventana Gen II, Ventana Medical Systems) as described (14). Prostate specimens were obtained from a previously described database that has been expanded to ∼130 specimens (14). Slides were read and scored by two pathologists in a blinded fashion. Scores of 0–3 were assigned according to the percentage of positive cells (0 = 0%; 1 = <25%; 2 = 25–50%; 3 = >50%) and the intensity of staining (0 = 0; 1 = 1+; 2 = 2+; 3 = 3+). The two scores were multiplied to give an overall score of 0–9.
λ phage clones containing the human PSCA gene were obtained by screening a human genomic library (Stratagene) with a human PSCA cDNA probe (15). BAC (bacterial artificial chromosome) clones containing the murine PSCA gene were obtained by screening a murine BAC library (Genome Systems, St. Louis). A 14-kb human NotI fragment and a 10-kb murine EcoRI fragments were subcloned into pBluescript (Stratagene) and subjected to restriction mapping. Fluorescence in situ chromosomal analysis was performed as described by using overlapping human λ phage clones (16).
Biochemical Studies.
Rabbit polyclonal antiserum was generated against the synthetic peptide TARIRAVGLLTVISK and affinity-purified by using a PSCA–glutathione S-transferase fusion protein. 293T cells were transiently transfected with pCDNA II (Invitrogen) expression vectors containing PSCA, CD59, E25, or vector alone by calcium phosphate precipitation. Immunoprecipitation was performed as described (17). Briefly, cells were labeled with 500 μCi of trans35S label (ICN) for 6 h. Cell lysates and conditioned medium were incubated with 1 μg of purified rabbit anti-PSCA antibody and 20 μl of protein A-Sepharose CL-4B (Pharmacia Biotech) for 2 h. For deglycosylation, immunoprecipitates were treated overnight at 37°C with 1 unit of N-glycosidase F (Boehringer Mannheim) or 0.1 unit of neuraminidase (Sigma) for 1 h followed by an overnight incubation in 2.5 milliunits of O-glycosidase (Boehringer Mannheim).
For flow cytometric analysis of PSCA cell surface expression, single cell suspensions were stained with purified anti-PSCA antibody (2 μg/ml) and a 1:500 dilution of phycoerythrin-labeled anti-rabbit IgG (Jackson ImmunoResearch). Data was acquired on a FACScan (Becton Dickinson) and analyzed by using lysis ii software. Control samples were stained with rabbit IgG (Jackson ImmunoResearch; 2 μg/ml) followed by secondary antibody. The glycosylphosphatidylinositol (GPI) linkage was analyzed by digestion of 2 × 106 cells with 0.5 unit of phosphatidylinositol-specific phospholipase C (PLC, Boehringer Mannheim) for 90 min at 37°C. Cells were analyzed prior to and after digestion by either flow cytometry scanning or immunoblotting.
RESULTS
RDA Identifies PSCA.
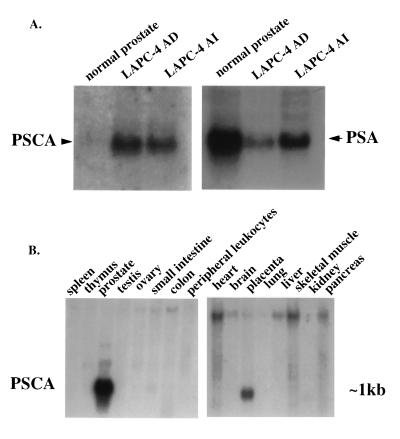
RDA was used to isolate cDNAs up-regulated in an androgen-independent subline of the LAPC-4 prostate cancer xenograft (12). In the course of this work, one 660-bp fragment (clone 15) was identified that was found to be highly overexpressed in xenograft tumors when compared with normal prostate but was not differentially expressed between androgen-dependent and -independent LAPC-4 tumors (Fig. 1A). Comparison of the expression of this clone to that of PSA in normal prostate and xenograft tumors suggested that clone 15 was relatively cancer-specific (Fig. 1A).
Figure 1.
Northern blot analysis of PSCA expression. (A) Total RNA from normal prostate and LAPC-4 androgen-dependent (AD) and -independent (AI) prostate cancer xenografts were analyzed by using PSCA- or PSA-specific probes. Equivalent RNA loading and RNA integrity were demonstrated separately by ethidium staining for 18S and 28S RNA and are not shown. (B) Human multiple tissue Northern blot analysis of PSCA. The filter was obtained from CLONTECH and contains 2 μg of poly(A) RNA in each lane.
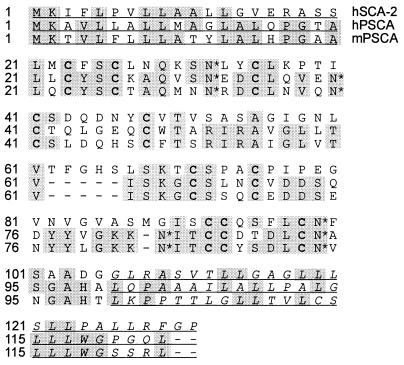
Sequence analysis revealed that clone 15 had no exact match in the databases but shared 30% nucleotide homology with stem cell antigen 2 (SCA-2), a member of the Thy-1/Ly-6 superfamily of GPI-anchored cell surface antigens. Clone 15 encodes a 123-amino acid protein that is 30% identical to SCA-2 (also called RIG-E) and contains a number of highly conserved cysteine residues characteristic of the Ly-6/Thy-1 gene family (Fig. 2) (18). Consistent with its homology to a family of GPI-anchored proteins, clone 15 contains both an amino-terminal hydrophobic signal sequence and a carboxyl-terminal stretch of hydrophobic amino acids preceded by a group of small amino acids defining a cleavage/binding site for GPI linkage (19). It also contains four predicted N-glycosylation sites. Because of its strong homology to SCA-2, clone 15 was renamed PSCA.
Figure 2.
Amino acid sequences of human SCA-2 (hSCA-2), human PSCA (hPSCA), and mouse PSCA (mPSCA). Shaded regions highlight conserved amino acids, and conserved cysteines are indicated by boldface type. Four predicted N-glycosylation sites in PSCA are indicated by asterisks. The underlined amino acids at the beginning and end of the protein represent amino-terminal hydrophobic signal sequences and C-terminal GPI-anchoring sequences, respectively.
The human PSCA cDNA was used to search murine expressed sequence tag (EST) databases to identify homologues for potential transgenic and knockout experiments. One EST obtained from fetal mouse and another from neonatal kidney were 70% identical to the human cDNA at both the nucleotide and amino acid levels. The homology between the mouse clones and human PSCA included regions of divergence between human PSCA and its GPI-anchored homologues, indicating that these clones likely represented the mouse homologue of PSCA. Alignment of these ESTs and 5′ extension by using rapid amplification of cDNA ends coupled to PCR provided the entire coding sequence (see Fig. 2).
PSCA Expression Is Prostate-Specific.
The distribution of PSCA mRNA in normal human tissues was examined by Northern blot analysis. PSCA is expressed predominantly in prostate, with a lower level of expression present in placenta (Fig. 1B). Small amounts of mRNA can be detected in kidney and small intestine after prolonged exposure and are approximately 1% of the level seen in prostate. RT-PCR analysis of PSCA expression in normal human tissues produced similar results (data not shown). The major PSCA transcript in normal prostate is ∼1 kb (Fig. 1B). Mouse PSCA expression was analyzed by RT-PCR in mouse spleen, liver, lung, prostate, kidney, and testis (data not shown). Like human PSCA, murine PSCA is expressed predominantly in prostate. Expression can also be detected in kidney at a level similar to that seen for placenta in human tissues. These data indicate that PSCA expression is largely prostate-specific.
PSCA Is Expressed by a Subset of Basal Cells in Normal Prostate.
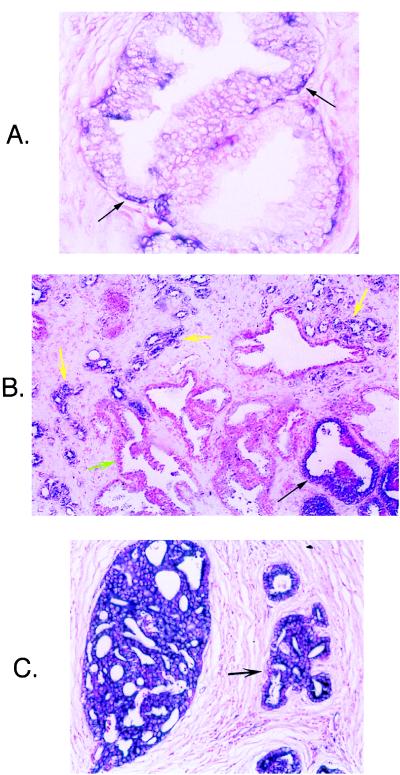
Normal prostate contains two major epithelial cell populations—secretory luminal cells and subjacent basal cells. In situ hybridizations were performed on multiple sections of normal prostate by using an antisense RNA probe specific for PSCA to localize its expression. PSCA is expressed exclusively in a subset of normal basal cells (Fig. 3A). Little to no staining is seen in stroma, secretory cells, or infiltrating lymphocytes. Hybridization with sense PSCA RNA probes showed no background staining. Hybridization with an antisense probe for GAPDH confirmed that the RNA in all cell types was intact. Because basal cells represent the putative progenitor cells for the terminally differentiated secretory cells, these results suggest that PSCA may be a prostate-specific stem/progenitor cell marker (20).
Figure 3.
In situ hybridization with antisense RNA probe for human PSCA on normal and malignant prostate specimens. (A) PSCA is expressed by a subset of basal cells within the basal cell epithelium (black arrows) but not by the terminally differentiated secretory cells lining the prostatic ducts. (×400.) (B) PSCA is expressed strongly by a high-grade prostatic intraepithelial neoplasia (black arrow) and by invasive prostate cancer glands (yellow arrows) but is not detectable in normal epithelium (green arrow) at ×40 magnification. (C) Strong expression of PSCA in high-grade carcinoma. (×200.)
PSCA mRNA Is Overexpressed by a Majority of Human Prostate Cancers.
The initial analysis comparing PSCA expression in normal prostate and LAPC-4 xenograft tumors suggested that PSCA was overexpressed in prostate cancer. One hundred twenty-six paraffin-embedded prostate cancer specimens were analyzed by mRNA in situ hybridization for PSCA expression. Specimens were obtained from primary tumors removed by radical prostatectomy or transurethral resection in all cases except one. All specimens were probed with both a sense and antisense construct to control for background staining. Slides were assigned a composite score, with a score of 6 to 9 indicating strong expression and a score of 4 meaning moderate expression. One hundred two of 126 (81%) cancers stained strongly for PSCA, and another nine of 126 (7%) displayed moderate staining (Fig. 3 B and C). High-grade prostatic intraepithelial neoplasia, the putative precursor lesion of invasive prostate cancer, stained strongly positive for PSCA in 82% (97 of 118) of specimens (Fig. 3B) (21). Normal glands stained consistently weaker than malignant glands (Fig. 3B). Nine specimens were obtained from patients treated before surgery with hormone ablation therapy. Seven of nine (78%) of these residual presumably androgen-independent cancers overexpressed PSCA, a percentage similar to that seen in untreated cancers. Because such a large percentage of specimens expressed PSCA mRNA, no statistical correlations could be made between PSCA expression and pathological features such as tumor stage and grade. These results suggest that PSCA mRNA overexpression is a common feature of androgen-dependent and -independent prostate cancer.
PSCA expression was also detected in the androgen-independent androgen-receptor-negative prostate cancer cell lines PC3 and DU145 by RT–PCR analysis (data not shown). These data suggest that PSCA can be expressed in the absence of functional androgen receptor.
PSCA Is a GPI-Anchored Glycoprotein Expressed on the Cell Surface.
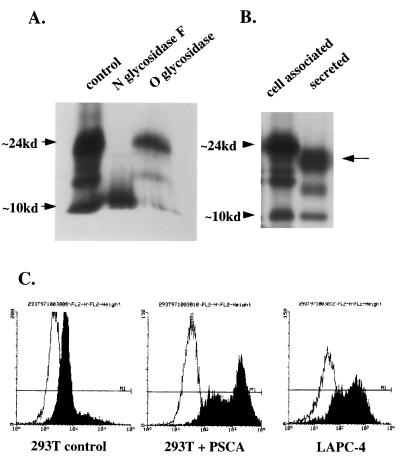
The deduced PSCA amino acid sequence predicts that PSCA is heavily glycosylated and anchored to the cell surface through a GPI mechanism. To test these predictions, we produced an affinity-purified polyclonal antibody raised against a unique PSCA peptide. This peptide contains no glycosylation sites and was predicted, based on a comparison to the three-dimensional structure of CD59 (another GPI-anchored PSCA homologue), to lie in an exposed portion of the mature protein (22). Recognition of PSCA by the affinity-purified antibody was demonstrated by immunoblot and immunoprecipitation analysis of a glutathione S-transferase–PSCA fusion protein and extracts of 293T cells transfected with PSCA. The polyclonal antibody immunoprecipitates predominantly a 24-kDa band from PSCA-transfected but not mock-transfected cells (Fig. 4A). Three smaller bands are also present, the smallest being ≈10 kDa. The immunoprecipitate was treated with N- and O-specific glycosidases to determine whether these bands represented glycosylated forms of PSCA. N-glycosidase F deglycosylated PSCA, whereas O-glycosidase had no effect (Fig. 4A). Some GPI-anchored proteins are known to have both membrane-bound and secreted forms (23, 24). Fig. 4B indicates that some PSCA is secreted in the 293T-overexpressing system. The secreted form of PSCA migrates at a lower molecular weight than the cell surface-associated form, perhaps reflecting the absence of the covalent GPI-linkage. This result may reflect the high level of expression in the 293T cell line and needs to be confirmed in prostate cancer cell lines and in vivo.
Figure 4.
Biochemical analysis of PSCA. (A) PSCA was immunoprecipitated from 293T cells transiently transfected with a PSCA construct and then digested with either N-glycosidase F or O-glycosidase. (B) PSCA was immunoprecipitated from 293T transfected cells and from conditioned medium from these cells. Cell-associated PSCA migrates higher than secreted PSCA on a 15% polyacrylamide gel. (C) Flow cytometry analysis of mock-transfected 293T cells, PSCA-transfected 293T cells, and LAPC-4 prostate cancer xenograft cells by using an affinity-purified polyclonal anti-PSCA antibody. Cells were not permeabilized to detect only surface expression. The y axis represents relative cell number and the x axis represents fluorescent staining intensity on a logarithmic scale.
Fluorescence-activated cell sorting analysis was used to localize PSCA expression to the cell surface. Nonpermeabilized mock-transfected 293T cells, PSCA-expressing 293T cells, and LAPC-4 cells were stained with affinity-purified antibody or secondary antibody alone. Fig. 4C shows cell surface expression of PSCA in PSCA-transfected 293T and LAPC-4 cells but not in mock-transfected cells. To confirm that this cell surface expression is mediated by a covalent GPI linkage, cells were treated with GPI-specific PLC. Release of PSCA from the cell surface by PLC was indicated by more than a one order of magnitude reduction in fluorescence intensity (data not shown). Recovery of PSCA in postdigest conditioned medium was also confirmed by immunoblotting (data not shown). The specificity of PLC digestion for GPI-anchored proteins was confirmed by performing the same experiment on 293T cells transfected with the GPI-linked antigen CD59 or the non-GPI-linked transmembrane protein E25a (25). PLC digestion reduced cell surface expression of CD59 to the same degree as PSCA but had no effect on E25. These results support the prediction that PSCA is a glycosylated GPI-anchored cell surface protein.
The PSCA Gene Maps to Chromosome 8q24.2.
Southern blot analysis of LAPC-4 genomic DNA revealed that PSCA is encoded by a single copy gene (data not shown). Other Ly-6 gene family members contain four exons, including a first exon encoding a 5′ untranslated region and three additional exons encoding the translated and 3′ untranslated regions. Genomic clones of human and murine PSCA containing all but the presumed 5′ first exon were obtained by screening λ phage libraries. Mouse and human PSCA clones had a similar genomic organization. The human clone was used to localize PSCA by fluorescence in situ hybridization analysis. Cohybridization of overlapping human PSCA λ phage clones resulted in specific labeling only of chromosome 8 (Fig. 5). Ninety-seven percent of detected signals localized to chromosome 8q24, of which 87% were specific for chromosome 8q24.2. These results show that PSCA is located at chromosome 8, band q24.2.
Figure 5.
In situ hybridization of biotin-labeled PSCA probes to human metaphase cells from phytohemagglutinin-stimulated peripheral blood lymphocytes. The chromosome 8 homologues are identified with arrows; specific labeling was observed at 8q24.2. (Inset) Partial karyotypes of two chromosome 8 homologues illustrating specific labeling at 8q24.2 (arrowheads). Images were obtained by using a Zeiss Axiophot microscope coupled to a cooled charge-coupled device camera. Separate images of 4′,6-diamidino-2-phenylindole-stained chromosomes and the hybridization signal were merged by using image analysis software (nu200 and image 1.57).
DISCUSSION
PSCA is a prostate cancer-associated tumor antigen. Northern blot and in situ data show that PSCA is predominantly prostate-specific in normal tissues and is overexpressed in greater than 80% of prostate cancers. The expression of PSCA in cancers appears to be the reverse of what is seen with PSA. Although PSA is expressed more strongly in normal than malignant prostate, PSCA is expressed more highly in prostate cancer. This relationship is clearly seen in the case of the LAPC-4 xenograft tumors and is inferred from the large number of in situ hybridizations performed. These data suggest that PSCA may be a useful marker for discriminating cancers from normal glands in prostatectomy specimens.
The cell surface location of PSCA makes it a putative target for therapy and diagnosis. One possibility is that antibodies directed against PSCA may be used to locate metastatic disease in patients considering local therapy or in those believed to have metastatic disease, analogous to the Prostascint test currently being developed by using an antibody against PSMA (26). Antibodies against PSCA might be used to deliver radioisotopes or other toxins specifically to prostate cancer cells. Finally, PSCA could be tested as an immunogen for various immunological therapies. Similar approaches are currently under development for PSMA (4, 27).
A preliminary survey of 126 prostate cancer specimens did not reveal any clear correlation of PSCA expression with cancer grade, stage, or hormone dependency but rather showed widespread overexpression of this marker. PSCA expression was maintained in most androgen-independent tumors and was also detected in androgen-receptor negative cell lines, suggesting that PSCA may a useful marker of androgen-independent tumors that have lost functional androgen receptor. It should be emphasized that the primary goal of this survey was to demonstrate expression of PSCA mRNA in vivo. Also, metastatic prostate cancers were not studied other than the LAPC-4 xenograft, which was derived from a lymph node metastasis. Additional studies looking at PSCA protein expression may provide important clinical information not attainable by in situ hybridization. In a recent study on thymosin β15 expression, RNA in situ analyses were consistently positive, whereas immunohistochemical analysis revealed a possible correlation between Gleason grade and level of thymosin β15 expression (28).
An intriguing finding in this study was the localization of PSCA mRNA to a subset of basal cells in normal prostate. The basal cell epithelium is believed to contain the progenitor cells for the terminally differentiated secretory cells (20). Recent studies using cytokeratin markers suggest that the basal cell epithelium contains at least two distinct cellular subpopulations, one expressing cytokeratins 5 and 14 and the other cytokeratins 5, 8, and 18 (29). The finding that PSCA is expressed by only a subset of basal cells suggests that PSCA may be a marker for one of these two basal cell lineages.
A number of investigators have hypothesized that prostate cancers may arise from transformation of basal cells (30). Verhagen et al. (31) identified a population of cells in prostate cancers that coexpress basal and secretory cell cytokeratins 5, 8, and 18. Bcl-2, c-met, and other genes whose expression in normal prostate is restricted to basal cells have also been associated with prostate cancer (11, 32). The expression of PSCA in prostate cancer supports the concept that cancers arise from transformation of basal cells (29, 30). Additional work clearly will be needed to prove this hypothesis. It should be particularly important to study PSCA expression at the protein level, because it remains possible that although PSCA mRNA is not detected in secretory cells, PSCA protein may be present. Differences in RNA and protein localization have been noted previously for androgen receptor and PSMA expression (33, 34).
The biological function of PSCA is not known. The Ly-6 gene family is involved in diverse cellular functions, including signal transduction and cell–cell adhesion. Signaling through SCA-2 has been demonstrated to prevent apoptosis in immature thymocytes (35). Thy-1 is involved in T cell activation and transmits signals through src-like tyrosine kinases (36). Ly-6 genes have been implicated both in tumorigenesis and in homotypic cell adhesion (37–39). From its restricted expression in basal cells and its homology to SCA-2, we hypothesize that PSCA may play a role in stem/progenitor cell functions such as self-renewal (antiapoptosis) and/or proliferation.
PSCA is highly conserved in mice and humans. The identification of a conserved gene that is predominantly restricted to prostate supports the hypothesis that PSCA may play an important role in normal prostate development and should be evaluated by creation of transgenic and knockout models.
PSCA maps to chromosome 8q24.2. Human SCA-2 (also called RIG-E) and another recently identified human Ly-6 homologue (i.e., E48) also localize to this region, suggesting that a large family of related genes may exist at this locus (18, 39). Intriguingly, chromosome 8q has been reported to be a region of allelic gain and amplification in a majority of advanced and recurrent prostate cancers (40). c-myc localizes proximal to PSCA at chromosome 8q24 and extra copies of c-myc (either through allelic gain or amplification) have been found in 68% of primary prostate tumors and 96% of metastases (41). Additional work will be necessary to see if PSCA overexpression is caused by amplification or chromosomal gain.
Acknowledgments
We thank Charles Sawyers and William Isaacs for advice and for critical reading of the manuscript and Jean B.deKernion for his support. This work was supported by grants from CaPCURE (R.E.R. and O.N.W.), the Jonsson Comprehensive Cancer Center (R.E.R.), National Institutes of Health Grant K08 CA74169 (R.E.R.), the STOP Cancer Foundation (R.E.R), and Public Health Service Grant CA40046 (M.M.L.). O.N.W. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- PSA
prostate-specific antigen
- PSMA
prostate-specific membrane antigen
- RDA
representational difference analysis
- RT-PCR
reverse transcriptase-coupled PCR
- SCA-2
stem cell antigen 2
- PLC
phospholipase C
- PSCA
prostate stem cell antigen
- GPI
glycosylphosphatidylinositol
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF043498).
References
- 1.Lalani E-N, Laniado M E, Abel P D. Cancer Metastasis Rev. 1997;16:29–66. doi: 10.1023/a:1005792206377. [DOI] [PubMed] [Google Scholar]
- 2.Klein K A, Reiter R E, Redula J, Moradi H, Zhu X L, Brothman A R, Lamb D J, Marcelli M, Belldegrun A, Witte O N, Sawyers C L. Nat Med. 1997;3:402–408. doi: 10.1038/nm0497-402. [DOI] [PubMed] [Google Scholar]
- 3.Darson M F, Pacelli A, Roche P, Rittenhouse H G, Wolfert R L, Young C Y, Klee G G, Tindall D J, Bostwick D G. Urology. 1997;49:857–862. doi: 10.1016/s0090-4295(97)00108-8. [DOI] [PubMed] [Google Scholar]
- 4.Liu H, Moy P, Kim S, Xia Y, Rajasekaran A, Navarro V, Knudsen B, Bander N H. Cancer Res. 1997;57:3629–3634. [PubMed] [Google Scholar]
- 5.Fair W R, Israeli R S, Heston W D. Prostate. 1997;32:140–148. doi: 10.1002/(sici)1097-0045(19970701)32:2<140::aid-pros9>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 6.Sadasvian R, Morgan R, Jennings S, Austenfield M, Van Veldhuizen P, Stephens R, Noble M. J Urol. 1993;150:126–131. doi: 10.1016/s0022-5347(17)35413-7. [DOI] [PubMed] [Google Scholar]
- 7.Lyne J C, Melhem M F, Finley G G, Wen D, Liu N, Deng D H, Salup R. Cancer J Sci Am. 1997;3:21–30. [PubMed] [Google Scholar]
- 8.Kuhn E J, Kurnot R A, Sesterhenn I A, Chang E H, Moul J W. J Urol. 1993;150:1427–1433. doi: 10.1016/s0022-5347(17)35799-3. [DOI] [PubMed] [Google Scholar]
- 9.Rabbani S A, Harakidas P, Davidson D J, Henkin J, Mazar A P. Int J Cancer. 1995;63:840–845. doi: 10.1002/ijc.2910630615. [DOI] [PubMed] [Google Scholar]
- 10.Crowley C W, Cohen R L, Lucas B K, Liu G, Shuman M A, Levinson A D. Proc Natl Acad Sci USA. 1993;90:5021–5025. doi: 10.1073/pnas.90.11.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pisters L L, Troncoso P, Zhau H E, Li W, von Eschenback A C, Chung L W. J Urol. 1995;154:293–298. [PubMed] [Google Scholar]
- 12.Hubank M, Schatz D G. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun B S, Frieden R, Lessnick S L, May W A, Denny C T. Mol Cell Biol. 1995;15:4623–4630. doi: 10.1128/mcb.15.8.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magi-Galluzzi C, Mishra R, Fiorentino M, Montironi R, Yao H, Capodieci P, Wishnow K, Kaplan I, Stork P J S, Loda M. Lab Invest. 1997;76:37–43. [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.Rowley J D, Diaz M O, Espinosa R, Patel Y D, van Melle E, Ziemin S, Taillon-Miller P, Lichter P, Evans G A, Kersey J D, et al. Proc Natl Acad Sci USA. 1990;87:9358–9362. doi: 10.1073/pnas.87.23.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 18.Mao M, Yu M, Tong J H, Ye J, Zhu J, Huang Q H, Fu G, Yu L, Zhao S Y, Waxman S, et al. Proc Natl Acad Sci USA. 1996;93:5910–5914. doi: 10.1073/pnas.93.12.5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Udenfriend S, Kodukula K. Annu Rev Biochem. 1995;64:563–591. doi: 10.1146/annurev.bi.64.070195.003023. [DOI] [PubMed] [Google Scholar]
- 20.Bonkhoff H, Stein U, Remberger K. Prostate. 1994;24:114–118. doi: 10.1002/pros.2990240303. [DOI] [PubMed] [Google Scholar]
- 21.Yang Y, Hao J, Liu X, Dalkin B, Nagle R B. Am J Pathol. 1997;150:693–703. [PMC free article] [PubMed] [Google Scholar]
- 22.Kiefer B, Driscoll P C, Campbell I D, Willis A C, van der Merwe P A, Davis S J. Biochemistry. 1994;33:4471–4482. [PubMed] [Google Scholar]
- 23.Fritz B A, Lowe A W. Am J Physiol. 1996;270:G176–G183. doi: 10.1152/ajpgi.1996.270.1.G176. [DOI] [PubMed] [Google Scholar]
- 24.Meri S, Lehto T, Sutton C W, Tyynela J, Baumann M. Biochem J. 1996;316:923–935. doi: 10.1042/bj3160923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deleersnijder W, Hong G, Cortvrindt R, Poirier C, Tylzanowski P, Pittois K, Van Marck E, Merregaert J. J Biol Chem. 1996;271:19475–19482. doi: 10.1074/jbc.271.32.19475. [DOI] [PubMed] [Google Scholar]
- 26.Sodee D B, Conant R, Chalfant M, Miron S, Klein E, Bahnson R, Spirnak J P, Carlin B, Bellon E M, Rogers B. Clin Nucl Med. 1996;21:759–767. doi: 10.1097/00003072-199610000-00002. [DOI] [PubMed] [Google Scholar]
- 27.Tjoa B A, Erickson S T, Bowes V A, Ragde H, Kenny G M, Cobb O E, Iretpon R C, Troychak M J, Boynton A L, Murphy G P. Prostate. 1997;32:272–278. doi: 10.1002/(sici)1097-0045(19970901)32:4<272::aid-pros7>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 28.Bao L, Loda M, Janmey P A, Stewart R, Anand-Apte B, Zetter B R. Nat Med. 1996;2:1322–1327. doi: 10.1038/nm1296-1322. [DOI] [PubMed] [Google Scholar]
- 29.Bonkhoff H, Remberger K. Prostate. 1996;28:98–106. doi: 10.1002/(SICI)1097-0045(199602)28:2<98::AID-PROS4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 30.Bonkhoff H. Eur Urol. 1996;30:201–205. doi: 10.1159/000474170. [DOI] [PubMed] [Google Scholar]
- 31.Verhagen A P M, Ramaekers F C S, Aalders T W, Schaafsma H E, Debruyne F M J, Schalken J A. Cancer Res. 1992;52:6182–6187. [PubMed] [Google Scholar]
- 32.Colombel M, Symmans F, Gil S, O’Toole K M, Chopin D, Benson M, Olsson C A, Korsmeyer S, Buttyan R. Am J Pathol. 1993;143:390–400. [PMC free article] [PubMed] [Google Scholar]
- 33.Magi-Galuzzi C, Xu X, Hlatky L, Hahnfeldt P, Kaplan I, Hsiao P-w, Chang C, Loda M. Modern Pathol. 1997;10:839. [PubMed] [Google Scholar]
- 34.Kawakami M, Nakayama J. Cancer Res. 1997;57:2321–2324. [PubMed] [Google Scholar]
- 35.Noda S, Kosugi A, Saitoh S, Narumiya S, Hamaoka T. J Exp Med. 1996;183:2355–2360. doi: 10.1084/jem.183.5.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas P M, Samuelson L E. J Biol Chem. 1992;267:12317–12322. [PubMed] [Google Scholar]
- 37.Bamezai A, Rock K L. Proc Natl Acad Sci USA. 1995;92:4294–4298. doi: 10.1073/pnas.92.10.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katz B Z, Eshel R, Sagi-Assif O, Witz I P. Int J Cancer. 1994;59:684–691. doi: 10.1002/ijc.2910590517. [DOI] [PubMed] [Google Scholar]
- 39.Brakenhoff R H, Gerretson M, Knippels E M, van Dijk M, van Essen H, Weghuis D O, Sinke R J, Snow G B, van Dongen G A. J Cell Biol. 1995;129:1677–1689. doi: 10.1083/jcb.129.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cher M L, MacGrogan D, Bookstein R, Brown J, Jenkins R B, Jensen R H. Genes Chromosomes Cancer. 1994;11:153–162. doi: 10.1002/gcc.2870110304. [DOI] [PubMed] [Google Scholar]
- 41.Jenkins R B, Qian J, Lieber M M, Bostwick D G. Cancer Res. 1997;57:524–531. [PubMed] [Google Scholar]