Abstract
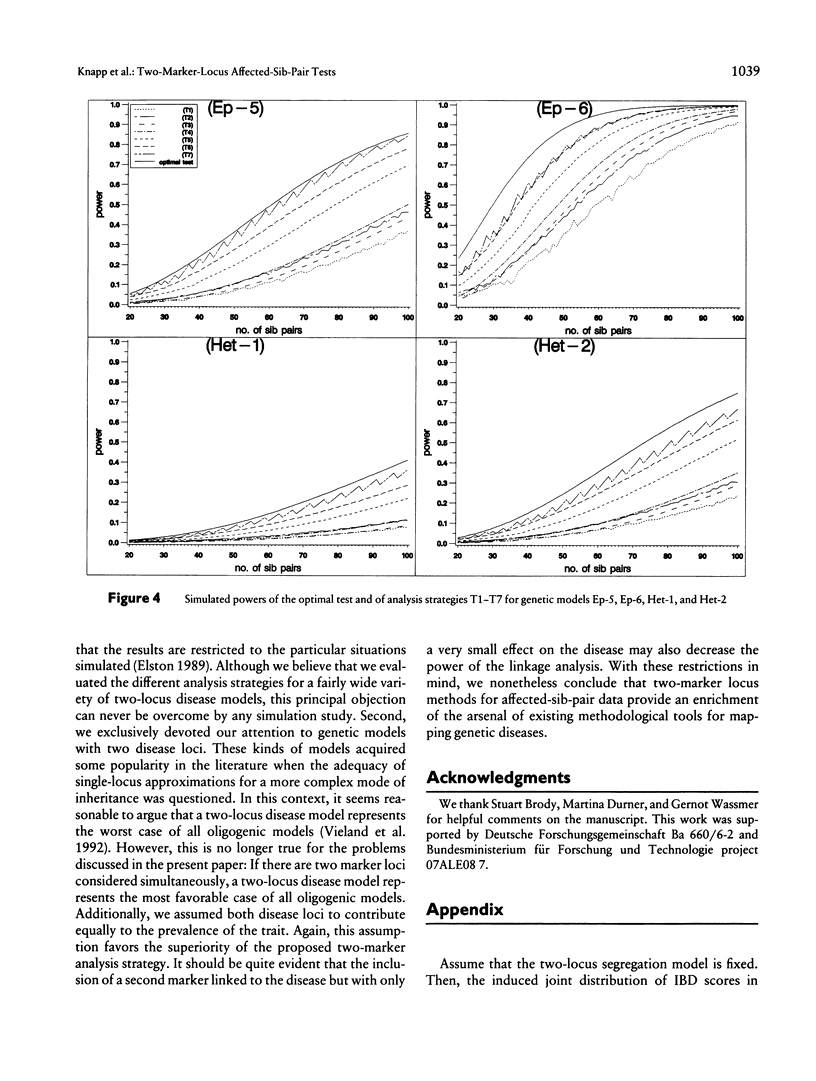
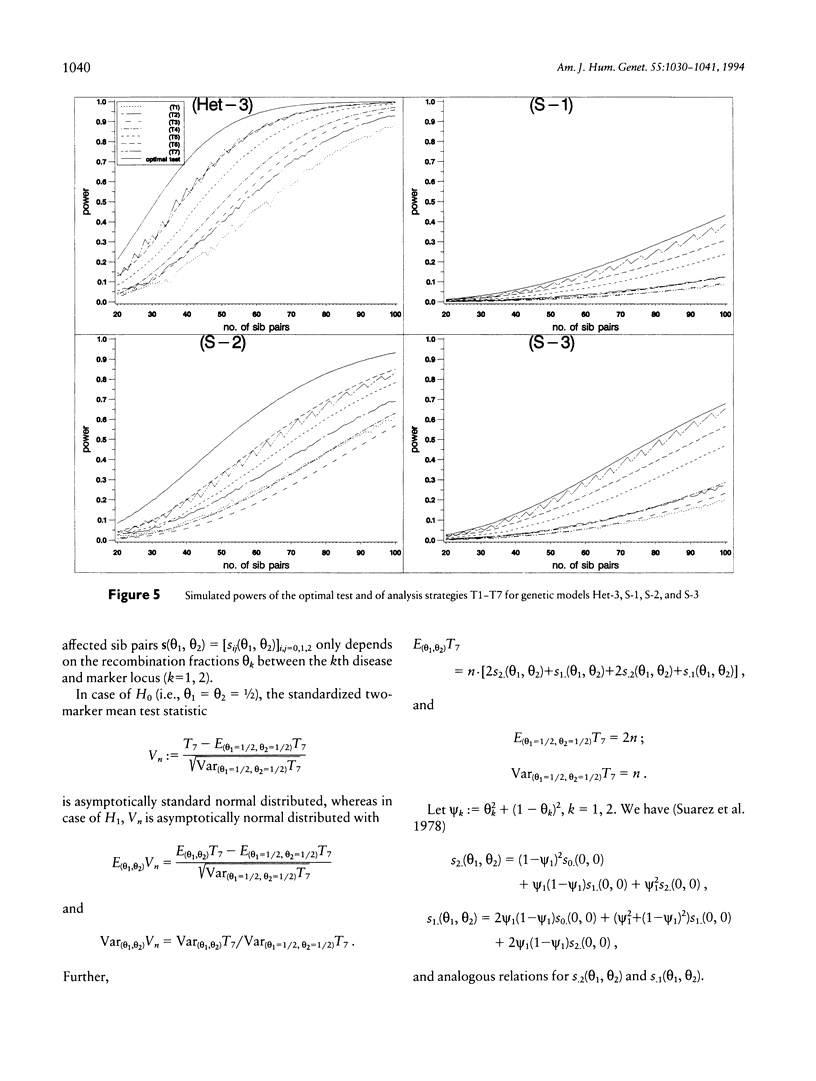
Recently, Schork et al. found that two-trait-locus, two-marker-locus (parametric) linkage analysis can provide substantially more linkage information than can standard one-trait-locus, one-marker-locus methods. However, because of the increased burden of computation, Schork et al. do not expect that their approach will be applied in an initial genome scan. Further, the specification of a suitable two-locus segregation model can be crucial. Affected-sibpair tests are computationally simple and do not require an explicit specification of the disease model. In the past, however, these tests mainly have been applied to data with a single marker locus. Here, we consider sib-pair tests that make it possible to analyze simultaneously two marker loci. The power of these tests is investigated for different (epistatic and heterogeneous) two-trait-locus models, each trait locus being linked to one of the marker loci. We compare these tests both with the test that is optimal for a certain model and with the strategy that analyzes each marker locus separately. The results indicate that a straightforward extension of the well-known mean test for two marker loci can be much more powerful than single-marker-locus analysis and that is power is only slightly inferior to the power of the optimal test.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Babron M. C., Martinez M., Bonaïti-Pellié C., Clerget-Darpoux F. Linkage detection by the Affected-Pedigree-Member method: what is really tested? Genet Epidemiol. 1993;10(6):389–394. doi: 10.1002/gepi.1370100610. [DOI] [PubMed] [Google Scholar]
- Dizier M. H., Clerget-Darpoux F. Two-disease locus model: sib pair method using information on both HLA and Gm. Genet Epidemiol. 1986;3(5):343–356. doi: 10.1002/gepi.1370030507. [DOI] [PubMed] [Google Scholar]
- Durner M., Greenberg D. A., Hodge S. E. Inter- and intrafamilial heterogeneity: effective sampling strategies and comparison of analysis methods. Am J Hum Genet. 1992 Oct;51(4):859–870. [PMC free article] [PubMed] [Google Scholar]
- Elston R. C. Man bites dog? The validity of maximizing lod scores to determine mode of inheritance. Am J Med Genet. 1989 Dec;34(4):487–488. doi: 10.1002/ajmg.1320340407. [DOI] [PubMed] [Google Scholar]
- Faraway J. J. Improved sib-pair linkage test for disease susceptibility loci. Genet Epidemiol. 1993;10(4):225–233. doi: 10.1002/gepi.1370100403. [DOI] [PubMed] [Google Scholar]
- Goldin L. R. Detection of linkage under heterogeneity: comparison of the two-locus vs. admixture models. Genet Epidemiol. 1992;9(1):61–66. doi: 10.1002/gepi.1370090107. [DOI] [PubMed] [Google Scholar]
- Hodge S. E. Some epistatic two-locus models of disease. I. Relative risks and identity-by-descent distributions in affected sib pairs. Am J Hum Genet. 1981 May;33(3):381–395. [PMC free article] [PubMed] [Google Scholar]
- Holmans P. Asymptotic properties of affected-sib-pair linkage analysis. Am J Hum Genet. 1993 Feb;52(2):362–374. [PMC free article] [PubMed] [Google Scholar]
- Knapp M. A powerful test of sib-pair linkage for disease susceptibility. Genet Epidemiol. 1991;8(2):141–143. doi: 10.1002/gepi.1370080207. [DOI] [PubMed] [Google Scholar]
- Knapp M., Seuchter S. A., Baur M. P. Linkage analysis in nuclear families. 1: Optimality criteria for affected sib-pair tests. Hum Hered. 1994 Jan-Feb;44(1):37–43. doi: 10.1159/000154187. [DOI] [PubMed] [Google Scholar]
- Neuman R. J., Rice J. P. Two-locus models of disease. Genet Epidemiol. 1992;9(5):347–365. doi: 10.1002/gepi.1370090506. [DOI] [PubMed] [Google Scholar]
- Risch N. Linkage strategies for genetically complex traits. III. The effect of marker polymorphism on analysis of affected relative pairs. Am J Hum Genet. 1990 Feb;46(2):242–253. [PMC free article] [PubMed] [Google Scholar]
- Schaid D. J., Nick T. G. Sib-pair linkage tests for disease susceptibility loci: common tests vs. the asymptotically most powerful test. Genet Epidemiol. 1990;7(5):359–370. doi: 10.1002/gepi.1370070506. [DOI] [PubMed] [Google Scholar]
- Schork N. J., Boehnke M., Terwilliger J. D., Ott J. Two-trait-locus linkage analysis: a powerful strategy for mapping complex genetic traits. Am J Hum Genet. 1993 Nov;53(5):1127–1136. [PMC free article] [PubMed] [Google Scholar]
- Suarez B. K., Rice J., Reich T. The generalized sib pair IBD distribution: its use in the detection of linkage. Ann Hum Genet. 1978 Jul;42(1):87–94. doi: 10.1111/j.1469-1809.1978.tb00933.x. [DOI] [PubMed] [Google Scholar]
- Vieland V. J., Greenberg D. A., Hodge S. E. Adequacy of single-locus approximations for linkage analyses of oligogenic traits: extension to multigenerational pedigree structures. Hum Hered. 1993 Nov-Dec;43(6):329–336. doi: 10.1159/000154155. [DOI] [PubMed] [Google Scholar]
- Vieland V. J., Hodge S. E., Greenberg D. A. Adequacy of single-locus approximations for linkage analyses of oligogenic traits. Genet Epidemiol. 1992;9(1):45–59. doi: 10.1002/gepi.1370090106. [DOI] [PubMed] [Google Scholar]



