Abstract
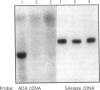
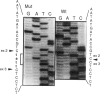
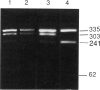
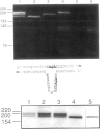
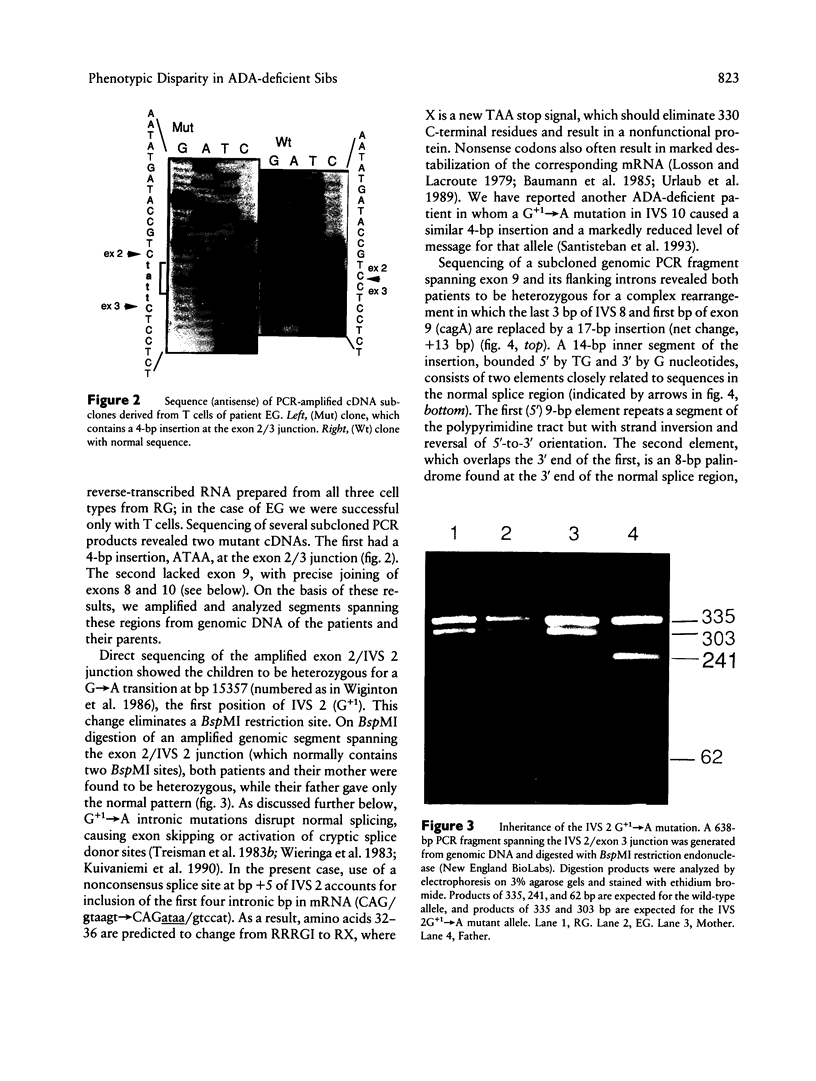
Adenosine deaminase (ADA) deficiency usually causes severe combined immune deficiency in infancy. Milder phenotypes, with delayed or late onset and gradual decline in immune function, also occur and are associated with less severely impaired deoxyadenosine (dAdo) catabolism. We have characterized the mutations responsible for ADA deficiency in siblings with striking disparity in clinical phenotype. Erythrocyte dAdo nucleotide pool size, which reflects total residual ADA activity, was lower in the older, more mildly affected sib (RG) than in her younger, more severely affected sister (EG). Cultured T cells, fibroblasts, and B lymphoblasts of RG had detectable residual ADA activity, while cells of EG did not. ADA mRNA was undetectable by northern analysis in these cells of both patients. Both sibs were found to be compound heterozygotes for the following novel splicing defects: (1) a G+1-->A substitution at the 5' splice site of IVS 2 and (2) a complex 17-bp rearrangement of the 3' splice site of IVS 8, which inserted a run of seven purines into the polypyrimidine tract and altered the reading frame of exon 9. PCR-amplified ADA cDNA clones with premature translation stop codons arising from aberrant pre-mRNA splicing were identified, which were consistent with these mutations. However, some cDNA clones from T cells of both patients and from fibroblasts and Epstein-Barr virus (EBV)-transformed B cells of RG, were normally spliced at both the exon 2/3 and exon 8/9 junctions. A normal coding sequence was documented for clones from both sibs. The normal cDNA clones did not appear to arise from either contamination or PCR artifact, and mosaicism seems unlikely to have been involved. These findings suggest (1) that a low level of normal pre-mRNA splicing may occur despite mutation of the invariant first nucleotide of the 5' splice donor sequence and (2) that differences in efficiency of such splicing may account for the difference in residual ADA activity, immune dysfunction, and clinical severity in these siblings.
Full text
PDF


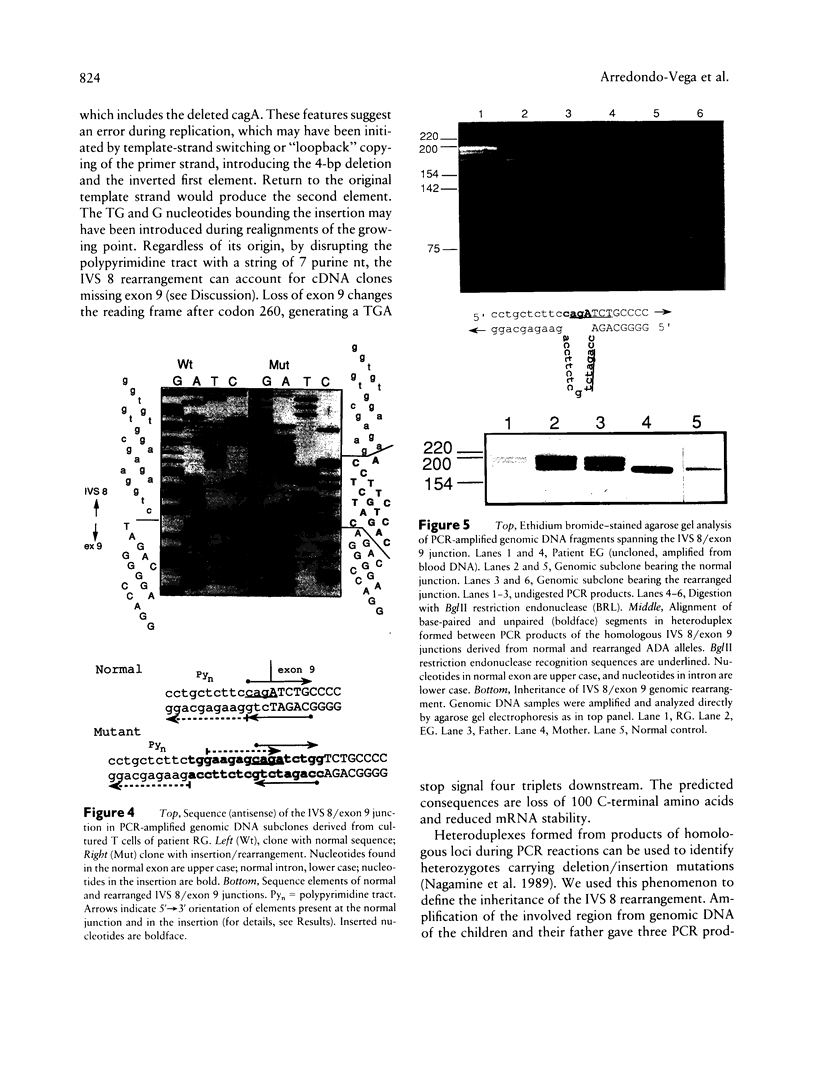






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Aebi M., Hornig H., Weissmann C. 5' cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5' splice region, not by the conserved 5' GU. Cell. 1987 Jul 17;50(2):237–246. doi: 10.1016/0092-8674(87)90219-4. [DOI] [PubMed] [Google Scholar]
- Akeson A. L., Wiginton D. A., Hutton J. J. Normal and mutant human adenosine deaminase genes. J Cell Biochem. 1989 Mar;39(3):217–228. doi: 10.1002/jcb.240390302. [DOI] [PubMed] [Google Scholar]
- Akeson A. L., Wiginton D. A., States J. C., Perme C. M., Dusing M. R., Hutton J. J. Mutations in the human adenosine deaminase gene that affect protein structure and RNA splicing. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5947–5951. doi: 10.1073/pnas.84.16.5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arredondo-Vega F. X., Kurtzberg J., Chaffee S., Santisteban I., Reisner E., Povey M. S., Hershfield M. S. Paradoxical expression of adenosine deaminase in T cells cultured from a patient with adenosine deaminase deficiency and combine immunodeficiency. J Clin Invest. 1990 Aug;86(2):444–452. doi: 10.1172/JCI114730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann B., Potash M. J., Köhler G. Consequences of frameshift mutations at the immunoglobulin heavy chain locus of the mouse. EMBO J. 1985 Feb;4(2):351–359. doi: 10.1002/j.1460-2075.1985.tb03636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonthron D. T., Markham A. F., Ginsburg D., Orkin S. H. Identification of a point mutation in the adenosine deaminase gene responsible for immunodeficiency. J Clin Invest. 1985 Aug;76(2):894–897. doi: 10.1172/JCI112050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Cole W. G., Chiodo A. A., Lamande S. R., Janeczko R., Ramirez F., Dahl H. H., Chan D., Bateman J. F. A base substitution at a splice site in the COL3A1 gene causes exon skipping and generates abnormal type III procollagen in a patient with Ehlers-Danlos syndrome type IV. J Biol Chem. 1990 Oct 5;265(28):17070–17077. [PubMed] [Google Scholar]
- Coulter-Karis D. E., Hershfield M. S. Sequence of full length cDNA for human S-adenosylhomocysteine hydrolase. Ann Hum Genet. 1989 May;53(Pt 2):169–175. doi: 10.1111/j.1469-1809.1989.tb01781.x. [DOI] [PubMed] [Google Scholar]
- Dominski Z., Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J. A., Kozakewich H. P., Pavelka K., Grier H. E., Shamberger R. C., Korf B., Morton C. C. Consistent cytogenetic aberrations in hepatoblastoma: a common pathway of genetic alterations in embryonal liver and skeletal muscle malignancies? Genes Chromosomes Cancer. 1991 Jan;3(1):37–43. doi: 10.1002/gcc.2870030107. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Geffner M. E., Stiehm E. R., Stephure D., Cowan M. J. Probable autoimmune thyroid disease and combined immunodeficiency disease. Am J Dis Child. 1986 Nov;140(11):1194–1196. doi: 10.1001/archpedi.1986.02140250120047. [DOI] [PubMed] [Google Scholar]
- Giblett E. R., Anderson J. E., Cohen F., Pollara B., Meuwissen H. J. Adenosine-deaminase deficiency in two patients with severely impaired cellular immunity. Lancet. 1972 Nov 18;2(7786):1067–1069. doi: 10.1016/s0140-6736(72)92345-8. [DOI] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Kawamoto H., Ito K., Kashii S., Monden S., Fujita M., Norioka M., Sasai Y., Okuma M. A point mutation in the 5' splice region of intron 7 causes a deletion of exon 7 in adenosine deaminase mRNA. J Cell Biochem. 1993 Mar;51(3):322–325. doi: 10.1002/jcb.240510311. [DOI] [PubMed] [Google Scholar]
- Kishimoto T. K., O'Conner K., Springer T. A. Leukocyte adhesion deficiency. Aberrant splicing of a conserved integrin sequence causes a moderate deficiency phenotype. J Biol Chem. 1989 Feb 25;264(6):3588–3595. [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T., Ruskin B., Green M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984 Apr;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Krawczak M., Reiss J., Cooper D. N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992 Sep-Oct;90(1-2):41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- Kuivaniemi H., Kontusaari S., Tromp G., Zhao M. J., Sabol C., Prockop D. J. Identical G+1 to A mutations in three different introns of the type III procollagen gene (COL3A1) produce different patterns of RNA splicing in three variants of Ehlers-Danlos syndrome. IV. An explanation for exon skipping some mutations and not others. J Biol Chem. 1990 Jul 15;265(20):12067–12074. [PubMed] [Google Scholar]
- Lamond A. I., Konarska M. M., Sharp P. A. A mutational analysis of spliceosome assembly: evidence for splice site collaboration during spliceosome formation. Genes Dev. 1987 Aug;1(6):532–543. doi: 10.1101/gad.1.6.532. [DOI] [PubMed] [Google Scholar]
- Levy Y., Hershfield M. S., Fernandez-Mejia C., Polmar S. H., Scudiery D., Berger M., Sorensen R. U. Adenosine deaminase deficiency with late onset of recurrent infections: response to treatment with polyethylene glycol-modified adenosine deaminase. J Pediatr. 1988 Aug;113(2):312–317. doi: 10.1016/s0022-3476(88)80271-3. [DOI] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markert M. L., Hershfield M. S., Schiff R. I., Buckley R. H. Adenosine deaminase and purine nucleoside phosphorylase deficiencies: evaluation of therapeutic interventions in eight patients. J Clin Immunol. 1987 Sep;7(5):389–399. doi: 10.1007/BF00917017. [DOI] [PubMed] [Google Scholar]
- Markert M. L., Hershfield M. S., Wiginton D. A., States J. C., Ward F. E., Bigner S. H., Buckley R. H., Kaufman R. E., Hutton J. J. Identification of a deletion in the adenosine deaminase gene in a child with severe combined immunodeficiency. J Immunol. 1987 May 15;138(10):3203–3206. [PubMed] [Google Scholar]
- Marton A., Delbecchi L., Bourgaux P. DNA nicking favors PCR recombination. Nucleic Acids Res. 1991 May 11;19(9):2423–2426. doi: 10.1093/nar/19.9.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes B., Potier M., Wakamatsu N., Melancon S. B., Klavins M. H., Tsuji S., Mahuran D. J. An unusual splicing mutation in the HEXB gene is associated with dramatically different phenotypes in patients from different racial backgrounds. J Clin Invest. 1992 Aug;90(2):306–314. doi: 10.1172/JCI115863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhans A., Vartanian J. P., Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990 Apr 11;18(7):1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan G., Levinsky R. J., Hugh-Jones K., Fairbanks L. D., Morris G. S., Simmonds H. A. Heterogeneity of biochemical, clinical and immunological parameters in severe combined immunodeficiency due to adenosine deaminase deficiency. Clin Exp Immunol. 1987 Dec;70(3):491–499. [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine C. M., Chan K., Lau Y. F. A PCR artifact: generation of heteroduplexes. Am J Hum Genet. 1989 Aug;45(2):337–339. [PMC free article] [PubMed] [Google Scholar]
- Newman A. J., Lin R. J., Cheng S. C., Abelson J. Molecular consequences of specific intron mutations on yeast mRNA splicing in vivo and in vitro. Cell. 1985 Aug;42(1):335–344. doi: 10.1016/s0092-8674(85)80129-x. [DOI] [PubMed] [Google Scholar]
- Ohshima Y., Gotoh Y. Signals for the selection of a splice site in pre-mRNA. Computer analysis of splice junction sequences and like sequences. J Mol Biol. 1987 May 20;195(2):247–259. doi: 10.1016/0022-2836(87)90647-4. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Parker R., Guthrie C. A point mutation in the conserved hexanucleotide at a yeast 5' splice junction uncouples recognition, cleavage, and ligation. Cell. 1985 May;41(1):107–118. doi: 10.1016/0092-8674(85)90065-0. [DOI] [PubMed] [Google Scholar]
- Peebles C. L., Belcher S. M., Zhang M., Dietrich R. C., Perlman P. S. Mutation of the conserved first nucleotide of a group II intron from yeast mitochondrial DNA reduces the rate but allows accurate splicing. J Biol Chem. 1993 Jun 5;268(16):11929–11938. [PubMed] [Google Scholar]
- Reed R., Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985 May;41(1):95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Reed R. The organization of 3' splice-site sequences in mammalian introns. Genes Dev. 1989 Dec;3(12B):2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- Roscigno R. F., Weiner M., Garcia-Blanco M. A. A mutational analysis of the polypyrimidine tract of introns. Effects of sequence differences in pyrimidine tracts on splicing. J Biol Chem. 1993 May 25;268(15):11222–11229. [PubMed] [Google Scholar]
- Rubinstein A., Hirschhorn R., Sicklick M., Murphy R. A. In vivo and in vitro effects of thymosin and adenosine deaminase on adenosine-deaminase-deficient lymphocytes. N Engl J Med. 1979 Feb 22;300(8):387–392. doi: 10.1056/NEJM197902223000802. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Role of the 3' splice site consensus sequence in mammalian pre-mRNA splicing. Nature. 1985 Oct 24;317(6039):732–734. doi: 10.1038/317732a0. [DOI] [PubMed] [Google Scholar]
- Santisteban I., Arredondo-Vega F. X., Kelly S., Mary A., Fischer A., Hummell D. S., Lawton A., Sorensen R. U., Stiehm E. R., Uribe L. Novel splicing, missense, and deletion mutations in seven adenosine deaminase-deficient patients with late/delayed onset of combined immunodeficiency disease. Contribution of genotype to phenotype. J Clin Invest. 1993 Nov;92(5):2291–2302. doi: 10.1172/JCI116833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. On the origin of RNA splicing and introns. Cell. 1985 Sep;42(2):397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- Shovlin C. L., Hughes J. M., Simmonds H. A., Fairbanks L., Deacock S., Lechler R., Roberts I., Webster A. D. Adult presentation of adenosine deaminase deficiency. Lancet. 1993 Jun 5;341(8858):1471–1471. doi: 10.1016/0140-6736(93)90910-9. [DOI] [PubMed] [Google Scholar]
- Soukup S. W., Lampkin B. L. Trisomy 2 and 20 in two hepatoblastomas. Genes Chromosomes Cancer. 1991 May;3(3):231–234. doi: 10.1002/gcc.2870030310. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Kretzner L., Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988 Aug;7(8):2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Rosbash M. Exon mutations uncouple 5' splice site selection from U1 snRNA pairing. Cell. 1990 Nov 2;63(3):619–629. doi: 10.1016/0092-8674(90)90457-p. [DOI] [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Treisman R., Proudfoot N. J., Shander M., Maniatis T. A single-base change at a splice site in a beta 0-thalassemic gene causes abnormal RNA splicing. Cell. 1982 Jul;29(3):903–911. doi: 10.1016/0092-8674(82)90452-4. [DOI] [PubMed] [Google Scholar]
- Umetsu D. T., Schlossman C. M., Ochs H. D., Hershfield M. S. Heterogeneity of phenotype in two siblings with adenosine deaminase deficiency. J Allergy Clin Immunol. 1994 Feb;93(2):543–550. doi: 10.1016/0091-6749(94)90365-4. [DOI] [PubMed] [Google Scholar]
- Urlaub G., Mitchell P. J., Ciudad C. J., Chasin L. A. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989 Jul;9(7):2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Hofer E., Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984 Jul;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Wiginton D. A., Adrian G. S., Friedman R. L., Suttle D. P., Hutton J. J. Cloning of cDNA sequences of human adenosine deaminase. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7481–7485. doi: 10.1073/pnas.80.24.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiginton D. A., Adrian G. S., Hutton J. J. Sequence of human adenosine deaminase cDNA including the coding region and a small intron. Nucleic Acids Res. 1984 Mar 12;12(5):2439–2446. doi: 10.1093/nar/12.5.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiginton D. A., Kaplan D. J., States J. C., Akeson A. L., Perme C. M., Bilyk I. J., Vaughn A. J., Lattier D. L., Hutton J. J. Complete sequence and structure of the gene for human adenosine deaminase. Biochemistry. 1986 Dec 16;25(25):8234–8244. doi: 10.1021/bi00373a017. [DOI] [PubMed] [Google Scholar]