Abstract
Human T cell leukemia virus I (HTLV-I) causes acute leukemic disease in a low percentage of infected individuals through obscure mechanisms. Our studies compare two rabbit HTLV-I-infected T cell lines: one, RH/K34, causes lethal experimental leukemia and the other, RH/K30, mediates asymptomatic infection. We show herein that the product of the protooncogene vav is constitutively Tyr-phosphorylated in RH/K34 but not in RH/K30. A role for the retrovirus in phosphorylation of Vav was assigned by transfection experiments with molecular clones of HTLV-I derived from the two lines. The HTLV-I molecular clone from RH/K30, but not that from RH/K34, down-regulates Vav phosphorylation in a Herpesvirus ateles-transformed T cell line. Use of recombinant virus clones revealed that a pX region sequence differing by two nucleotides between the two clones mediates this down-regulation. Because Vav is involved in T cell signaling and Vav phosphorylation occurs upon activation of T cells, control of the activation state of Vav by viral proteins may relate to the leukemogenic potential of certain HTLV-I-infected cells.
The retrovirus human T cell leukemia virus (HTLV-I) infects 10–20 million persons worldwide. Of those infected, the majority remain asymptomatic, whereas a small percentage develop acutely fatal adult T cell leukemia/lymphoma (ATLL), chronic cutaneous leukemia, the neurologic disorder HTLV-I-associated myelopathy/tropical spastic paraparesis, or one of several chronic diseases that have been linked to HTLV-I (1, 2). Mechanisms by which infection with this retrovirus may result in such diverse outcomes remain obscure. HTLV-I isolates vary little in primary structure and no associations between viral sequence and outcome of infection are evident.
HTLV-I readily infects certain animals, including monkeys, rats, and rabbits, and in some specific instances, infection leads to disease in experimental animals (3). Although the majority of rabbit cell lines derived by infection with HTLV-I human lines give rise to chronic asymptomatic infection, administration of certain HTLV-I-infected T cell lines leads to acutely fatal or smoldering disease that mimics human ATLL (4–6). Differences between such lines and those mediating asymptomatic infection can provide clues concerning the variable pathogenicity of HTLV-I. RH/K30 and RH/K34 are two HTLV-I-transformed T cell lines; RH/K30 mediates asymptomatic infection and RH/K34 causes a fatal leukemia-like disease accompanied by thymic depletion via apoptosis (6, 7). Despite the overwhelming differences in their in vivo behavior, these two γδ T cell lines have only minor differences in expression of surface markers (8). Although the integrated proviruses differ by only 18 nucleotides of their 9-kb sequence (9), molecular clones of the proviruses from these lines show differences in their ability to give rise to chronic infection both in vivo and in vitro and in levels of virus production after their use to transfect susceptible cells (9). Because the virus clones alone do not have the ability to cause disease in animals, it was reasoned that the properties of the HTLV-I-infected cells are critical to the pathogenic processes. Therefore, the RH/K30 and RH/K34 cell lines were examined for clues to their different in vivo activities.
Recent reports of constitutive phosphorylation of molecules involved in signaling pathways in HTLV-I lines (10, 11) prompted consideration of protein phosphorylation patterns of RH/K30 and RH/K34. Because both lines express high levels of cell surface CD25 and are interleukin 2 (IL-2)-independent (8), significant differences in their IL-2-mediated signaling pathways were not predicted. However, the role of protein phosphorylation is not limited to IL-2-mediated signaling but is also involved in normal cell proliferation, differentiation, and oncogenic transformation (12, 13). Investigations of phosphorylated proteins indicated a significant difference between the two cell lines in that the product of the protooncogene vav is Tyr-phosphorylated in RH/K34 but not in RH/K30.
The Vav protein, which was first identified by its transforming activity (14), is expressed only in hematopoietic cells and trophoblasts (14, 15). Vav plays a role in lymphoid cell maturation (16, 17). Involvement of Vav in signal transduction is implied by the fact that Tyr phosphorylation of Vav is induced by engagement of T cell, B cell, or Fc receptors (18–20); by interaction of various cytokine and other cell surface receptors with their ligands (21–23); and by the impairment of lymphocyte signaling in Vav-deficient mice (24–26). The Vav oncoprotein contains modular domains involved in protein–protein and possibly protein–lipid and protein–DNA interactions. Vav homology domains include pleckstrin PH, src SH2, SH3, and calponin CH (27–30). Furthermore, Vav contains a motif characteristic of guanidine nucleotide releasing factors (14, 31, 32), two Pro-rich domains, a Cys-rich region homologous to the diacylglycerol binding site of protein kinase C (16), and a Leu region similar to a helix–loop–helix (33). Vav is predicted to play an important role in linking Tyr signaling from activated receptors to the nucleus by virtue of interactions with other molecules involved in signal transduction via SH2, SH3, and other domains (34).
The present data indicate that the Vav protein is expressed in both HTLV-I lines studied but is constitutively Tyr-phosphorylated only in the RH/K34 cell line, which causes a fatal leukemia-like disease. A role for virus in this difference is shown herein by the fact that an HTLV-I molecular clone derived from the RH/K30 cell line, which mediates asymptomatic infection, down-regulates Vav phosphorylation in a Herpesvirus ateles-transformed T cell line after transfection. Although the virus cloned from RH/K34 causes transient infection of the cell line, it has little or no effect on the phosphorylation state of Vav. This difference in activity of the two viruses was localized to the HTLV-I pX region sequence, which differs between the two clones by 2 nucleotides. Viral regulation of the phosphorylation state of Vav may subvert normal T cell functions and has possible implication in HTLV-I leukemogenic potential.
MATERIALS AND METHODS
Cell Lines, HTLV-I Clone Construction, and Cell Transfection.
RH/K30 and RH/K34 are two rabbit γδ T cell lines transformed in vitro directly or indirectly by HTLV-I virus from an irradiated human MT-2 T cell line (8). MT-2 cell is a human T cell line transformed by HTLV-I (35). RL-5 is a rabbit αβ T cell line transformed in vivo with Herpesvirus ateles (36).
K30p and K34p DNA molecular clones were constructed from RH/K30 and RH/K34 cells, respectively, by DNA digestion with EcoRI and cloned into bacteriophage λEMBL4 and subsequently into plasmid vector pSv2-gpt. Chimeric HTLV-I clones were constructed by digestion of K30p or K34p clones with combinations of restriction enzymes (NotI, SalI, and MluI) and then shuffling the segments between K30p and K34p (37).
RL-5 was transfected by the HTLV-I DNA molecular clones from RH/K30 or RH/K34 or by the chimeric clones by using DEAE-dextran as described (37, 38). Cells were maintained in culture in RPMI 1640 complete medium containing 10% fetal calf sera (Intergen, Walkersville, MD), 2 mM l-glutamine, penicillin (100 units/ml), and streptomycin (100 μg/ml; BioWhittaker).
Antibodies and Chemicals.
Anti-Vav antibodies, the peptide used for its preparation, and the anti-phosphotyrosine [Tyr(P)] monoclonal antibody (4G10) were obtained from Upstate Biotechnology. Glutathione S-transferase (GST) molecules containing SH2 and SH3 domains were from Santa Cruz Biotechnology. Goat anti-rabbit and anti-mouse horseradish peroxidase-conjugated antibodies were from Bio-Rad. Protein A-Sepharose, Triton X-100, phenylmethylsulfonyl fluoride, 7-amino-1-chloro-3-tosylamido-2-heptanone, iodoacetamide, aprotinin, and leupeptin were purchased from Sigma. Louis, MO). Immobilon P membrane was from Millipore, and the enhanced chemiluminescence substrate was from Amersham.
Immunoprecipitation and Western Blotting.
Cell lines (107 cells per ml) were washed in PBS and treated with lysis buffer (10 mM Tris⋅HCl, pH 7.5/150 mM NaCl/1% Triton X-100/0.02% NaN3/1 mM phenylmethylsulfonyl fluoride/10 mM 7-amino-1-chloro-3-tosylamido-2-heptanone/5 mM iodoacetamide/2 mM aprotinin/2 mM leupeptin). For Vav immunoprecipitation, samples were precleared with normal rabbit serum and protein A-Sepharose and incubated overnight with either normal rabbit IgG or anti-Vav antibodies, then the complex was precipitated with protein A-Sepharose for 1 hr, and washed three times with lysis buffer and once with 10 mM Tris⋅HCl buffer. Before separation, samples were mixed with 100 μl of Laemmli sample buffer and heated at 100°C for 3 min, and 10-μl aliquots (equivalent to 106 cells) were separated by SDS/PAGE and then transferred to an Immobilon P membrane. Blots were developed with anti-Vav or anti-Tyr(P) 4G10 antibodies and with peroxidase-labeled goat anti-Ig antibodies, by using the enhanced chemiluminescence substrate. For Tyr(P) immunoprecipitations, samples were precleared with normal mice serum and reacted with agarose-conjugated anti-Tyr(P) 4G10 antibody; washing and subsequent steps were as described above.
For precipitations with GST containing SH2 and SH3 domains, cell lysates (107 cells) were first precleared with GST-glutathione-agarose conjugate and then precipitated with agarose-immobilized GST containing different SH2 (Grb2, Ras-Gap, PLCg1, SH-PTP2, LCK, Fyn, and PI3) and SH3 (Grb2 N and C, Ras-Gap, Lck, Fyn, and PLCg1) domains. Samples were washed as described and separated by SDS/PAGE (10% gel). Separated proteins were transferred onto an Immobilon P membrane and developed with rabbit anti-Vav antibody, followed by peroxidase-labeled goat anti-rabbit Ig.
Developed films were scanned by using the PDSI transmission densitometer (Molecular Dynamics).
RESULTS
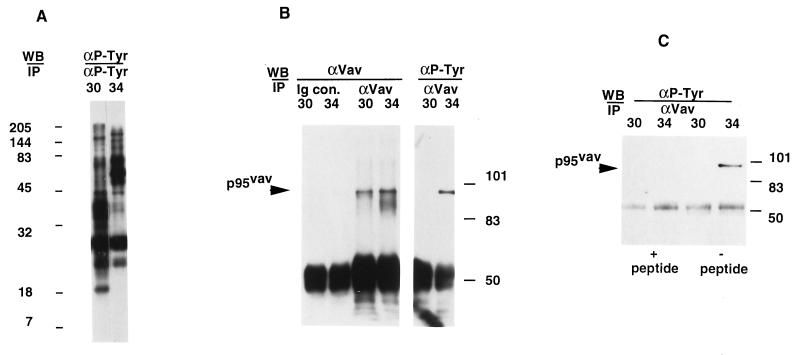
The pattern of Tyr phosphorylation of proteins was investigated in two HTLV-I-infected cell lines, RH/K30 and RH/K34, that exhibit differences in their ability to mediate leukemia-like disease. Lysates of the two cell lines were precipitated with anti-Tyr(P) and analyzed by immunoblots for overall differences in proteins containing Tyr(P). Differences in the distribution of high and low molecular weight proteins between the cells was observed (Fig. 1A). One of the differentially phosphorylated molecules was identified as the product of the protooncogene vav by immunoprecipitation with anti-Vav antibody. Despite the fact that the two cell lines express similar levels of Vav, this molecule was Tyr-phosphorylated only in the RH/K34 cell line (Fig. 1B). Precipitation with anti-Vav removed all detectable phosphorylated Vav from lysates (data not shown), and the specificity of the reaction with anti-Vav was confirmed by depletion of reactivity with lysates from both cells in the presence of the peptide used to produce the antibody (Fig. 1C). These data demonstrate that Vav is present in both cell lines, but Vav is phosphorylated only in RH/K34.
Figure 1.
Analysis of Tyr phosphorylation profiles of HTLV-I-transformed cell lines RH/K30 and RH/K34 and identification of phosphorylated Vav in RH/K34. (A) Rabbit HTLV-I-transformed T cell lines RH/K30 (3) and RH/K34 (9) (107 cells) were washed in PBS and treated with lysis buffer. Lysates were precleared with normal mouse serum and immunoprecipitated with agarose conjugated anti-Tyr(P) 4G10 antibody. Samples were mixed with 100 μl of Laemmli sample buffer, 10-μl aliquots were separated by SDS/PAGE on a 14% gel and transferred onto Immobilon P membrane, and blots were developed with peroxidase-labeled 4G10 antibody, using the enhanced chemiluminescence substrate. Numbers to the left of the blot indicate migration of molecular size standards from a Kaleidoscope (Bio-Rad) standard. (B) RH/K30 and RH/K34 cells (107 cells) were washed then lysed, precleared with normal rabbit serum, immunoprecipitated with normal rabbit Ig or with rabbit anti-Vav antibody, separated by SDS/PAGE (8% gel), and transferred, and blots were developed with anti-Vav antibody or with anti-Tyr(P) 4G10 antibody and corresponding peroxidase-labeled antibody. (C) Precleared RH/K30 and RH/K34 cell lysates (107 cells) were immunoprecipitated with anti-Vav antibody in presence or in absence of specific peptide (20 μg/ml) corresponding to amino acid residues 523–547 of Vav, separated by SDS/PAGE (10% gel), transferred, and blotted with 4G10 antibody.
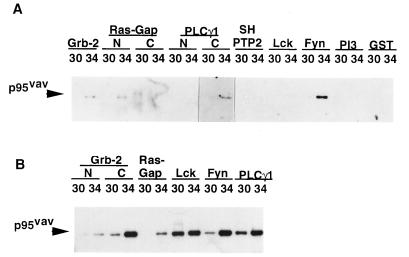
Vav is predicted to play an important role in linking Tyr signaling from activated receptors to the nucleus by virtue of interactions with other molecules involved in signal transduction via SH2, SH3, and other domains (34). To test the ability of Vav from RH/K30 and RH/K34 to associate with other signaling molecules, we analyzed in vitro the interaction of cell lysates with immobilized GST molecules containing SH2 domains from Grb2, Ras-Gap, PLCg1, SH-PTP2, LCK, Fyn, and PI3 and containing SH3 domains from Grb2, Ras-Gap, PLCg1, LCK, and Fyn. As expected, associations were observed between the phosphorylated Vav in RH/K34 and certain SH2 domains including Grb2, N-terminal (but not C-terminal) Ras-Gap, and the PLCg1 C-terminal (but not N-terminal) SH2; in addition, strong reactivity with the Fyn SH2 domain was observed (Fig. 2A). Use of GST molecules with SH3 domains, which interact with Pro-rich domains, revealed quantitative differences between the amounts of protein precipitated with different SH3 domains. In most cases, greater amounts of RH/K34 Vav were precipitated by the SH3 domains than by RH/K30 Vav (Fig. 2B). In addition, it was observed that interaction with the Ras-Gap SH3 domain occurred only with Tyr-phosphorylated Vav from the cell line RH/K34.
Figure 2.
In vitro association of Vav with SH2 and SH3 domains from different molecules involved in signal transduction. (A) RH/K30 and RH/K34 cell lysates (107 cells) were precleared with GST-glutathione-agarose conjugate, then precipitated with agarose-immobilized GST containing different SH2 domains corresponding to Grb2, Ras-Gap, PLCg1, SH-PTP2, LCK, Fyn, and PI3, separated by SDS/PAGE (10% gel), transferred onto an Immobilon P membrane, and developed with rabbit anti-Vav antibody and peroxidase-labeled goat anti-rabbit Ig. (B) Precleared cell lysates were precipitated with agarose-immobilized GST containing different SH3 domains corresponding to Grb2, N and C Ras-Gap, Lck, Fyn, and PLCg1 and processed as in A.
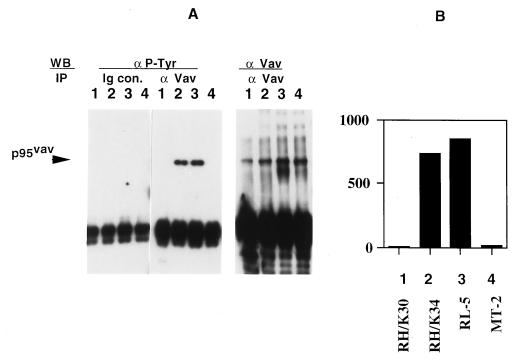
Demonstration of these differences in the state of Vav phosphorylation and Vav reactivity between RH/K30 and RH/K34 raises the question of whether this difference is controlled by variations in the HTLV-I genomes present in the two cell lines. It remains possible that the protein phosphorylation patterns were an inherent property of the cells that was retained upon infection and transformation with HTLV-I and was not influenced by differences in the structure of the HTLV-I provirus. To test this possibility, two cell lines, a rabbit T cell line, RL-5, which is transformed with Herpesvirus ateles (36), and the HTLV-I transformed human cell line MT-2 were assessed for the presence of phosphorylated Vav and compared with the two lines, RH/K30 and RH/K34. Only RL-5 and RH/K34 contained Tyr-phosphorylated Vav (Fig. 3 A and B). Vav was present, but not phosphorylated, in the human HTLV-I cell line MT-2 that was used for derivation of both rabbit cell lines, RH/K30 and RH/K34 (8).
Figure 3.
Vav phosphorylation in different T cell lines. (A) Precleared cell lysates (107 cells) from RH/K30, RH/K34, RL-5, and MT-2 cell lines (designated as lanes 1–4, respectively) were immunoprecipitated with normal rabbit Ig (Ig con) or anti-Vav, transferred onto Immobilon P membranes, and immunoblotted with anti-Tyr(P) or with rabbit anti-Vav antibodies. (B) Densitometric analysis of the blot obtained with anti-Tyr(P) in A by using arbitrary densitometry units. Identity of cell line is shown.
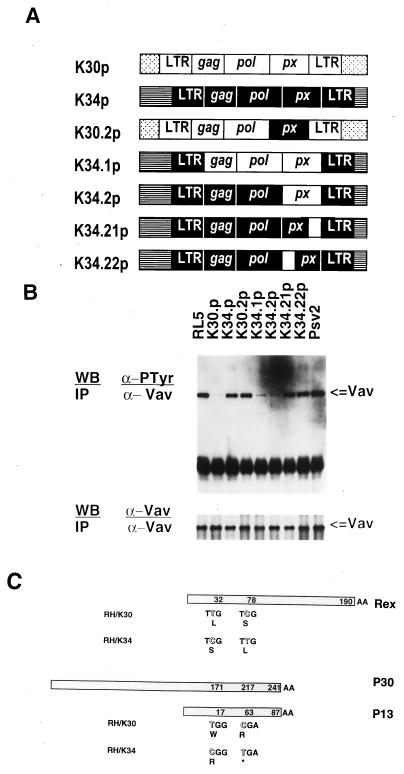
Experiments to directly link the virus to the phosphorylation differences were carried out with molecular HTLV-I clones derived from the RH/K30 and RH/K34 cell lines; the clones are designated K30p and K34p, respectively (Fig. 4A). The clones, which differ by 18 nucleotides over the length of the virus, were transfected into the RL-5 cell line, which contains constitutively phosphorylated Vav. Although HTLV-I virions are produced in RL-5 cells upon transfection with either K30p or K34p (37, 38), transfection with K30p led to the nearly complete down-regulation of Vav phosphorylation (Fig. 4B). By contrast, transfections with K34p had a lesser or no effect on the phosphorylation state of Vav in RL-5 cells (Fig. 4). The alteration of the Vav phosphorylation state by introduction of the K30p clone into a T cell suggests an active role for the virus in determination of the phosphorylation state of this cell protein.
Figure 4.
Modulation of Vav phosphorylation in RL-5 cells by transfection with HTLV-I DNA clones. (A) Schematic representation of the K30p and K34p HTLV-I molecular clones and chimeric clones used to transfect the RL-5 cell line. Open and solid boxes correspond to the genome segments of HTLV-I clones K30 and K34, respectively. For example, the clones designated K34.21p and K34.22p correspond to the K34 with single pX region substitutions characteristic of K30. (B Upper) Precleared cell lysates (107 cells) from RL-5 cells and RL-5 transfected with molecular clone K30p or K34p, with chimeric clones, or with control pSv2 plasmid were immunoprecipitated with anti-Vav antibody, transferred onto Immobilon P membrane, and immunoblotted with anti-Tyr(P) antibody. (Lower) Duplicate samples immunoblotted with anti-Vav. (C) Schematic representation of HTLV-I molecular clones K30p and K34p showing predicted amino acid sequence differences in their pX region for the proteins Rex, p13, and p30.
To specify viral genes responsible for the down-regulation of Vav phosphorylation, chimeric molecular HTLV-I clones were constructed by shuffling regions containing sequence differences between K30p and K34p proviruses (37, 38). Clones chosen for these experiments (Fig. 4A) differed in their long terminal repeat and pX region sequences. RL-5 cells were transfected and tested for the presence of phosphorylated Vav. Data indicated that Vav phosphorylation was unchanged in RL-5 cells transfected with recombinant clones containing substitutions characteristic of the pX region of K34p. By contrast, Vav phosphorylation was down-regulated in RL-5 cells transfected with clones having the K30p sequence. Differences in the long terminal regions had no effect (Fig. 4B). These results localized HTLV-I genes involved in down-regulation of Vav phosphorylation in transfected cells to the pX region, which encodes the proteins Rex, Tax (p40), p12, p13, and p30. The two clones used herein encode identical Tax and p12 proteins. The region involved in the down-regulation event contains two nucleotide substitutions between K30p and K34p; these substitutions result in differences in Rex, p13, and p30 (Fig. 4C). It appears from the data in Fig. 4 that both of the K30p substitutions are required for the down-regulation event; clones with a single pX region substitution did not alter the state of Vav phosphorylation.
DISCUSSION
Identification of differences between HTLV-I cell lines that cause leukemia-like disease and those mediating asymptomatic infection provides a means to understand mechanisms by which infection with this virus can give rise to diverse pathological consequences. The present study identifies differences in the Vav oncoprotein that is phosphorylated in the RH/K34 cell line, which causes leukemia-like disease, but not in RH/K30, which mediates asymptomatic infection. The active role of virus in the down-regulation of Vav phosphorylation was demonstrated and the region of the viral genome responsible for this event was narrowed to two substitutions in the pX region.
The viral sequence implicated in Vav dephosphorylation has two mutations resulting in two amino acid replacements, Leu → Ser at position 32 and Ser → Leu at position 78 in the Rex proteins of K30 and K34, respectively. The two clones used herein encode identical Tax and p12 proteins. The pX region sequence (see Fig. 4C) also encodes the less-well-characterized proteins p13 and p30, which derive from transcripts of different lengths in the same reading frame. One substitution results in a Trp → Arg replacement at position 17 of p13 and position 171 of p30, whereas the second gives rise to a premature in-frame stop codon resulting in truncation of both K34 proteins at position 63 of p13 and position 217 of p30. Results with the chimeric constructs indicate that both substitutions are needed for control of Vav phosphorylation. Determination of precise mechanisms by which these proteins control phosphorylation awaits further characterization of the activities of Rex and p13/30.
Interaction of phosphorylated Vav from RH/K34 in vitro with different SH2 domains (Grb2, Ras-Gap, PLCg1, and Fyn) can provide insight into the activated signaling pathways present in the leukemogenic cell but absent in RH/K30. More limited differences in the reactivity of RH/K30 and RH/K34 Vav with SH3 domains was seen; notably, only Vav from the RH/K34 cell binds the Ras-Gap CH3 domain. This confirms reports that only phosphorylated Vav can function as a guanidine nucleotide releasing factor for one or more members of the Ras-like family of small GTP-binding proteins (39, 40). The quantitative differences in SH3 reactivity with Vav from both cell lines may be related to the possibility that interaction with the SH3 domain is influenced by the state of Vav phosphorylation. The regulation and association of Vav with Zap-70 described by different groups was not observed in our system (41–43). Immunoprecipitations using several different anti-Zap 70 antibodies indicated that both the RH/K30 and RH/K34 cell lines were negative for phosphorylated Zap (W.M., unpublished results).
The precise role of Vav phosphorylation in the determination of the leukemic cell phenotype is not known, but there is presumptive evidence for involvement of Vav in related phenomena. Vav increases GTP binding to Ras after Tyr phosphorylation or by its binding to diglycerides (39, 44); both pathways are involved in cell activation (45, 46) and apoptosis induction (47). Furthermore, it has been reported that only phosphorylated Vav can catalyze GDP/GTP exchange on Rac-1, a protein implicated in cell proliferation and cytoskeletal organization (40). These properties associated with Vav activation may be used by a leukemogenic cell to persist and induce its pathological effect. More recently, it was reported that Vav is constitutively phosphorylated in autoimmune mice (MRL lpr/lpr) and that cells from these mice are refractory to various stimuli (48); this autoimmune syndrome shares certain similarities with autoimmunity linked to HTLV-I (49).
Vav has transformation potential, either alone or in cooperation with other oncogene products (14, 50); the significance of this to the present situation is uncertain, because all HTLV-I cells are transformed. Despite the fact that Tax and p12 have been reported as transforming elements of HTLV-I (51–53), their identity in the K30p and K34p clones (9) precludes a direct role for them in control of Vav phosphorylation. Constitutive Vav phosphorylation was observed in certain HTLV-I-transformed cell lines. Vav is not phosphorylated in MT-2 cells, which were used to derive both rabbit T cell lines studied herein. MT-2, a human cell line derived in vitro by coculture with a patient blood sample, contains at least eight genomic copies of HTLV-I provirus (35). The presence of these virus elements may cause Vav to be dephosphorylated. Finally, Vav phosphorylation in the HTLV-I transformed cell line RH/K34 and in the Herpesvirus ateles-transformed T cell line RL-5 suggests that Vav may be targeted by diverse potential oncogenes and its activation may play some part in the cell transformation. A possible role for interaction of Vav with members of the Herpesvirus family is supported by our observation that Epstein–Barr virus-transformed B cell lines also have phosphorylated Vav (W.M., unpublished results).
Although assignment of causality in leukemia-like disease mediated by RH/K34 to the observed difference in Vav phosphorylation is premature, it is reasonable to suggest that cell activation state is linked to leukemogenic potential. Furthermore, it is known that virus production by cells from human leukemia patients is often low or undetectable. Our previous studies with K30p and K34p HTLV-I chimeric clones implicated the same pX region substitutions identified herein in Vav activation as important to the control of the level of virus production, both in vitro and in vivo (37). The RH/K34 cell line and transfectants (including RL-5 transfectants) made with clones containing the K34p pX sequence, produce significantly less virus than do RH/K30 cells and transfectants made with clones containing K30p pX sequences (37). Control of activation state and viral production levels by rex and/or p13/30 genes is consistent with a role for these genes in pathogenicity. Any hypothesis must take into account the fact that virus from the cell giving rise to asymptomatic infection mediates high levels of virion production and has the ability to down-regulate Vav phosphorylation, at least in the RH/K30 and RL-5 rabbit T cell lines.
To ascribe leukemogenic potential to the control of Vav phosphorylation alone remains an oversimplification, in light of the complexity of the human HTLV-I leukemogenic process and experimental observations with the rabbit infection model. Such complexity is demonstrated in HTLV-I-infected individuals by the long incubation period for ATLL, by the fact that the acutely fatal disease mediated by RH/K34 cells is not caused by inoculation with K34-transfected RL-5 cells that have phosphorylated Vav, or by inoculation with the K34p molecular clone as naked DNA (37). Nonetheless, observations of thymic atrophy in ATLL patients (54) and in rabbits experimentally infected with the RH/K34 line (5, 6) argue strongly that T cell activation and apoptotic processes are influenced in HTLV-I-associated leukemia. The present data suggest a role for viral genes in control of these processes and provide evidence for relevant differences between an HTLV-I-infected line giving rise to asymptomatic infection and one causing acute disease.
Acknowledgments
We thank Dr. Paul Naccache for his contributions to this manuscript.
ABBREVIATIONS
- HTLV-I
human T cell leukemia virus I
- ATLL
adult T cell leukemia/lymphoma
- Tyr(P)
phosphotyrosine
- GST
glutathione S-transferase
Footnotes
References
- 1.Cann A J, Chen I S Y. In: Virology. Fields B N, editor. New York: Raven; 1990. pp. 1520–1527. [Google Scholar]
- 2.de Thé G, Bomford R. AIDS Res Hum Retroviruses. 1993;9:381–386. doi: 10.1089/aid.1993.9.381. [DOI] [PubMed] [Google Scholar]
- 3.Sawasdikosol S, Kindt T J. In: AIDS Research Review. Koff W, Wong-Staal F, Kennedy R C, editors. Vol. 2. New York: Mercel Dekker; 1992. pp. 211–233. [Google Scholar]
- 4.Seto A, Kawanishi M, Matsuda S, Ogawa K, Eguchi T, Miyoshi I. Jpn J Cancer Res. 1987;78:1150. [PubMed] [Google Scholar]
- 5.Simpson M R, Leno M, Hubbard B S, Kindt T J. J Infect Dis. 1996;173:722–726. doi: 10.1093/infdis/173.3.722. [DOI] [PubMed] [Google Scholar]
- 6.Simpson M R, Zhao T M, Hubbard B S, Sawasdikosol S, Kindt T J. Lab Invest. 1996;74:696–710. [PubMed] [Google Scholar]
- 7.Leno M, Simpson R M, Bowers F S, Kindt T J. J Exp Med. 1995;181:1575–1580. doi: 10.1084/jem.181.4.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sawasdikosol S, Hague B H, Zhao T M, Bowers F S, Simpson R M, Robinson M A, Kindt T J. J Exp Med. 1993;178:1337–1345. doi: 10.1084/jem.178.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao T M, Robinson M A, Sawasdikosol S, Simpson R M, Kindt T J. Virology. 1993;195:271–274. doi: 10.1006/viro.1993.1373. [DOI] [PubMed] [Google Scholar]
- 10.Migone T S, Lin J X, Ceresto A, Mulloy J C, O’Shea J J, Franchini G, Leonard W J. Science. 1995;269:79–81. doi: 10.1126/science.7604283. [DOI] [PubMed] [Google Scholar]
- 11.Xu X, Kang S H, Heidenreich O, Okerholm M, O’Shea J, Nerenberg M I. Clin Invest. 1995;96:1548–1555. doi: 10.1172/JCI118193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantley L C, Auger K R, Carpenter C, Duckworth A, Graziani R, Kapeller R, Soltoff S. Cell. 1991;64:281–302. doi: 10.1016/0092-8674(91)90639-g. [DOI] [PubMed] [Google Scholar]
- 13.Chan A C, Desai D M, Weiss A. Annu Rev Immunol. 1994;12:255–292. doi: 10.1146/annurev.iy.12.040194.003011. [DOI] [PubMed] [Google Scholar]
- 14.Katzav S, Martin-Zanca D, Barbacid M. EMBO J. 1989;8:2283–2290. doi: 10.1002/j.1460-2075.1989.tb08354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higuchi T, Kanzaki H, Fujimoto M, Hatayama H, Watanabe H, Fukumoto M, Kaneko Y, Higashitsuji H, Kishishita M, Mori T, Fujita J. Biol Reprod. 1995;53:840–846. doi: 10.1095/biolreprod53.4.840. [DOI] [PubMed] [Google Scholar]
- 16.Wulf G M, Arda C N, Limet B. EMBO J. 1993;12:5065–5074. doi: 10.1002/j.1460-2075.1993.tb06200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zmuidzinas A, Fisher K, Lira S A, Forrester L, Bryant S, Bernstein A, Barbacid M. EMBO J. 1995;14:1–11. doi: 10.1002/j.1460-2075.1995.tb06969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bustelo X R, Barbacid M. Science. 1992;256:1196–1199. doi: 10.1126/science.256.5060.1196. [DOI] [PubMed] [Google Scholar]
- 19.Bustelo X R, Ledbeter J A, Barbacid M. Nature (London) 1992;356:69–71. doi: 10.1038/356068a0. [DOI] [PubMed] [Google Scholar]
- 20.Margolis B, Hu P, Katzav S, Li W, Olivier J M, Ulrich A, Weiss A, Schlessinger J. Nature (London) 1992;356:71–74. doi: 10.1038/356071a0. [DOI] [PubMed] [Google Scholar]
- 21.Evans G A, Howard O M Z, Erwin R, Farrar W L. Biochem J. 1993;294:339–342. doi: 10.1042/bj2940339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Platanais L C, Sweet M E. J Biol Chem. 1994;269:3143–3146. [PubMed] [Google Scholar]
- 23.Weng W K, Jarvis L, Lebien T W. J Biol Chem. 1994;269:32514–32521. [PubMed] [Google Scholar]
- 24.Tarakhovsky A, Turner M, Schaal S, Mee J P, Duddy L P, Rajewsky K, Tybulewiz V L J. Nature (London) 1995;374:467–470. doi: 10.1038/374467a0. [DOI] [PubMed] [Google Scholar]
- 25.Zhang R, Alt F W, Davidson L, Orkin S H, Swat W. Nature (London) 1995;374:470–473. doi: 10.1038/374470a0. [DOI] [PubMed] [Google Scholar]
- 26.Fischer K, Zmuidzinas A, Gardner S, Barbacid M, Bernstein A, Guidos C. Nature (London) 1995;374:474–477. doi: 10.1038/374474a0. [DOI] [PubMed] [Google Scholar]
- 27.Hasalm R, Kolde H B, Hemmings B. Nature (London) 1993;363:309–310. doi: 10.1038/363309b0. [DOI] [PubMed] [Google Scholar]
- 28.Musacchio A, Gibson T, Rich P, Thompson J, Saraste M. Trends Biochem Sci. 1993;18:343–348. doi: 10.1016/0968-0004(93)90071-t. [DOI] [PubMed] [Google Scholar]
- 29.Koch A, Anderson D, Moran M F, Ellis C, Pawson T. Science. 1991;252:668–674. doi: 10.1126/science.1708916. [DOI] [PubMed] [Google Scholar]
- 30.Castresana J, Saraste M. FEBS Lett. 1995;374:149–151. doi: 10.1016/0014-5793(95)01098-y. [DOI] [PubMed] [Google Scholar]
- 31.Adams J M, Houston H, Allen J, Lints T, Harvey R. Oncogene. 1992;7:611–618. [PubMed] [Google Scholar]
- 32.Galland F, Katzav S, Birnbaum D. Oncogene. 1992;7:585–587. [PubMed] [Google Scholar]
- 33.Ono Y, Fuji T, Igrashi K, Kuno T, Kikkawa U, Nishizuka Y. Proc Natl Acad Sci. 1989;86:4868–4871. doi: 10.1073/pnas.86.13.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu P, Margolis B, Schlessinger J. BioEssays. 1993;15:179–183. doi: 10.1002/bies.950150306. [DOI] [PubMed] [Google Scholar]
- 35.Miyoshi I, Kubonishi S, Yoshimoto T, Akagi Y, Ohtsuki Y, Nagata K, Hinuma Y. Nature (London) 1981;294:770–771. doi: 10.1038/294770a0. [DOI] [PubMed] [Google Scholar]
- 36.Daniel M D, Melendez L V, Hunt R D, King N W, Anver M, Fraser C E O, Baranona H, Baggs R B. J Natl Cancer Inst. 1974;53:1803–1807. [PubMed] [Google Scholar]
- 37.Zhao T M, Robinson M A, Bowers F S, Kindt T J. Proc Natl Acad Sci USA. 1996;93:6653–6658. doi: 10.1073/pnas.93.13.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao T M, Robinson M A, Bowers F S, Kindt T J. J Virol. 1995;69:2024–2030. doi: 10.1128/jvi.69.4.2024-2030.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gulbins E, Schlottman K, Brenner B, Lang F, Coggershall K M. Biochem Biophys Res Commun. 1995;217:876–885. doi: 10.1006/bbrc.1995.2853. [DOI] [PubMed] [Google Scholar]
- 40.Crespo P, Schuebel K E, Ostrom A A, Gutkind J S, Bustelo R. Nature (London) 1997;385:169–172. doi: 10.1038/385169a0. [DOI] [PubMed] [Google Scholar]
- 41.Katzav S, Sutherland M, Packham G, Yi T, Weiss A. J Biol Chem. 1994;269:32579–32585. [PubMed] [Google Scholar]
- 42.Raab M, da Silva A, Findell P R, Rudd C E. Immunity. 1995;6:155–164. doi: 10.1016/s1074-7613(00)80422-7. [DOI] [PubMed] [Google Scholar]
- 43.Deckert M, Tartare-Deckert S, Couture C, Altman A. Immunity. 1996;5:591–604. doi: 10.1016/s1074-7613(00)80273-3. [DOI] [PubMed] [Google Scholar]
- 44.Gulbins E, Coggershall K M, Baier G, Telford D, Langlet C, Baier-Bitterlich G, Bonnefoy-Berard N, Burn P, Wittinghofer A, Altman A. Mol Cell Biol. 1994;14:4749–4758. doi: 10.1128/mcb.14.7.4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barbacid M. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 46.Hall A. Science. 1990;249:635–640. doi: 10.1126/science.2116664. [DOI] [PubMed] [Google Scholar]
- 47.Gulbins E, Bissonnette R, Mahboubi A, Martin S, Nishioka W, Brunner T, Baier G, Bitterlich-Baier G, Byrd C, Lang F, Resnick R, Altman A, Green D. Immunity. 1995;2:341–351. doi: 10.1016/1074-7613(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 48.Mimura T, Minota S, Nojima Y, Morino N, Hamaskai K, Fururya H, Yazaki Y. J Immunol. 1997;158:2977–2983. [PubMed] [Google Scholar]
- 49.Nishioka K, Sumida T, Hasunuma T. Arthritis Rheum. 1996;39:1410–1418. doi: 10.1002/art.1780390821. [DOI] [PubMed] [Google Scholar]
- 50.Bustelo X R, Suen K L, Leftheris K, Meyers C A, Barbacid M. Oncogene. 1994;9:2405–2413. [PubMed] [Google Scholar]
- 51.Cross S L, Feinberg M B, Wolf J B, Holbrook N J, Wong-Staal F, Leonard W J. Cell. 1987;49:47–56. doi: 10.1016/0092-8674(87)90754-9. [DOI] [PubMed] [Google Scholar]
- 52.Fujii M, Sassone-Corsi P, Verma I M. Proc Natl Acad Sci USA. 1988;85:8526–8530. doi: 10.1073/pnas.85.22.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mulloy J C, Crowley R W, Fullen J, Leonard W J, Franchini G. J Virol. 1996;70:3599–3605. doi: 10.1128/jvi.70.6.3599-3605.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanaoka M, Sasaki M, Matsumoto H, Tankawa H, Yamabe H, Tomimoto T, Tasaka C, Fujiwara H, Uchiyama T, Takatsuki K. Acta Pathol Jpn. 1979;29:723–738. doi: 10.1111/j.1440-1827.1979.tb00940.x. [DOI] [PubMed] [Google Scholar]