Abstract
Background
Currently, there is little data available regarding the role of gender-specific gene expression on synonymous codon usage (translational selection) in most organisms, and particularly plants. Using gender-specific EST libraries (with > 4000 ESTs) from Zea mays and Triticum aestivum, we assessed whether gender-specific gene expression per se and gender-specific gene expression level are associated with selection on codon usage.
Results
We found clear evidence of a greater bias in codon usage for genes expressed in female than in male organs and gametes, based on the variation in GC content at third codon positions and the frequency of species-preferred codons. This finding holds true for both highly and for lowly expressed genes. In addition, we found that highly expressed genes have greater codon bias than lowly expressed genes for both female- and male-specific genes. Moreover, in both species, genes with female-specific expression show a greater usage of species-specific preferred codons for each of the 18 amino acids having synonymous codons. A supplemental analysis of Brassica napus suggests that bias in codon usage could also be higher in genes expressed in male gametophytic tissues than in heterogeneous (flower) tissues.
Conclusion
This study reports gender-specific bias in codon usage in plants. The findings reported here, based on the analysis of 1 497 876 codons, are not caused either by differences in the biological functions of the genes or by differences in protein lengths, nor are they likely attributable to mutational bias. The data are best explained by gender-specific translational selection. Plausible explanations for these findings and the relevance to these and other organisms are discussed.
Background
Although tissue-specific gene expression has been associated with bias in codon usage in certain multicellular organisms including humans, Drosophila melanogaster, and Arabidopsis thaliana [1-5], very little data currently exists for most organisms, particularly regarding the role of gender-specific tissues and gametes. One of the few studies addressing the effect of gender, in Drosophila, suggests that genes having a higher ratio of female to male expression have a greater bias in codon usage [4,6]. A largely unstudied biological system where gender-specific gene expression could significantly alter codon usage is plants. Recent findings, in A. thaliana, have indicated that male gametes pass on a greater number of induced harmful mutations to their offspring, suggesting that mutations are subject to less selection in male tissues/gametes than in female tissues/gametes [7]. Such findings at the population level (short-term), suggest that gender could also impact the selective processes that alter molecular evolution in plants, including the usage of synonymous codons. Given that gender-specific selective pressures on codon usage could alter gene evolution and structure, and thereby influence population genetics, disease, and/or reproductive biology, and given the general lack of data to date, further investigation is warranted. Here, we focus on the evaluation of gender-specific codon usage in plants.
Nonrandom use of synonymous codons is a prevalent phenomenon observed in a diverse range of organisms [1,8-17]. A bias in codon usage occurs when synonymous codons are not all used at the same frequency in coding DNA [14,18,19]. Such bias in codon usage could result from mutational pressure, as indicated by a positive correlation between the nucleotide content of third codon positions and adjacent introns [20-23], or from selective pressure. Selective pressure has been supported by two findings. Firstly, greater levels of bias in codon usage are generally associated with a greater frequency of "preferred" codons (those used most frequently in the most biased genes) [24], a trend that corresponds to the abundance and/or gene number of tRNA in bacteria, yeast, C. elegans, Drosophila, Arabidopsis and other organisms [1,8,9,12-15,25-27]. Secondly, bias in codon usage has been well correlated to the level of gene expression, with the greatest bias occurring in highly expressed genes [8,10,12,28,29]. Each of these findings suggests that the use of preferred codons confers fitness benefits that enhance translational efficiency, a phenomenon particularly advantageous for the highly expressed genes [8,12]. In this regard, gene expression level is an essential component of understanding gender-specific influences on codon usage.
The main challenges for comparing male and female codon usage relative to gene expression for plant species, where the availability of genomic DNA sequences is often limited, are obtaining sufficient coding DNA data to assess codon usage in those tissues and determining the level of gene expression. EST datasets provide an effective solution to both issues. In particular, EST data have proven to be an effective means of quantifying gene expression in a range of tissues as the extent of redundancy in ESTs reflects the abundance of mRNA in the tissue or cells from which the library was obtained [8,10,12,30-32]. In addition, the increased availability of EST data, the long sequence length (200 to 800 bp) and the level of accuracy of the sequence data (from efficient sequence techniques and base-calling software, and the general use of only high quality reads) [33-35] makes it possible to study codon usage directly from EST sequences, even before the corresponding genomic sequences are available [36,37]. In this regard, the recent availability of male- and female-specific EST libraries in plants provide an effective resource to better understand selection on codon usage and how it may be influenced by gender.
In the present study, the main goal was to assess whether gender-specific gene expression per se and gender-specific gene expression level are correlated with codon usage in Zea mays and Triticum asestivum. As a supplemental analysis, we compared the bias in codon usage for genes expressed only in gametophytic tissue (male germline cells, microspores) and in flower tissue (composed of the somatic and reproductive tissues) at both high and low expression levels in Brassica napus. Given that gene function and protein length [8,10], have previously been found to influence codon usage, we also evaluated the role of those parameters within our analysis.
Results
In order to compare codon usage relative to gender-specific expression, we collected data from sperm and egg EST libraries for Z. mays and anther and ovary libraries for T. aestivum (Table 1). Microspore and flower libraries were obtained for B. napus. In brief, we obtained gender/tissue-specific datasets in the following manner: 1) clustering and assembly of ESTs from each library using CAP3 [38], 2) identification of unigenes (from now forward referred to as "genes") having translation products that matched known or hypothetical proteins in A. thaliana (using BLASTX [39-41]), 3) extraction of genes with tissue-specific expression by comparisons between the two compared tissues for each species (using MEGABLAST) [41], and 4) determination of the expression level per gene (based on the number of ESTs, see Methods). The six tissue-specific sequence datasets obtained are: Z. mays sperm-specific genes (N = 955), Z. mays egg-specific genes (N = 946), T. aestivum anther-specific genes (N = 3326), T. aestivum ovary-specific genes (N = 1489), B. napus microspore-specific genes (N = 1675) and B. napus flower-specific genes (N = 3181). Note that gender-specific genes represent those that are specific to a particular tissue or gamete (e.g. sperm) when compared to only one other tissue or gamete (e.g., egg,) and not relative to all tissues from the plant. Thus, these gene sets are larger than one would have found if the ESTs had been compared to all tissues of a plant. Subsequently, the GC content at third nucleotide positions (GC3) and the frequency of preferred codons (Fpr), each of which have been shown to be effective indicators of bias in codon usage [1,8,31], were determined for every gene from each of the tissue-specific datasets. Bias in codon usage was quantified using a single EST sequence to represent each gene (i.e., genes are represented by the longest EST in the contig or a singleton EST, see Methods). The data show, as described in detail below, that genes specific to female tissues and gametes have a greater bias in codon usage in both Z. mays and T. aestivum than genes expressed in male tissues and gametes. As well, male microspores have a greater bias than the heterogeneous tissues of the flower in B. napus.
Table 1.
The cDNA libraries used in the present investigation. The number of ESTs from each library that match genes in Arabidopsis thaliana, and the associated number of contigs and singletons are provided.
| Species and tissue or cell type | Relevant properties of tissue or cells | Name of library in NCBI and contributor(s) | Total number of ESTs per library (March 2006) | Number of ESTs matching genes in Arabidopsis (e < 10-7) | Number of contigs identified using CAP3 | Number of singletons identified using CAP3 | Unigenes (contigs plus singletons) |
|---|---|---|---|---|---|---|---|
| Zea mays | |||||||
| Sperm | Male | Zea mays sperm cell cDNA library (S. McCormick) Engel et al.[64] | 5174 | 2457 | 384 | 781 | 1165 |
| Egg | Female | Zea mays egg cell cDNA library (H. Yang and S. McCormick) Yang et al. [65] | 4085 | 1957 | 264 | 863 | 1127 |
| Triticum aestivum | |||||||
| Anther-meiotic | Male | Wheat meiotic anther cDNA library (O. Anderson) | 9139 | 6118 | 860 | 3058 | 3918 |
| Ovary | Female | Ov (Genoplante) | 5311 | 3699 | 813 | 1295 | 2108 |
| Brassica napus | |||||||
| Microspores (from 3 to 4 mm buds) | Male | 65JKBNM0 (M. Malik and J. Krochko) | 6539 | 5325 | 781 | 1620 | 2401 |
| Buds and young flowers | Heterogeneous (male, female and somatic) | ADIS-MPIZ 021 (B. Weisshaar) | 8747 | 6164 | 995 | 2665 | 3660 |
The GC content at third nucleotide positions and the frequency of preferred codons were each statistically significantly higher for genes expressed specifically in eggs as compared to sperm in Z. mays and for genes expressed in ovary as compared to anther in T. aestivum (Figure 1). This result was statistically significant for genes expressed at both high and at low levels (high > 5 ESTs per 10 000; low ≤ 5 ESTs per 10 000) as well as across all genes. Statistically significant higher values for GC3 were detected in microspore-specific genes as compared to the flower-specific genes in B. napus for genes expressed at low levels, but not for highly expressed genes (only GC3 was determined for B. napus as preferred codons have not been described yet). Within the male-specific and the female-specific genes, GC3 and Fpr were statistically significantly greater for the highly expressed as opposed to the lowly expressed genes in both Z. mays and T. aestivum and for the male-specific specific as compared to flower-specific genes in B. napus. Notably, lowly-expressed female-specific genes have a statistically significant higher GC3 and Fpr than the highly expressed male-specific genes in Z. mays and no difference was detected between these two groups for T. aestivum. Most of the statistically significant comparisons (of 35 pairwise comparisons in total) remained significant after Bonferroni correction except for some contrasts among genes for high versus low expression within a gender (i.e., male-specific tissue/gametes in Z. mays and T. aestivum and within the flower in B. napus) and a single between gender comparison for Fpr at high expression levels in T. aestivum (GC3 for this comparison remained statistically significant). All other between gender comparisons remained statistically significant.
Figure 1.
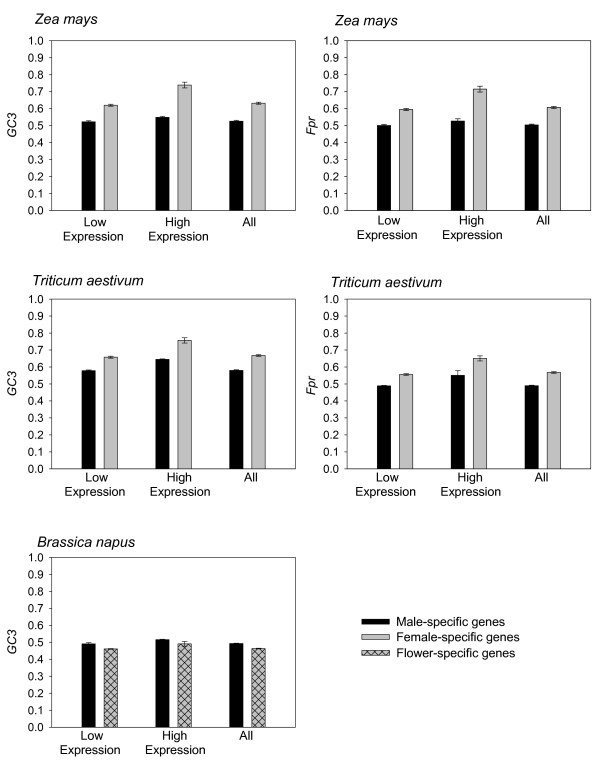
The GC content at third codon positions and the frequency of preferred codons for genes expressed specifically in male and female gametes or tissues in Zea mays and Triticum aestivum and the GC content of male-specific and flower-specific genes in Brassica napus relative to gene expression level. P-values between male and female (flower for B. napus) expressed genes for GC3 and for Fpr are as follows (respectively, in parenthesis):Z. mays: high (< 1 × 10-16**, < 1 × 10-16**), low (< 1 × 10-16 **, < 1 × 10-16**), all expression levels (< 1 × 10-16**, < 1 × 10-16**); T. aestivum: high (< 1 × 10-16*, 0.001*), low (< 1 × 10-16**, < 1 × 10-16**), all expression levels (< 1 × 10-16**, < 1 × 10-16**); B. napus (GC3 only): high (0.141), low (< 1 × 10-16**) and all expression levels (< 1 × 10-16**). P-values for high versus low expression level within gender (or flower): Z. mays: Female (< 1 × 10-16**, < 1 × 10-16**), Male (0.012*, 0.010*); T. aestivum: Female (< 1 × 10-16**, < 1 × 10-16**), Male (0.036*, 0.025*); B. napus: Male (1 × 10-16**), Flower (0.018*). P-values for lowly expressed female-specific genes versus highly expressed male-specific genes: Z. mays (< 1 × 10-16**, < 1 × 10-16**) and T. aestivum (0.867, 0.553). P-values for highly expressed female-specific genes versus lowly expressed male-specific genes: Z. mays (< 1 × 10-16**, < 1 × 10-16**) and T. aestivum (< 1 × 10-16**, < 1 × 10-16**). P-value for highly expressed male-specific genes versus lowly expressed genes in flower in B. napus (0.760). P-value for highly expressed male-specific genes versus lowly expressed genes in the flower in B. napus (< 1 × 10-16).* Indicates statistical significance (P < 0.05), **Indicates that P-value remains statistically significant after the Bonferroni correction.
The relative synonymous codon usage (RSCU) represents the observed frequency of a codon divided by the expected frequency (i.e., if all synonymous codons were used equally) [19]. Values different from 1, thus indicate the presence of bias. Analysis of RSCU for genes with male-specific and female-specific expression (concatenated across all genes) indicated that the bias in codon usage towards species-specific preferred codons was consistently higher in the female than in male tissues/gametes for Z. mays and T. aestivum (Table 2). In particular, 26 of 27 of the species-specific preferred codons for 18 amino acids in Z. mays (i.e, 18 have synonymous codons, Table 2) [42] were more frequent in female-specific than in male-specific genes. This represents a higher usage of at least one species-specific preferred codon for every amino acid with synonymous codons (noting that some amino acids have more than one preferred codon). For T. aestivum, female-specific genes had a greater usage of all 23 of the species-specific preferred codons (for 22 of 23 comparisons, the difference in RSCU was greater than 0.1). In addition, hierarchical clustering was conducted using Pearson correlation coefficients between RSCU values for each combination of species and gender-specific tissues/gametes for Z. mays and T. aestivum [43]. The results indicate that these groups cluster by gender rather than by species, consistent with gender being a major parameter in shaping codon usage (Figure 2). The RSCU data also show that B. napus demonstrates a preference towards GC ending codons in male-specific as compared to flower-specific genes (Table 2). The entire dataset across all species consisted of 1 497 876 codons.
Table 2.
Relative synonymous codon usage (RSCU) as determined from concatenated EST sequences of female- and male-specific genes in Zea mays and Triticum aestivum and male-specific and flower-specific sequences in Brassica napus.
| Amino acid | Codon | RSCUa | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zea mays | Triticum aestivum | Brassica napus | ||||||||||
| Female | Male | Difference (female minus male) b | Codon(s) previously identified as Preferredc | Female | Male | Difference (female minus male) | Codon(s) previously identified as Preferredc | Male | Flower | Difference (male minus flower)d | ||
| Phe | UUU | 0.65 | 0.86 | -0.21 | 0.60 | 0.82 | -0.22 | 0.83 | 0.90 | -0.07 | ||
| UUC | 1.35 | 1.14 | +0.21 | * | 1.40 | 1.18 | +0.22 | * | 1.17 | 1.10 | +0.07 | |
| Leu | UUA | 0.26 | 0.43 | -0.17 | 0.27 | 0.42 | -0.15 | 0.49 | 0.61 | -0.12 | ||
| UUG | 0.87 | 1.07 | -0.20 | * | 0.79 | 0.98 | -0.19 | 1.26 | 1.27 | -0.01 | ||
| CUU | 1.13 | 1.30 | -0.17 | 0.98 | 1.28 | -0.30 | 1.44 | 1.46 | -0.02 | |||
| CUC | 1.61 | 1.23 | +0.38 | * | 1.82 | 1.39 | +0.43 | * | 1.46 | 1.29 | +0.17 | |
| CUA | 0.47 | 0.56 | -0.09 | 0.39 | 0.49 | -0.10 | 0.52 | 0.60 | -0.08 | |||
| CUG | 1.66 | 1.40 | +0.26 | * | 1.77 | 1.44 | +0.33 | 0.82 | 0.76 | +0.06 | ||
| Ile | AUU | 0.92 | 1.11 | -0.19 | 0.84 | 1.07 | -0.23 | 1.06 | 1.05 | +0.01 | ||
| AUC | 1.57 | 1.22 | +0.35 | * | 1.65 | 1.27 | +0.38 | * | 1.39 | 1.26 | +0.13 | |
| AUA | 0.51 | 0.67 | -0.16 | 0.52 | 0.65 | -0.13 | 0.55 | 0.69 | -0.14 | |||
| Val | GUU | 0.96 | 1.25 | -0.29 | 0.89 | 1.19 | -0.30 | 1.43 | 1.46 | -0.03 | ||
| GUC | 1.26 | 1.07 | +0.19 | * | 1.30 | 1.10 | +0.20 | * | 0.95 | 0.89 | +0.06 | |
| GUA | 0.35 | 0.46 | -0.11 | 0.33 | 0.43 | -0.10 | 0.39 | 0.47 | -0.08 | |||
| GUG | 1.44 | 1.22 | +0.22 | * | 1.49 | 1.28 | +0.21 | * | 1.22 | 1.17 | +0.05 | |
| Ser | UCU | 0.98 | 1.20 | -0.22 | 0.88 | 1.18 | -0.30 | 1.52 | 1.54 | -0.02 | ||
| UCC | 1.23 | 1.02 | +0.21 | * | 1.37 | 1.13 | +0.24 | * | 1 | 0.88 | +0.12 | |
| UCA | 0.93 | 1.26 | -0.33 | 0.88 | 1.13 | -0.25 | 1 | 1.11 | -0.11 | |||
| UCG | 0.75 | 0.61 | +0.14 | * | 0.90 | 0.63 | +0.27 | 0.75 | 0.68 | +0.07 | ||
| AGU | 0.61 | 0.82 | -0.21 | 0.57 | 0.77 | -0.2 | 0.77 | 0.84 | -0.07 | |||
| AGC | 1.51 | 1.09 | +0.42 | * | 1.40 | 1.16 | +0.24 | * | 0.96 | 0.95 | +0.01 | |
| Pro | CCU | 1.07 | 1.27 | -0.20 | 0.93 | 1.18 | -0.25 | 1.55 | 1.49 | +0.06 | ||
| CCC | 0.99 | 0.73 | +0.26 | * | 1.07 | 0.84 | +0.23 | * | 0.63 | 0.57 | +0.06 | |
| CCA | 1.04 | 1.35 | -0.31 | 0.92 | 1.21 | -0.29 | 1.08 | 1.19 | -0.11 | |||
| CCG | 0.90 | 0.65 | +0.25 | * | 1.08 | 0.78 | +0.30 | * | 0.73 | 0.76 | -0.03 | |
| Thr | ACU | 0.92 | 1.13 | -0.21 | 0.80 | 1.06 | -0.26 | 1.26 | 1.25 | +0.01 | ||
| ACC | 1.41 | 1.05 | +0.36 | * | 1.51 | 1.17 | +0.34 | * | 1.07 | 0.96 | +0.11 | |
| ACA | 0.91 | 1.21 | -0.30 | 0.83 | 1.15 | -0.32 | 0.93 | 1.09 | -0.16 | |||
| ACG | 0.77 | 0.60 | +0.17 | * | 0.86 | 0.62 | +0.24 | * | 0.74 | 0.71 | +0.03 | |
| Ala | GCU | 1.13 | 1.34 | -0.21 | 0.87 | 1.19 | -0.32 | 1.74 | 1.69 | +0.05 | ||
| GCC | 1.23 | 0.99 | +0.24 | * | 1.44 | 1.12 | +0.32 | * | 0.76 | 0.72 | +0.04 | |
| GCA | 0.88 | 1.07 | -0.19 | 0.74 | 1 | -0.26 | 0.81 | 0.96 | -0.15 | |||
| GCG | 0.77 | 0.60 | +0.17 | * | 0.96 | 0.68 | +0.28 | 0.68 | 0.64 | +0.04 | ||
| Tyr | UAU | 0.65 | 0.88 | -0.23 | 0.63 | 0.82 | -0.19 | 0.78 | 0.83 | -0.05 | ||
| UAC | 1.35 | 1.12 | +0.23 | * | 1.37 | 1.18 | +0.19 | * | 1.22 | 1.17 | +0.05 | |
| His | CAU | 0.85 | 1.08 | -0.23 | 0.77 | 0.99 | -0.22 | 0.99 | 1.07 | -0.08 | ||
| CAC | 1.15 | 0.92 | +0.23 | * | 1.23 | 1.01 | +0.22 | * | 1.01 | 0.93 | +0.08 | |
| Gln | CAA | 0.63 | 0.77 | -0.14 | 0.57 | 0.73 | -0.16 | 0.89 | 0.98 | -0.09 | ||
| CAG | 1.37 | 1.23 | +0.14 | * | 1.43 | 1.27 | +0.16 | * | 1.11 | 1.02 | +0.09 | |
| Asn | AAU | 0.68 | 0.97 | -0.29 | 0.71 | 0.92 | -0.21 | 0.76 | 0.79 | -0.03 | ||
| AAC | 1.32 | 1.03 | +0.29 | * | 1.29 | 1.08 | +0.21 | * | 1.24 | 1.21 | +0.03 | |
| Lys | AAA | 0.50 | 0.70 | -0.20 | 0.49 | 0.64 | -0.15 | 0.72 | 0.88 | -0.16 | ||
| AAG | 1.50 | 1.30 | +0.20 | * | 1.51 | 1.36 | +0.15 | * | 1.28 | 1.12 | +0.16 | |
| Asp | GAU | 0.87 | 1.09 | -0.22 | 0.82 | 1.04 | -0.22 | 1.18 | 1.21 | -0.03 | ||
| GAC | 1.13 | 0.91 | +0.22 | * | 1.18 | 0.96 | +0.22 | * | 0.82 | 0.79 | +0.03 | |
| Glu | GAA | 0.63 | 0.83 | -0.20 | 0.60 | 0.75 | -0.15 | 0.81 | 0.90 | -0.09 | ||
| GAG | 1.37 | 1.17 | +0.20 | * | 1.40 | 1.25 | +0.15 | * | 1.19 | 1.10 | +0.09 | |
| Cys | UGU | 0.58 | 0.77 | -0.19 | 0.51 | 0.73 | -0.22 | 0.98 | 1.01 | -0.03 | ||
| UGC | 1.42 | 1.23 | +0.19 | * | 1.49 | 1.27 | +0.22 | * | 1.02 | 0.99 | +0.03 | |
| Arg | CGU | 0.66 | 0.77 | -0.11 | 0.60 | 0.72 | -0.12 | 1.08 | 0.98 | +0.10 | ||
| CGC | 1.29 | 1.06 | +0.23 | * | 1.57 | 1.21 | +0.36 | * | 0.68 | 0.54 | +0.14 | |
| CGA | 0.41 | 0.55 | -0.14 | 0.34 | 0.46 | -0.12 | 0.54 | 0.58 | -0.04 | |||
| CGG | 0.89 | 0.77 | +0.12 | 1.06 | 0.91 | +0.15 | 0.45 | 0.51 | -0.06 | |||
| AGA | 0.90 | 1.17 | -0.27 | 0.84 | 1.13 | -0.29 | 1.71 | 1.92 | -0.21 | |||
| AGG | 1.84 | 1.68 | +0.16 | * | 1.58 | 1.56 | +0.02 | * | 1.54 | 1.48 | +0.06 | |
| Gly | GGU | 0.86 | 1.03 | -0.17 | 0.74 | 0.97 | -0.23 | 1.27 | 1.22 | +0.05 | ||
| GGC | 1.41 | 1.13 | +0.28 | * | 1.65 | 1.30 | +0.35 | * | 0.66 | 0.64 | +0.02 | |
| GGA | 0.86 | 1.02 | -0.16 | 0.71 | 0.92 | -0.21 | 1.34 | 1.41 | -0.07 | |||
| GGG | 0.88 | 0.83 | -0.05 | 0.90 | 0.82 | +0.08 | 0.72 | 0.73 | -0.01 | |||
Positive differences in RSCU greater than 0.1 in the tissue comparisons (highlighting codon preferences in female tissues) are in bold. Start and termination codons and the single codon for tryptophan have been excluded. Preferred codon data is not available for B napus.
a The total number of codons used to estimate RSCU: Zea mays 116 919 (sperm) and 94 794 (egg), Triticum aestivum 453 286 (anther) and 199 397 (ovary) and Brassica napus 232 516 (microspore) and 400 964 (flower). RSCU indicates the difference from a 1:1 ratio for all available codons within synonymous codon groups.
b +, higher RSCU in female-specific genes; -, higher RSCU in male-specific genes.
c Previously described as preferred codons in Z. mays and T. aestivum as indicated by their frequencies in high-versus low-biased genes by Kawabe and Miyashita (2003) [42]. Asterisk (*) indicates a preferred codon. No values available for B. napus.
d +, higher RSCU in male-specific genes; -, higher RSCU in flower-specific genes.
Figure 2.

Hierarchical clustering based on Pearson correlation matrixes on relative synonymous codon usage values derived from concatenated EST sequences for each combination of species and gender (Zea mays and Triticum aestivum male-specific and female-specific genes).
Given that bias in codon usage has been inversely associated with protein length in certain eukaryotic organisms [e.g., [8]], we assessed whether it played a role in the observed gender-specific bias described above. For this, we identified the protein length for each gene under study (number of amino acids) in the Arabidopsis thaliana protein sequence database (i.e., genes previously identified as a match, see above). We used protein lengths from Arabidopsis because our DNA sequence data was derived from ESTs (that are partial sequences, not containing gene or protein length information) and because, unlike well-studied model organisms, the complete and/or annotated genomic DNA or protein sequences are not yet available for most of the genes in these plants. Protein lengths tend to be highly conserved among eukaryotes [44]. The data indicates that the mean protein length was greater for male-specific genes than female-specific genes in Z. mays and T. aestivum and for flower-specific genes as compared to male-specific genes in B. napus (mean protein length (± Standard Error): Z. mays egg 409.1 (± 13.2), Z. mays sperm 493.2 (± 13.3), T. aestivum ovary 426.3 (± 8.1), T. aestivum anther 571.3 (± 6.7), B. napus microspore 370 (± 6.7), B. napus flower 441.3 (± 5.6)). The higher mean lengths generally resulted from the presence of a relatively few genes that encoded very long proteins (between 1000 and 5000 amino acids). The number of genes encoding proteins with more than 1000 amino acids relative to the total number of genes are: Zea mays egg 38/946 = 4.0%,. Z. mays sperm 80/955 = 8.3%, T. aestivum ovary 69/1489 = 4.6%, T. aestivum anther 368/3326 = 11.1%, B. napus microspore 51/1675 = 3.0%, B. napus flower 176/3181 = 5.5%. We thus determined RSCU values for genes encoding proteins of similar lengths, in order to assess whether this protein length variation was related to our findings of gender-specific biases in codon usage. For this, male-specific and female-specific genes from each species under study (and B. napus flower-specific genes) were classified as being of either short (≤200 amino acids), medium (>200 and ≥400 amino acids) or of long length (>400 amino acids).
Comparisons were conducted for RSCU values relative to gender for Z. mays and T. aestivum (and male-versus flower-specific genes in B. napus) for the concatenated EST sequences (longest EST per contig) within each protein length category. The results show that female-specific genes consistently have higher values for RSCU for the preferred codons (as indicated by "+" sign in each of the columns) within each of these three protein length categories for Z. mays and T. aestivum (Table 3) [see Additional data file 1; Tables 1 to 3)] indicating that protein length variation does not explain the gender-specific bias observed for specific codons described in Table 2 (see Table 3). The bias in codon usage for male-specific versus flower-specific genes in B. napus was most evident for genes encoding long proteins.
Table 3.
Difference in the relative synonymous codon usage (RSCU) for short genes (less than or equal to 200 amino acids in the homologous Arabidopsis thaliana protein), medium length genes (more than 200 amino acids and less than or equal to 400) and long genes (more 400 amino acids) as determined from concatenated EST sequences of female- and male-specific sequences in Zea mays and Triticum aestivum and male-specific and flower-specific sequences in Brassica napus.
| Amino acid | Codon | Difference in RSCU (Female-specific genes minus male-specific genes)a | Difference in RSCU (Male-specific genes minus flower-specific genes)b | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zea maysc | Triticum aestivumc | Brassica napusc | ||||||||||
| Short | Medium | Long | Codon(s) previously identified as preferredd | Short | Medium | Long | Codon(s) previously identified as preferredd | Short | Medium | Long | ||
| Phe | UUU | -0.15 | -0.19 | -0.23 | -0.06 | -0.12 | -0.24 | +0.05 | -0.01 | -0.21 | ||
| UUC | +0.15 | +0.19 | +0.23 | * | +0.06 | +0.12 | +0.24 | * | -0.05 | +0.01 | +0.21 | |
| Leu | UUA | -0.07 | -0.15 | -0.19 | -0.06 | -0.09 | -0.14 | -0.03 | -0.01 | -0.28 | ||
| UUG | -0.22 | -0.20 | -0.16 | * | -0.01 | -0.13 | -0.15 | +0.13 | -0.05 | -0.02 | ||
| CUU | -0.22 | -0.10 | -0.18 | -0.17 | -0.20 | -0.30 | +0.09 | -0.09 | -0.06 | |||
| CUC | +0.50 | +0.34 | +0.27 | * | +0.24 | +0.24 | +0.32 | * | -0.18 | -0.05 | +0.59 | |
| CUA | -0.06 | -0.10 | -0.07 | -0.03 | -0.04 | -0.10 | -0.06 | +0.02 | -0.17 | |||
| CUG | +0.07 | +0.23 | +0.32 | * | +0.04 | +0.23 | +0.37 | +0.05 | +0.18 | -0.04 | ||
| Ile | AUU | -0.14 | -0.21 | -0.18 | -0.16 | -0.14 | -0.22 | +0.08 | +0.06 | -0.08 | ||
| AUC | +0.24 | +0.33 | +0.35 | * | +0.12 | +0.25 | +0.33 | * | -0.09 | -0.07 | +0.46 | |
| AUA | -0.09 | -0.13 | -0.17 | +0.03 | -0.12 | -0.11 | 0 | +0.01 | -0.38 | |||
| Val | GUU | -0.32 | -0.22 | -0.30 | -0.20 | -0.14 | -0.30 | +0.06 | -0.07 | -0.05 | ||
| GUC | +0.25 | +0.22 | +0.08 | * | +0.06 | +0.13 | +0.15 | * | -0.13 | +0.01 | +0.20 | |
| GUA | -0.17 | -0.08 | -0.09 | -0.04 | -0.06 | -0.10 | -0.02 | +0.03 | -0.20 | |||
| GUG | +0.23 | +0.08 | +0.31 | * | +0.18 | +0.07 | +0.25 | * | +0.08 | +0.03 | +0.05 | |
| Ser | UCU | -0.19 | -0.14 | -0.28 | -0.13 | -0.17 | -0.32 | -0.01 | -0.06 | -0.01 | ||
| UCC | +0.19 | +0.11 | +0.22 | * | +0.19 | +0.19 | +0.13 | * | -0.06 | +0.02 | +0.30 | |
| UCA | -0.33 | -0.38 | -0.25 | -0.09 | -0.28 | -0.15 | +0.04 | +0.05 | -0.34 | |||
| UCG | +0.11 | +0.10 | +0.15 | * | +0.13 | +0.23 | +0.25 | +0.02 | -0.02 | +0.15 | ||
| AGU | -0.19 | -0.18 | -0.20 | -0.14 | -0.13 | -0.17 | +0.09 | -0.02 | -0.21 | |||
| AGC | +0.41 | +0.47 | +0.36 | * | +0.04 | +0.16 | +0.25 | * | -0.09 | +0.03 | +0.10 | |
| Pro | CCU | -0.28 | -0.19 | -0.14 | -0.10 | -0.14 | -0.24 | +0.15 | +0.03 | +0.05 | ||
| CCC | +0.29 | +0.26 | +0.18 | * | +0.22 | +0.19 | +0.13 | * | -0.01 | +0.05 | +0.10 | |
| CCA | -0.25 | -0.23 | -0.33 | -0.22 | -0.24 | -0.21 | -0.03 | +0.03 | -0.25 | |||
| CCG | +0.23 | +0.17 | +0.28 | * | +0.10 | +0.19 | +0.31 | * | -0.10 | -0.12 | +0.10 | |
| Thr | ACU | -0.21 | -0.18 | -0.21 | -0.01 | -0.26 | -0.25 | +0.09 | +0.01 | -0.01 | ||
| ACC | +0.29 | +0.33 | +0.31 | * | +0.13 | +0.25 | +0.28 | * | -0.10 | -0.06 | +0.37 | |
| ACA | -0.17 | -0.30 | -0.30 | -0.11 | -0.19 | -0.29 | +0.01 | +0.04 | -0.43 | |||
| ACG | +0.10 | +0.14 | +0.20 | * | -0.01 | +0.19 | +0.27 | * | 0 | +0.01 | +0.07 | |
| Ala | GCU | -0.11 | -0.21 | -0.22 | -0.19 | -0.27 | -0.28 | +0.18 | 0 | +0.02 | ||
| GCC | +0.18 | +0.19 | +0.25 | * | +0.22 | +0.27 | +0.22 | * | -0.15 | 0 | +0.16 | |
| GCA | -0.10 | -0.19 | -0.19 | -0.11 | -0.18 | -0.23 | -0.04 | +0.01 | -0.33 | |||
| GCG | +0.03 | +0.19 | +0.17 | * | +0.08 | +0.18 | +0.29 | +0.02 | -0.01 | +0.15 | ||
| Tyr | UAU | -0.13 | -0.28 | -0.19 | -0.11 | -0.08 | -0.21 | +0.06 | +0.07 | -0.24 | ||
| UAC | +0.13 | +0.28 | +0.19 | * | +0.11 | +0.08 | +0.21 | * | -0.06 | -0.07 | +0.24 | |
| His | CAU | -0.27 | -0.22 | -0.16 | -0.03 | -0.13 | -0.20 | +0.04 | -0.01 | -0.21 | ||
| CAC | +0.27 | +0.22 | +0.16 | * | +0.03 | +0.13 | +0.20 | * | -0.04 | +0.01 | +0.21 | |
| Gln | CAA | -0.11 | -0.10 | -0.15 | -0.10 | -0.10 | -0.14 | -0.09 | -0.03 | -0.17 | ||
| CAG | +0.11 | +0.10 | +0.15 | * | +0.10 | +0.10 | +0.14 | * | +0.09 | +0.03 | +0.17 | |
| Asn | AAU | -0.32 | -0.32 | -0.21 | -0.01 | -0.11 | -0.19 | +0.05 | +0.06 | -0.15 | ||
| AAC | +0.32 | +0.32 | +0.21 | * | +0.01 | +0.11 | +0.19 | * | -0.05 | -0.06 | +0.15 | |
| Lys | AAA | -0.14 | -0.17 | -0.20 | -0.03 | -0.10 | -0.12 | -0.09 | -0.08 | -0.27 | ||
| AAG | +0.14 | +0.17 | +0.20 | * | +0.03 | +0.10 | +0.12 | * | +0.09 | +0.08 | +0.27 | |
| Asp | GAU | -0.06 | -0.21 | -0.24 | -0.09 | -0.13 | -0.20 | +0.04 | -0.01 | -0.10 | ||
| GAC | +0.06 | +0.21 | +0.24 | * | +0.09 | +0.13 | +0.20 | * | -0.04 | +0.01 | +0.10 | |
| Glu | GAA | -0.08 | -0.20 | -0.20 | -0.05 | -0.07 | -0.13 | -0.08 | -0.02 | -0.16 | ||
| GAG | +0.08 | +0.20 | +0.20 | * | +0.05 | +0.07 | +0.13 | * | +0.08 | +0.02 | +0.16 | |
| Cys | UGU | -0.08 | -0.14 | -0.20 | -0.10 | -0.18 | -0.17 | +0.04 | +0.02 | -0.14 | ||
| UGC | +0.08 | +0.14 | +0.20 | * | +0.10 | +0.18 | +0.17 | * | -0.04 | -0.02 | +0.14 | |
| Arg | CGU | -0.01 | -0.16 | -0.07 | -0.14 | -0.12 | -0.07 | +0.12 | -0.05 | +0.22 | ||
| CGC | +0.14 | +0.17 | +0.14 | * | +0.06 | +0.33 | +0.20 | * | +0.02 | -0.01 | +0.22 | |
| CGA | +0.01 | -0.18 | -0.12 | -0.03 | -0.08 | -0.12 | -0.11 | +0.03 | -0.03 | |||
| CGG | +0.03 | +0.08 | +0.18 | +0.18 | +0.10 | +0.18 | -0.05 | -0.01 | -0.10 | |||
| AGA | -0.21 | -0.18 | -0.26 | -0.03 | -0.15 | -0.27 | +0.04 | +0.03 | -0.57 | |||
| AGG | +0.04 | +0.28 | +0.12 | * | -0.05 | -0.07 | +0.07 | * | -0.02 | -0.01 | +0.20 | |
| Gly | GGU | -0.11 | -0.11 | -0.21 | -0.16 | -0.11 | -0.23 | +0.08 | +0.05 | +0.03 | ||
| GGC | +0.34 | +0.16 | +0.31 | * | +0.20 | +0.29 | +0.29 | * | -0.15 | +0.01 | +0.12 | |
| GGA | -0.22 | -0.08 | -0.16 | -0.07 | -0.19 | -0.18 | -0.04 | -0.06 | -0.06 | |||
| GGG | 0 | +0.04 | +0.07 | +0.04 | +0.01 | +0.12 | +0.11 | 0 | -0.09 | |||
Start and termination codons and the single codon for tryptophan have been excluded. Preferred codon data is not available for B napus. Data for codons previously identified as having codon biases (bold in Table 2) are in bold. [See Additional data file 1 (Tables 1 to 3) for the full data].
a +, higher RSCU in female-specific genes; -, higher RSCU in male-specific genes.
b +, higher RSCU in male-specific genes; -, higher RSCU in flower-specific genes.
c The total number of short genes and the average gene length (± Standard error), respectively, used to estimate RSCU: Z. mays sperm 149, 137.9 (± 3.2), Z. mays egg 204, 135.3 (± 2.9), T. aestivum anther 340, 142.3 (± 2.0), T. aestivum ovary 309, 138.8 (± 2.3), B. napus microspore 485, 144.8 (± 1.5), B. napus flower 598, 140.6 (± 1.6). The total number of medium length genes and the average gene length, respectively, used to estimate RSCU: Z. mays sperm 330, 295.2 (± 3.1), Z. mays egg 352, 296.2 (± 3.2), T. aestivum anther 893, 309.0 (± 1.9), T. aestivum ovary 520, 298.4 (± 2.6), B. napus microspore 608, 294.3 (± 2.4), B. napus flower 1127, 297.5 (± 1.7). The total number of long genes and the average gene length, respectively, used to estimate RSCU: Z. mays sperm 476, 741.6 (± 20.7), Z. mays egg 390, 655.4 (± 18.9), T. aestivum anther 2093, 753.4 (± 8.3), T. aestivum ovary 660, 661.7 (± 13.0), B. napus microspore 582, 639.5 (± 12.6), B. napus flower 1456, 676.1 (± 8.6).
d Previously described as preferred codons in Z. mays and T. aestivum as indicated by their frequencies in high-versus low-biased genes by Kawabe and Miyashita (2003) [42]. Asterisk (*) indicates a preferred codon. No values available for B. napus.
Given that the codons showing bias generally ended in G or C for each of the species examined (Table 2), we also compared the GC3 content between each of the two tissue/gamete types per species for each of the protein length categories (Table 4). The results of pairwise comparisons show that the GC3 values are statistically significantly higher for female-specific than male-specific genes in Z. mays and T. aestivum and for male-specific than flower-specific genes in B. napus within each of the protein length categories, consistent with a gender-bias on codon usage. Examination of only those genes encoding very long proteins (equal to or more than 1000 amino acids) showed similar trends for each of these species. Notably, GC3 values were inversely correlated with protein length within each of the six species and tissue-specific datasets (i.e., for the male-specific, female-specific and for flower-specific datasets, Table 4) [see also Additional data file 1; Table 4], consistent with the relationship between codon usage and protein length reported in other species [8].
Table 4.
Mean GC3 values for short genes (less than or equal to 200 amino acids in the homologous Arabidopsis thaliana protein), medium length genes (more than 200 amino acids and less than or equal to 400) and long genes (more 400 amino acids), and for genes encoding very long proteins only (equal to or more than 1000 amino acids) for female-specific and male-specific genes in Zea mays and Triticum aestivum and for male-specific and flower-specific genes in Brassica napus.
| Short | Medium | Long | Very long onlya | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Female | Male | P-value | Female | Male | P-value | Female | Male | P-value | Female | Male | P-value | |
| Zea mays | 0.702 | 0.632 | <1.0 × 10-16 | 0.635 | 0.542 | <1.0 × 10-16 | 0.593 | 0.483 | <1.0 × 10-16 | 0.505 | 0.656 | 0.044 |
| Triticum aestivum | 0.768 | 0.729 | 0.015 | 0.677 | 0.617 | <1.0 × 10-16 | 0.612 | 0.526 | <1.0 × 10-16 | 0.518 | 0.436 | <1.0 × 10-16 |
| Brassica napus | Male | Flower | Male | Flower | Male | Flower | Male | Flower | ||||
| 0.523 | 0.501 | 0.002 | 0.489 | 0.472 | 0.0002 | 0.469 | 0.448 | <1.0 × 10-16 | 0.441 | 0.423 | 0.340 | |
P-values are those for the Mann-Whitney Rank Sum Test (t-tests yielded similar P-values). P-values less than 0.05 are in bold. P-values not remaining statistically significant after Bonferroni correction among these contrasts are bold italics. For P-values of comparisons between protein length categories [see Additional data file 1 (Table 4)].
a, Sample sizes are relatively small for this group, influencing P-values for Z. mays and B. napus; the direction and magnitude of the differences are maintained for each comparison (see text). Mean protein lengths (± Standard Error) for this category are as follows. Z. mays sperm 1556.0 (± 63), Z. mays egg 1485.2 (± 112), T. aestivum anther 1377.0 (± 24.7), T. aestivum ovary 1334.7 (± 72.8), B. napus microspore 1370.5 (± 70.6), B. napus flower 1361.4 (± 34.0). Other mean protein length values are presented in Table 3.
Biological function has been proposed as a potential factor altering certain molecular evolutionary processes [10]; therefore we examined the gene profiles for each of the contrasting tissues for each species under study. The profile of biological functions of genes expressed only in the female tissues/gametes and male tissues/gametes were nearly identical for Z. mays and for T. aestivum (Figure 3). Similarly, the biological functions for genes specifically expressed in male microspores and those expressed in flowers were strongly associated in B. napus.
Figure 3.
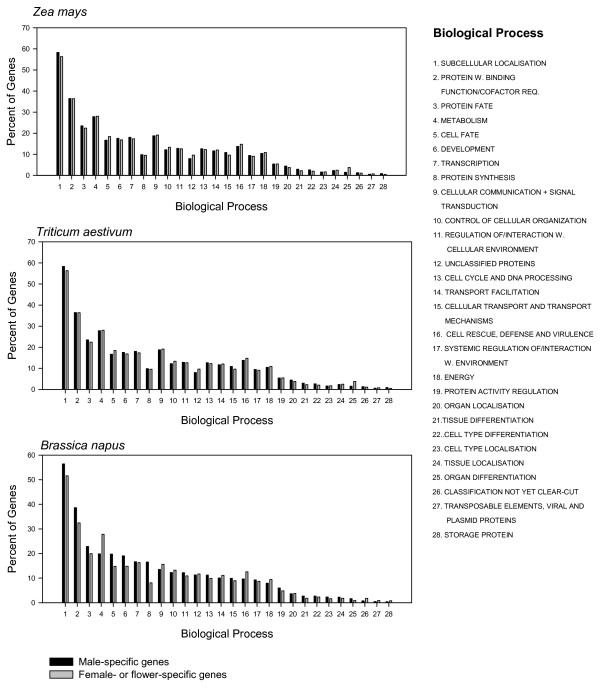
Biological functions of genes expressed only in the sperm and eggs in Zea mays, in the anther and ovary in Triticum aestivum and in the microspore and flower in Brassica napus. Sum of percentages may exceed 100 as some genes belong to more than one category.
Discussion
Gender differences in codon usage
The statistically significant higher GC content at third codon positions and the greater frequency of preferred codons for genes expressed specifically in eggs as compared to sperm in Z. mays and for genes expressed in the ovary as compared to anther in T. aestivum, provide strong evidence that there is a greater bias in codon usage for genes expressed in female tissues than in male tissues and/or gametes (Figure 1). These findings were similar when genes were classified as either high expressing or low expressing, suggesting that gender has a substantial impact on synonymous codon use. We can infer that these differences are likely due to selective pressure because the bias is associated with gene expression (tissue-specific gene expression or high versus low expression level, Figure 1). Furthermore, the data indicate that the gender effect cannot be attributed to variation in protein lengths or to gene function (Table 3, Table 4, Figure 3). Overall, these results, across a broad range of genes, provide evidence that codon usage is altered by gender-specific pressures in plants.
Genes expressed in eggs have a higher relative synonymous codon usage value than those expressed in sperm for 26 of the 27 previously identified preferred codons for Z. mays, and indicates, remarkably, that selective pressure in eggs specifically acts to enhance the frequency of preferred codons for each of the 18 amino acids that have synonymous codons (i.e., egg-specific genes have a greater frequency of at least one of the preferred codons per amino acid, Table 2). Similar findings for T. aestivum, showing that all 23 of the preferred codons for this species are enhanced in genes expressed in the ovary as compared to the anther, also demonstrate that a selective pressure inherent to these female organs and gametes is acting to enhance the incidence of preferred codons across all synonymous codon groups. In addition, the fact that the female-bias was detected for each gene length category (i.e., among short genes, among medium length genes, and among long genes, Tables 3 and 4) and that the gender-specific gene expression was the major determinant of hierarchical clustering (relative to RSCU values), rather than species (Figure 2), supports the notion that the codon usage bias demonstrated here is greatly influenced by gender-specific factors. It is notable for T. aestivum that in three cases the RSCU was greater for female-than male-specific genes (bold values, no asterisk, Table 2) for G or C ending codons that had not been previously identified as preferred, but had been described as preferred in Z. mays and other plant species [42]. The overall results presented here, showing greater use of preferred codons in genes expressed in female organs and gametes, suggests that these codons are probably also preferred in T. aestivum, at least for genes expressed in the reproductive tissues and gametes.
The greater bias in codon usage among genes expressed in female organs and gametes as compared to male organs and gametes, reflecting an increased propensity for translational selection, could be caused by several factors. In particular, it is possible that protein products of genes expressed in female organs and gametes experience a more diverse biochemical environment than their male counterparts, a phenomenon that could lead to greater selective constraint on proteins [45,46], and thus, on their translation. It is also possible that mutations at third codon positions in genes expressed in female organs and gametes may on average have greater effects on fitness, as has been proposed for genes expressed across a broad array of tissues (higher selection coefficients) [8,9]. This could occur, for example, if translational inefficiency in female organs and gametes alters the cellular energy resources or interferes with essential biological processes in a manner not prevalent in male regions. Mutations affecting female regions could also have greater fitness effects because of the general uncertainty in the pollination process, which makes it highly advantageous for each ovary, ovule and/or egg to be fully functional (thereby mutations in female regions may affect fitness overall more than for anthers, pollen or sperm) [47] and because maternal traits can have a much greater impact on seed production (seed number, size, and dispersal) and survival (and thus, on overall fitness) [48]. Another possible explanation for the observed results is that there are differences in gene function between female and male organs and gametes, a theory that has been proposed as a potential factor altering amino acid substitution rates [10]. As shown in Figure 3, however, this is not the likely explanation in this study, as there is remarkable similarity in the biological functions represented by the male-specific and female-specific genes in both Z. mays and T. aestivum. Nonetheless, subtle differences in gene function (e.g., specific genes that influence codon usage) or other, unidentified, functional differences between the male and female tissues/gametes could play a role [10]. An additional potential contributing factor worth consideration is that genes that have greater breadth of expression throughout the entire plant (that can have greater bias in codon usage) [1], are coincidentally also more commonly expressed in female organs and gametes than in males. Although this possibility cannot be definitively excluded, it seems unlikely given the similarity between the functional profiles of female- and male-specific genes. Altogether, it seems that the best explanations are differences in the amount of selective pressure for effective translation due to different cellular environments and/or a greater impact of mutations on female tissues and gametes. Further studies will nonetheless be needed to ascertain the mechanisms underlying the greater bias in codon usage in female organs and gametes in these plant species.
The relationship between gender-specific gene expression and codon usage in Z. mays and T. aestivum is consistent with the very limited data currently available for other organisms. It has been shown in humans, for example, that genes expressed in ovaries have likely been under slightly greater selective constraint than testes for codon usage following the divergence of humans and mice [3]. The trend notably corresponds to the generally high rates of protein evolution (and thus reduced selective pressure) reported in genes involved in spermatogenesis in primates [49]. In Drosophila, it has been found that the relative expression of genes in females versus males (the female : male ratio of gene expression) is well correlated to bias in codon usage [4]. In addition, in Arabidopsis, previous findings have indicated that more induced harmful mutations are passed to progeny by the sperm than by the eggs, consistent with the relatively lower selective pressure on mutations in male than in female tissues and gametes [7]. The present results extend these findings to include gender-specific selection on codon usage. Each of these gender-specific trends, in humans, Drosophila, and Arabidopsis are consistent with the findings we report here, and suggest that the higher bias in codon usage for genes expressed in female tissues could be inherent to a range of organisms. Further studies will be needed to better understand the full range of organisms for which gender-specific gene expression is associated with a bias in codon usage.
Gene expression level
Gene expression level has been shown to be positively correlated with bias in codon usage in many organisms [1,8-15,24-27,31]. Selection is the best explanation for this finding because higher levels of gene expression lead to greater opportunity for selection to alter codon usage [8,10,12] and because mutational bias has only rarely been associated with gene expression level (in certain microorganisms) [8,50,51]. In Drosophila, a positive relationship between gene expression and bias in codon usage has been reported for female tissues, but a relatively weak negative correlation was detected for male tissues [4]. Our findings of greater values for GC3 and Fpr for highly expressed genes than for lowly expressed genes for both male-specific and female-specific genes from Z. mays and T. aestivum suggests that gene expression level is positively correlated to bias in codon usage for genes expressed in male and in female regions for these plants.
It is notable nonetheless that we found that the differences in the bias in codon usage between highly and lowly expressed genes were not as marked the male as in female tissues and gametes, as evidenced by the fact that the Bonferroni correction excluded the statistical significance of this comparison for both Z. mays and T. aestivum. In fact, the lowly expressed female-specific genes had statistically significantly higher bias in codon usage than the highly expressed male-specific genes in Z. mays and no difference was detected between these two groups for T. aestivum (Figure 1). It thus seems that female tissues/gametes maintain substantial selective pressure on codon usage even for genes with reduced expression, in a manner not characteristic of male tissues/gametes.
Selection and gender-bias
The greater bias in codon usage for female-specific than for male-specific genes is currently best explained by selection. This is for the following reason. The male-specific and female-specific gene sets examined here were determined based on calculations that these genes were solely or primarily expressed in one tissue type and not in the other (i.e., the lack of ESTs in the contrasting tissue indicates that the mRNA was very rare or absent). Thus, the observed effects are associated with gender-specific gene expression (gene expression allows opportunity for selection, and is not usually associated with mutational bias [8,10,12]). In addition, our data indicate that gene function and protein length variation between male and female tissues/gametes do not explain the observed bias in codon usage between the gender-specific gene sets. Gene expression level differences cannot be implicated because the gender bias was detected for genes expressed at similar levels (high versus high expression and low versus low expression, regardless of protein length, Figure 1,) [see Additional data file 1, Table 5]. Notably, because we examined ESTs for the present analysis, which rarely contain introns, and studied plant species where annotated genomic DNA (containing the introns) is not yet available (NCBI, personal communication), we do not include an analysis of the GC (or AT) content of introns versus third codon positions in our genes, an approach sometimes used to exclude mutational bias [8,12,24,42,45,52]. Nevertheless, each of these trends, taken in their entirety, suggest that the bias in codon usage associated with gender-specific expression is best explained by differential selective pressure on genes expressed in male-specific tissues/gametes versus female-specific tissues/gametes.
Male-specific versus flower-specific genes in Brassica napus
Although the differences in bias in codon usage between the microspore and flower in B. napus were generally lower in magnitude than the previous between gender comparisons for Z. mays and T. aestivum (Figure 1, Table 2), the data overall indicates that genes expressed in the two B. napus tissues have specific patterns of codon usage. Specifically, the higher GC content in B. napus microspore-specific genes than in flower-specific genes suggests that the male portion of the flower may be under more selective pressure for codon usage than the flower as a whole. In particular, given that the flower and flower bud EST library should represent genes from the male, female and vegetative (somatic) tissue, one can infer that the combined vegetative and female tissue is under less selective pressure than the microspore. Given that the vegetative tissue usually represents the greatest fraction of the flower tissue (petals, sepals) [53], then it could, in turn, be inferred that the somatic region is likely under reduced selective pressure for codon usage than the microspore (with no inference regarding the pressure in female tissues). The fact that the GC content at third codon positions of the genes specifically expressed in the microspores varied markedly among synonymous codon groups (Table 2), and was found to be positively correlated to gene expression level (Figure 1), further supports the notion that translational selection is enhanced in the microspore component of the flower. Moreover, from examination of Table 2, it is evident that for six of seven comparisons where the differences in RSCU between tissues were greater than 0.1, the microspore had enhanced usage of G or C ending codons (which were notably also the preferred codons in Z. mays and T. aestivum), a trend consistent with greater selective pressure. Notably, analysis of RSCU values relative to protein length suggests that differences between male-specific and flower-specific genes are greatest for genes encoding longer proteins (>400 amino acids, Table 3) in the B. napus tissue comparisons as these genes have substantially greater usage of G or C ending codons in this category ("+" signs in Table 3). This effect could be partially caused by the greater percentage of genes encoding very long proteins in the flower-specific dataset or by greater male-specific effects on codon usage for genes encoding longer proteins. Nonetheless, all three of the gene length categories demonstrate higher GC3 values for male-specific genes (Table 4). One possible interpretation of all of these findings in B. napus, when combined with the data from Z. mays and T. aestivum, is that the translational selection increases in the following order: flower-specific (heterogeneous) genes, male-specific genes, female-specific genes. Because these analyses are in different species, however, further evaluation of this possible relationship will be needed. Altogether, the totality of the findings here suggest that genes expressed in reproductive tissues may be under greater translational level selection than those expressed in vegetative (somatic) tissues, a factor consistent with the key role of reproductive success in fitness.
Protein length and gene expression
The analysis of protein lengths indicates that genes encoding shorter proteins tend to generally have greater bias in codon usage, as indicated by GC3, for the species examined here (Table 4) [see also Additional data file 1; Table 4]. This is consistent with the trends reported in other organisms to date such as Arabidopsis, Drosophila, C. elegans and yeast [e.g., [8,54]]. We also found marked evidence that the gene expression level in the species studied here is inversely correlated to protein length for each of the six datasets examined (across all genes per dataset), a result consistent with trends reported in humans, Drosophila and Populus tremula [2,55,56]. In particular, the Pearson correlation coefficients were: Z. mays male (R = -0.135, P = 2.6 × 10-5), Z. mays female (R = -0.080, P = 0.010), T. aestivum male (R = -0.049, 4.3 × 10-3), T. aestivum female (R = -0.18, P = 1.1 × 10-12), B. napus male (R = -0.149, P = 1.9 × 10-9), and B. napus flower (R = -0.081, P = 4.6 × 10-6). This suggests that the tendency of shorter genes to have greater bias in codon usage (Table 4), at least for the genes examined here, may be due to greater levels of gene expression and an associated selective pressure [54]. Notably, in a complementary analysis to Figure 1, we found that the gender-bias in codon usage was evident among highly and among lowly expressed genes within each the three different protein length categories (short, medium, and long) [see Additional data file 1; Table 5].
Thus, the gender-specific biases in Z. mays and T. aestivum (and flower-specific differences in B. napus) at high and low levels of gene expression observed in Figure 1 cannot be explained by differences in protein lengths. In addition to the inverse association between protein length and bias in codon usage, it is also evident from Table 4 that the three species examined here tend to have different values for GC3, with decreasingly lower values occurring from T. aestivum, to Z. mays and to B. napus. Altogether, it is evident from our entire analysis that the gender-specific effects on codon usage can be detected across a range of protein lengths, gene expression levels, and for different plant species, thereby demonstrating that gender-specific factors play a significant role in genome evolution.
Notable issues
It should be noted that the B. napus material used for the microspore cDNA library was grown at low temperatures (10°C/5°C) that could potentially alter some fraction of the gene expression in the microspore if stress-mediation genes were enhanced. In fact, we found less than 1% of the total genes in B. napus microspores were stress-related (data not shown). Another issue worth considering is the implication of previous findings of gender-specific mutation rates in plants, a trend that was based on the detection of higher evolutionary rates at silent sites, including third codon positions, in male gametes [57]. Higher mutation rates in sperm, however, should act to enhance per generational mutation rates across the entire genome, including those genes expressed in females and in males, and thus, not impact the observed bias in codon usage.
Nonetheless, it should be noted that the differential male/female inheritance of organelles (and the underlying mechanisms; pre- or post-zygotic), could influence whether these genes are expressed in male or female tissues/gametes, and potentially contribute to the codon usage for organellar genes and their substitution rates [57,58]). It is also notable that the abundance of tRNA of the preferred codons could be greater for female than the male tissues/gametes and contribute to the gender differences in codon usage [3,4]. This seems relatively unlikely given that the abundance of tRNAs would have to be higher in female-tissues/gametes for every single preferred codon and that such differences have been shown not to explain the gender-specific codon usage in Drosophila [4]. It should also be noted that the reproducibility of the results for GC3 and Fpr observed in this study is consistent with the notion that GC3 content alone could be an effective indicator of codon bias in some species [31].
Conclusion
This study reports findings of female-specific bias in codon usage in plants. The remarkable consistency of the increased GC content of third codon positions and the increased frequency of preferred codons for both Z. mays and T. aestivum, even across different gene expression levels and protein lengths, combined with the enhanced usage of species-specific preferred codons for each of the 18 amino acids having synonymous codons, strongly indicate that gender plays a key role in codon usage. The findings in B. napus suggest that the tissues of the reproductive system, including both male and female organs, have a greater impact on codon usage than somatic regions. Overall, it is apparent that gender needs to be a key player in furthering our understanding of translational level selection. Further study will be needed to ascertain whether this is a generalized phenomenon, inherent to other organisms, as it could play a key role in DNA and protein sequence changes relevant to epidemiology, population genetics and to molecular evolution.
Methods
Sequence data
Sperm and egg EST libraries from Z. mays and anther and ovary libraries for T. aestivum were extracted from Genbank using Entrez Nucleotide available at the National Center for Biotechnology Information [41] (Table 1). We chose these data because of the availability of large gender-specific EST libraries (>4000) in Genbank. When more than one library was available we chose the one most likely to reflect gamete expression (e.g., an ovary library was selected over a pistil library in T. aestivum). In addition to these libraries, we also collected B. napus sequences from an in-house cDNA library representing isolated late-uninucleate and early-binucleate microspores (male germline cells) and from a publicly available flower library (Table 1). The EST datasets used represent those that were available to us as of March 2006.
Expression profiles and preparation of sequence data
Each of the EST sequences from each of the six libraries was compared against the A. thaliana protein sequence database using BLASTX [39-41]. Only EST sequences having an e-value of less than 1 × 10-7 to known or hypothetical proteins of A. thaliana were kept for further analysis. Notably, this process automatically excludes all rRNAs from the analysis as they would not be in the protein database. Using these datasets, we clustered and assembled the ESTs for each library into contigs and singletons using the software program CAP3 [38] (Table 1). The expression profile for each gene was determined from the number of ESTs per contig, and this value was 1 for singleton ESTs. For each gene in each of the six datasets, we standardized the expression level by dividing these values by the total number of ESTs in the original and complete EST library (Table 1), an approach that has been previously demonstrated to be an effective measure of expression level [8,12,31]. These values were multiplied by 10 000 (to obtain ESTs per 10 000) and the expression level for each gene was categorized as either high (>5 ESTs per 10 000) or low (≤5 ESTs per 10 000). Although many of the putative unigenes used here have not yet been definitively described as genes for those species, we nonetheless refer to them as "genes" here and in the text.
We identified genes that were expressed only in the male library and only in the female library (or flower for B. napus) as follows. Beginning with the male EST library from each species, the longest EST sequence per contig was identified and chosen as the representative for that gene. Each singleton represents its own gene. Each of these ESTs was then submitted to MEGABLAST [41] as a query against the original and redundant female-specific EST dataset (or flower library for B. napus, Table 1). The original female EST dataset was used in order to be conservative in the identification of male-specific ESTs. ESTs having more than 95% similarity were considered a match, which represents a level of similarity rigorous enough to distinguish among genes in conserved gene families [32]. The genes not having matches were categorized as male-specific. The process was then repeated for the female set of genes (or flower-specific for B. napus), after removing the sequences that were identified as matches to the male EST library. Specifically, the longest EST per contig and each singleton was queried against the original and redundant male-specific EST library in MEGABLAST. Genes that did not have matches were considered female-specific. The final datasets were: Z. mays sperm-specific genes (N = 955), Z. mays egg-specific genes (N = 946), T. aestivum anther-specific genes (N = 3326), T. aestivum ovary-specific genes (N = 1489), B. napus microspore-specific genes (N = 1675) and B. napus flower-specific genes (N = 1637). Genes with high expression (>5 ESTs per 10 000) represented approximately one quarter (or less) of each of these datasets. Notably, these gender-specific genes represent those that are specific to a particular tissue or gamete (e.g. sperm) when compared to only one other tissue or gamete (e.g., egg), and thus the tissue/gamete-specific genes identified here for each species are more numerous than one would observe if these had been compared to all the available libraries for that species (as is the standard analysis for ESTs). As well, the description of a gene as tissue-specific (e.g., female-specific) does not necessarily indicate that the gene is not expressed in the opposing tissue (e.g., male-specific) but rather that there were no ESTs present in the publicly available EST dataset used for comparison. It should be noted that although the genomic DNA sequences were available for a small portion of the ESTs for each species, we only used EST sequences for this entire analysis for consistency.
The open reading frame for each singleton and an EST representing every contig (longest EST per contig) was identified for every gene from each of the six reduced-sized gender-specific sequence datasets (see above) using alignments from BLASTX against the A. thaliana protein database. The BLASTX amino acid based algorithm provides alignments of the six-frame translated EST relative to the protein database and thereby is more sensitive to elements of functionality and homology than DNA alignments and accurately reveals reading frames [41,59]. Using the amino acid alignments between the translated ESTs and the A. thaliana homologues, we identified and extracted that portion of each EST sequence representing the reading frame (these generally did not include the start or termination codons). Most edited sequences were between 200 and 700 bp in length. Gaps in the alignments were rare, but when identified these regions were excluded from the EST, as were any occasional missense codons (resulting from the less than 1% sequencing errors in most large scale EST sequencing projects; in-house ESTs had PHRED scores [33] of greater than 20, representing less than a 1% error). All DNA sequence editing was conducted using BioEdit 7.0.5.3 [60].
Data analysis
The GC content at third codon positions (GC3) and the frequency of preferred codons (Fpr) for each gene was determined using CodonW [61]. GC3 content has been shown to be well-correlated with the degree of biased codon usage for A. thaliana and other plant species [1,31]. For the determination of Fpr, we used the preferred codons (sometimes called favoured codons) for Z. mays and T. aestivum previously identified by Kawabe and Miyashita (2003) [41]. Preferred codons are those that are most frequently used in the most highly-biased genes (as compared to lowly-biased genes) per degenerate codon group, and have been well-correlated to the optimal codons for many species (those codons used in the most highly expressed genes) [8]. Fpr was not determined for B. napus as the preferred codon data have not been described yet (and cannot be determined here as then the same data would then have to be used to determine both the preferred codons and Fpr).
A series of pairwise comparisons were conducted with respect to each combination of gender and gene expression level using GC3 and Fpr (including the flower for B. napus). Additional pairwise contrasts were conducted to assess the impact of gene expression within each gender-specific dataset for each species (and the flower in B. napus) for a total of 35 contrasts. Tests were conducted using the non-parametric Mann-Whitney Rank Sum test (as normality was not detected in some contrasts; t-tests nonetheless yielded similar results). Statistical significance required P < 0.05. A Bonferroni correction was applied across all contrasts. All statistical analyses were conducted in SigmaPlot 10.0 and SigmaStat 3.5 for Windows (Systat© software 2006). In addition to these pairwise tests, we also determined the relative synonymous codon usage (RSCU) for the concatenated reading frames for the entire dataset of male-specific and for female-specific (and the flower for B. napus) genes for each species using CodonW [61]. See main text for the description of gender-specific RSCU, GC3, and gene expression relative to protein length. Hierarchical clustering was conducted based on the Pearson correlation coefficients between RSCU values for each combination of species and gender-specific tissues/gametes for Z. mays and T. aestivum (Systat, 2004)[43].
As a means to assess whether the profile of gene functions conducted by male-specific and female-specific genes differed for Z. mays and for T. aestivum, each of these gender-specific sequence datasets (open reading frames) were submitted to MIPs [62]. A similar analysis was conducted for male-specific and flower-specific genes in B. napus. The gene functions were determined by comparison to proteins characterized in the A. thaliana database (MATDB)[62] with annotation from The Arabidopsis Information Resource (TAIR) and as implemented by the software Classification Superviewer [63].
Authors' contributions
CAW conceived the project, conducted the majority of the bioinformatics and comparative analysis, and prepared the manuscript. MRM conducted laboratory work for the B. napus microspore EST library and participated in the analysis and the editing of the manuscript. JEK is the research group supervisor and contributed to the conception of the study, managed the data from the B. napus microspore library, and was involved in the data analysis and the editing of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
That includes five Tables that support the data described in the main text.
Contributor Information
Carrie-Ann Whittle, Email: carrie-ann.whittle@nrc-cnrc.gc.ca.
Meghna R Malik, Email: meghna.malik@nrc-cnrc.gc.ca.
Joan E Krochko, Email: joan.krochko@nrc-cnrc.gc.ca.
Acknowledgements
The authors thank three anonymous reviewers for their valuable comments on the manuscript. We also thank the researchers listed in Table 1 for making their EST data publicly available. This work was funded by the National Research Council Canada – Genomics and Health Initiative III. This paper is National Research Council Canada publication No. 48420.
References
- Chiapello H, Lisacek F, Caboche M, Henaut A. Codon usage and gene function are related in sequences of Arabidopsis thaliana. Gene. 1998;209:GC1–GC38. doi: 10.1016/s0378-1119(97)00671-9. [DOI] [PubMed] [Google Scholar]
- Urrutia AO, Hurst LD. Codon usage bias covaries with expression breadth and rate of synonymous evolution in humans, but this is not evidence for selection. Genetics. 2001;159:1191–1199. doi: 10.1093/genetics/159.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, Robins H, Levine AJ. Tissue-specific codon usage and the expression of humans genes. Proc Natl Acad Sci USA. 2004;101:12588–12591. doi: 10.1073/pnas.0404957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambuch TM, Parsch J. Patterns of synonymous codon usage in Drosophila melanogaster genes with sex-biased expression. Genetics. 2005;170:1691–1700. doi: 10.1534/genetics.104.038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlar D, Lavner Y. The action of selection on codon bias in humans is related to frequency, complexity and chronology of amino acids. BMC Genomics. 2006;7:67. doi: 10.1186/1471-2164-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hambuch TM, Parsh J. Molecular evolution of sex-biased genes in Drosophila. Mol Biol Evol. 2004;21:2130–2139. doi: 10.1093/molbev/msh223. [DOI] [PubMed] [Google Scholar]
- Whittle CA, Johnston MO. Male-biased transmission of deleterious mutations to the progeny in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2003;100:4055–4059. doi: 10.1073/pnas.0730639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Expression pattern and, surprisingly, gene length shade codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci USA. 1999;96:4482–4487. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L. tRNA gene number and codon usage in the C. elegans genome are coadapted for optimal translation of highly expressed genes. Trends Genet. 2000;16:287–289. doi: 10.1016/s0168-9525(00)02041-2. [DOI] [PubMed] [Google Scholar]
- Akashi H. Gene expression and molecular evolution. Curr Opin Genet Dev. 2001;11:660–666. doi: 10.1016/s0959-437x(00)00250-1. [DOI] [PubMed] [Google Scholar]
- Akashi H. Translational selection and yeast proteome evolution. Genetics. 2003;164:1291–1303. doi: 10.1093/genetics/164.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Yau CB, Loosely M, Myers C. Effects of gene expression on molecular evolution in Arabidopsis thaliana and Arabidopsis lyrata. Mol Biol Evol. 2004;21:1719–1726. doi: 10.1093/molbev/msh191. [DOI] [PubMed] [Google Scholar]
- Guoy M, Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982;10:7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li W-H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol. 1986;24:28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- Stenico M, Lloyd T, Sharp PM. Codon usage in Caenorhabditis elegans: delineation of translational selection and mutational biases. Nucleic Acids Res. 1994;22:2437–2446. doi: 10.1093/nar/22.13.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva MD. Synonymous codon usage in bacteria. Curr Issues Mol Biol. 2001;3:91–97. [PubMed] [Google Scholar]
- Chen SL, Lee W, Hottes AK, Shapiro L, Harley H. Codon usage between genomes is constrained by genome-wide mutational processes. Proc Natl Acad Sci USA. 2004;101:3480–3485. doi: 10.1073/pnas.0307827100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Hayashida H. Extraordinarily high evolutionary rate of pseudogenes: evidence for the presence of selective pressure against changes between synonymous codons. Proc Natl Acad Sci. 1981;78:5739–5743. doi: 10.1073/pnas.78.9.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The codon adaptation index – a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa S, Ohama T, Yamao F, Muto, Jukes TH, Ozeki H, Umesono K. Directional mutation pressure and transfer RNA in choice of the third nucleotide of synonymous two-codon sets. Proc Natl Acad Sci USA. 1988;85:1124–1128. doi: 10.1073/pnas.85.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. Directional mutation pressure and neutral molecular evolution. Proc Natl Acad Sci USA. 1988;85:2653–2657. doi: 10.1073/pnas.85.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano A, Andachi Y, Ohama T, Osawa S. Novel anticodon composition of transfer RNAs in Micrococcus luteus, a bacterium with a high genomic G + C content: Correlation with codon usage. J Mol Biol. 1991;221:387–401. doi: 10.1016/0022-2836(91)80061-x. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Averof M, Lloyd AT, Matassi G, Peden JF. DNA sequence evolution: The sounds of silence. Philos Trans R Soc Lond B Biol Sci. 1995;349:241–247. doi: 10.1098/rstb.1995.0108. [DOI] [PubMed] [Google Scholar]
- Wright SI. The "effective number of codons" used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J Mol Evol. 2002;53:290–298. doi: 10.1007/s002390010219. [DOI] [PubMed] [Google Scholar]
- Moriyama EN, Powell JR. Codon usage bias and tRNA abundance in Drosophila. J Mol Evol. 1997;45:514–523. doi: 10.1007/pl00006256. [DOI] [PubMed] [Google Scholar]
- Grantham R, Gautier C, Guoy M, Jacobzone M, Mercier M. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981;9:r43–r49. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray EE, Lotzer J, Eberle M. Codon usage in plant genes. Nucleic Acids Res. 1989;17:477–498. doi: 10.1093/nar/17.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: expression patterns affects selection intensity but not mutation rate. Mol Biol Evol. 2000;17:68–74. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- Wright SI, Lauga B, Charlesworth D. Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol Biol Evol. 2002;19:1407–1420. doi: 10.1093/oxfordjournals.molbev.a004204. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics. 2004;168:373–381. doi: 10.1534/genetics.104.028944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using PHRED. I Accuracy Assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Staden R, Beal KF, Bonfield JK. In: Bioinformatics Methods and Protocols. Misener S, Krawetz SA, editor. Totowa, NJ: The Humana Press; 1999. The Staden Package, 1998; pp. 115–130. [Google Scholar]
- Adzhubei AA, Laerdahl JK, Vlasova AV. preAssemble: A tool for automatic sequencer trace data processing. BMC Bioinformatics. 2006;17:17–22. doi: 10.1186/1471-2105-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin P, Hahn MW. Coding sequence divergence between two closely related plant species: Arabidopsis thailiana and Brassica rapa ssp. pekinensis. J Mol Evol. 2002;54:746–753. doi: 10.1007/s00239-001-0074-1. [DOI] [PubMed] [Google Scholar]
- Mitreva M, Wendl MC, Martin J, Wylie T, Yin Y, Larson A, Parkinson J, Waterston RH, McCarter JP. Codon usage patterns in Nematoda: analysis based on over 25 million codons in thirty-two species. Genome Biol. 2006;7:R75. doi: 10.1186/gb-2006-7-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers WE, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov
- Kawabe A, Miyashita N. Patterns of codon usage bias in three dicot and four monocot plant species. Genes Genet Syst. 2003;78:343–352. doi: 10.1266/ggs.78.343. [DOI] [PubMed] [Google Scholar]
- Systat 11 Statistics, Vol. I. Systat Software, Inc., Richmond, CA; 2004. [Google Scholar]
- Wang D, Hsieh M, Li W-H. A general tendency for conservation of protein lengths across eukaryotic Kingdoms. Mol Biol Evol. 2004;22:142–147. doi: 10.1093/molbev/msh263. [DOI] [PubMed] [Google Scholar]
- Maside X, Lee AW, Charlesworth B. Selection on codon usage in Drosophila americana. Curr Biol. 2004;14:150–154. doi: 10.1016/j.cub.2003.12.055. [DOI] [PubMed] [Google Scholar]
- Hastings KE. Strong evolutionary conservation of broadly expressed protein isoforms in the troponin I gene family and other vertebrate gene families. J Mol Evol. 1996;42:631–640. doi: 10.1007/BF02338796. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. Why do plants produce so many more ovules than seeds? Nature. 1989;338:21–22. [Google Scholar]
- Mazer SJ, Snow AA, Stanton ML. Fertilization dynamics and parental effects upon fruit development in Raphanus raphanistrum: consequences for seed size variation. Am J Bot. 1986;73:500–511. [Google Scholar]
- Wyckoff G, Wang W, Wu CI. Rapid evolution of male reproductive genes in the descent of man. Nature. 2000;403:304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- Datta A, Jink-Roberton S. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- Beletskii A, Bhagwat AS. Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:13919–13924. doi: 10.1073/pnas.93.24.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman RM, Hey J. The effects of mutation and natural selection on codon bias in the genes of Drosophila. Genetics. 1994;137:1049–1056. doi: 10.1093/genetics/137.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polowick PL, Sawhney VK. A scanning electron microscopic study on the initiation and development of floral organs of Brassica napus cv. Westar. Am J Bot. 1986;73:254–263. [Google Scholar]
- Moriyama EN, Powell JR. Gene length and codon usage bias in Drosophila melanogaster, Saccharomyces cerevisiae and Escherichia coli. Nucleic Acids Res. 1998;26:3188–3193. doi: 10.1093/nar/26.13.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Bettencourt BR, Meiklejohn CD, Hartl DL. Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of protein-protein interactions. Mol Biol Evol. 2005;22:1345–1354. doi: 10.1093/molbev/msi122. [DOI] [PubMed] [Google Scholar]
- Ingvarsson PK. Gene expression and protein length influence codon usage and rates of sequence evolution in Populus tremula. Mol Biol Evol. 2007;24:836–844. doi: 10.1093/molbev/msl212. [DOI] [PubMed] [Google Scholar]
- Whittle CA, Johnston MO. Male-driven evolution of mitochondrial and chloroplastidial DNA sequences in plants. Mol Biol Evol. 2002;19:938–949. doi: 10.1093/oxfordjournals.molbev.a004151. [DOI] [PubMed] [Google Scholar]
- Birky CW. Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc Natl Acad Sci USA. 1995;92:11331–11338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish W, States DJ. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- Hall T. BioEdit: A user-friendly biological sequence alignment program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Peden J. PhD Thesis. Department of Genetics, University of Nottingham, UK; 1999. Analysis of codon usage. [Google Scholar]
- Mewes HW, Frishman D, Guldener U, Mannhaupt G, Mayer K, Mokrejs M, Morgenster B, Munsterkkotter M, Rudd S, Weil B. MIPS: A database for genomes and protein sequences. Nucleic Acids Res. 2002;30:31–34. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provart N. Classification Superviewer. 2006. http://bbc.botany.utoronto.ca
- Engel ML, Charboud A, Dumas C, McCormick S. Sperm cells of Zea mays have a complex complement of mRNAs. Plant J. 2003;34:697–707. [PubMed] [Google Scholar]
- Yang H, Kaur N, Kiriakopolos S, McCormick S. EST generation and analyses towards identifying female gametophyte-specific genes in Zea mays L. Planta. 2006;224:1004–1014. doi: 10.1007/s00425-006-0283-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
That includes five Tables that support the data described in the main text.


