Abstract
Background
Corynebacterium glutamicum has several anaplerotic pathways (anaplerosis), which are essential for the productions of amino acids, such as lysine and glutamate. It is still not clear how flux changes in anaplerotic pathways happen when glutamate production is induced by triggers, such as biotin depletion and the addition of the detergent material, Tween 40. In this study, we quantitatively analyzed which anaplerotic pathway flux most markedly changes the glutamate overproduction induced by Tween 40 addition.
Results
We performed a metabolic flux analysis (MFA) with [1-13C]- and [U-13C]-labeled glucose in the glutamate production phase of C. glutamicum, based on the analysis of the time courses of 13C incorporation into proteinogenic amino acids by gas chromatography-mass spectrometry (GC-MS). The flux from phosphoenolpyruvate (PEP) to oxaloacetate (Oxa) catalyzed by phosphoenolpyruvate carboxylase (PEPc) was active in the growth phase not producing glutamate, whereas that from pyruvate to Oxa catalyzed by pyruvate carboxylase (Pc) was inactive. In the glutamate overproduction phase induced by the addition of the detergent material Tween 40, the reaction catalyzed by Pc also became active in addition to the reaction catalyzed by PEPc.
Conclusion
It was clarified by a quantitative 13C MFA that the reaction catalyzed by Pc is most markedly increased, whereas other fluxes of PEPc and PEPck remain constant in the glutamate overproduction induced by Tween 40. This result is consistent with the previous results obtained in a comparative study on the glutamate productions of genetically recombinant Pc- and PEPc-overexpressing strains. The importance of a specific reaction in an anaplerotic pathway was elucidated at a metabolic level by MFA.
Background
A coryneform bacterium, Corynebacterium glutamicum, which was isolated in the 1950s [1], is particularly able to produce large amounts of amino acids, such as glutamate and lysine. In glutamate fermentation, it is well known that there are some triggers of glutamate overproduction in C. glutamicum: the depletion of biotin, which is required for cell growth [2], the addition of detergent [3], the addition of lactam antibiotics, such as penicillin [4], and the temperature upshift of fermentation [5]. With these triggers a marked change at the final branch point, 2-oxoglutarate in the TCA cycle, was observed owing to a decrease in 2-oxoglutarate dehydrogenase complex (ODHC) activity [6-8]. Recently, it has been reported that the deletion of odhA causes glutamate overproduction [9]. Furthermore, it has been clear that the pentose phosphate pathway activity decreases when glutamate production is induced [10].
However, the mechanism of the flux redistribution of 2-oxoglutarate in the TCA cycle is not sufficient for explaining a change in the production activity of glutamate. To overproduce glutamate, oxaloacetate has to be supplied through anaplerotic pathways. Therefore, the changes in fluxes in the anaplerotic pathways are also important for clarifying the mechanism of glutamate overproduction. C. glutamicum has a complex set of anaplerotic pathways for supplying oxaloacetate, i.e., reactions from phosphoenolpyruvate (PEP) to oxaloacetate catalyzed by phosphoenolpyruvate carboxylase (PEPc) [11,12], from pyruvate (Pyr) to oxaloacetate catalyzed by pyruvate carboxylase (Pc) [13], from oxaloacetate to PEP catalyzed by PEP carboxykinase (PEPck) [14], and from malate to Pyr catalyzed by malic enzymes [15,16]. Peters-Wendisch et al. observed that the ability of C. glutamicum to produce glutamate is highly dependent on the Pc activity, using recombinant strains with the overexpression and deficiency of the Pc activity [17]. By comparing the final amount of glutamate production of the recombinant strain of Pc with that of the parental strain, Pc was considered important from a molecular biological view [18]. However, the metabolic fluxes of anaplerotic pathways under glutamate-overproducing conditions have not yet directly been measured.
For the flux analysis of complex pathways, such as anaplerosis, a precise and quantitative metabolic flux analysis involving a 13C isotope labeling experiment (13C MFA) is performed. For this experiment, the 13C-labeled glucose is generally added into a medium and cells are harvested in the log phase. The fractional 13C-labeling of proteinogenic amino acids is performed by gas chromatography-mass spectrometry (GC-MS) and nuclear magnetic resonance (NMR). Using this method, metabolic fluxes including anaplerosis can be estimated under the conditions of a continuous culture or mid-log phase, where it is easily possible to establish a steady state and harvest cells [16,18-23]. In the lysine production of C. glutamicum, 13C MFAs have successfully been performed to analyze the intracellular fluxes in continuous cultures [24,25].
However, there are no reports on the detailed quantitative analyses of anaplerosis based on the 13C MFA in the glutamate production phase of C. glutamicum, owing to the difficulties of setting the experimental conditions up. After the induction of glutamate production by triggering effects, cell growth is almost stopped in wild-type strains [8]. Thus, even when cells are cultured in a medium with a 13C-labeled substrate, the fractional 13C-labelings of proteinogenic amino acids from cells in the production phase do not always represent the metabolic flux distribution in the production phase. In this case, it mainly represents the metabolic flux distribution in the log growth phase before the triggering effects, since intracellular proteins are predominantly synthesized in the growth phase. Therefore, to extract pure information about metabolic fluxes in the glutamate production phase, a 13C-labeled substrate must be added into the medium after cells enter the glutamate production phase.
In this study, the direct quantitative analysis of metabolic fluxes by 13C MFA was performed in the glutamate production phase of C. glutamicum. The important points for the 13C MFA in the glutamate production phase are as follows: (1) 13C of metabolites should be converted from the 13C-labeled substrate (glucose) taken up in the glutamate production phase only and (2) reaching the plateau of the incorporation rate of 13C into proteinogenic amino acids in this phase should be confirmed. We established an experimental and analytical method for the 13C MFA in the glutamate production phase induced by Tween 40 addition as follows.
C. glutamicum was cultivated on natural glucose in the growth phase, and then glutamate production was induced by Tween 40 addition in the mid-log growth phase. The amount of Tween 40 added was precisely adjusted to achieve both stable growth and glutamate production, as 13C was incorporated into proteinogenic amino acids in the production phase. Subsequently, 13C-labeled glucose was added in the glutamate production phase, and the time course of the 13C labeling pattern of each amino acid was measured by GC-MS. It was confirmed that the 13C labeling patterns of the amino acids reach a plateau and the steady state by comparing them with that at each sampling time. A computational algorithm for precise 13C MFAs [26] was applied to the determination of metabolic fluxes by fitting 13C labeling patterns.
In this study, two different glutamate production activities were induced by the addition of two different amounts of Tween 40. The 13C MFA results between the growth and two different glutamate production were comparatively analyzed. From the obtained results, the roles of anaplerotic pathways (Pc, PEPc, and PEPck) in the glutamate overproduction induced by Tween 40 addition in C. glutamicum were clarified.
Results
Glutamate productions of C. glutamicum under two Tween 40 addition conditions
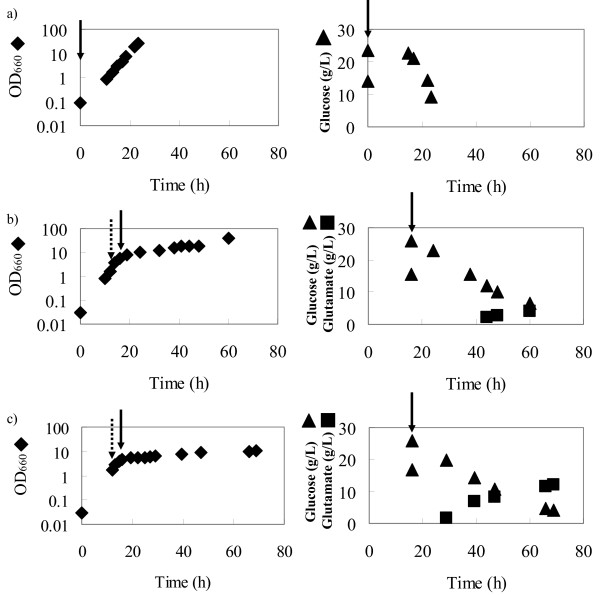
The fermentation of C. glutamicum was performed under two Tween 40 addition conditions: final concentrations of 0.5 and 0.8 mg/mL. The obtained results are shown in Fig. 1. It was obvious that the glutamate production in the fermentation under the 0.8 mg/mL of Tween 40 addition condition (Fig. 1-c) is larger than in the fermentation under 0.5 mg/mL of the Tween 40 addition condition (Fig. 1-b). In the fermentation without Tween 40 addition, no glutamate was produced into the medium (Fig. 1-a). Specific rates in the growth and the glutamate production phases are shown in Table 1 (all units: per hour). The glutamate production fluxes in the fermentations under the two Tween 40 addition conditions were 20 (Fig. 1-b) and 68 (Fig. 1-c) with respect to the glucose uptake of 100. The amount of glutamate overproduced was proportional to that of Tween 40 added.
Figure 1.
Time courses of cell growth (OD660), glucose consumption, and glutamate production of C. glutamicum. Figs. 1-a, -b, and -c indicate the fermentations with Tween 40 additions using the final concentrations of 0 (without addition), 0.5, and 0.8 mg/mL, respectively. Diamonds, triangles, and squares indicate OD660, glucose, and glutamate, respectively. Dark and dotted arrows indicate the points of 13C glucose ([1-13C] and [U-13C] glucose) and Tween 40 additions, respectively.
Table 1.
Specific rates in growth and two glutamate production phases of C. glutamicum.
| Amount of Tween 40 addition (final concentration (mg/ml)) | |||
| 0 | 0.5 | 0.8 | |
| growth rate (μ) | 0.25 | 0.04 | 0.01 |
| consumption rate (ν) | 0.55 | 0.14 | 0.17 |
| production rate (ρ) | - | 0.03 | 0.12 |
Observation of 13C incorporation into each proteinogenic amino acid by GC-MS
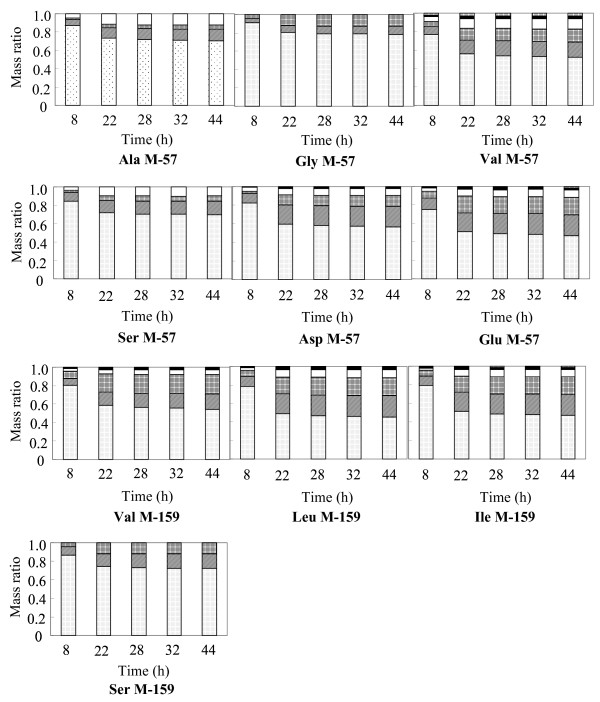
For the 13C MFA of C. glutamicum in the glutamate production phase, it was necessary to obtain the information of 13C of each amino acid converted in the glutamate production phase. In this study, the time course of 13C incorporation into each amino acid was measured by GC-MS analysis. The obtained result is shown in Fig. 2, where the glutamate production flux was 20 with respect to the glucose uptake of 100 (Fig. 1-b). The GC-MS data of all the amino acids were not changed after 44 h of 13C-labeled glucose addition (60 h in the fermentation) and reached a plateau. Therefore, these GC-MS data were used for the 13C MFA in the glutamate production phase. Also, for the 13C MFA in the production phase in the fermentation with a glutamate production flux of 68 (Fig. 1-c), the GC-MS data were used after 53 h since 13C-labeled glucose had been added (69 h in the fermentation).
Figure 2.
Time courses of mass isotopomer distributions of proteinogenic amino acids in fermentation after addition of 13C-labeled glucose, where glutamate production was 20 with respect to glucose uptake of 100. The data (m0, m1 ..., mn indicating the mass isotopomer distributions obtained from mass spectral intensity ratios, as previously described [22]) at each sampling time were summed up to unity from the bottom. Time indicates that after 13C glucose addition. The M-57 and M-159 of each amino acid indicate mass ion fragment groups with high intensities, and thus, these data were used for 13C MFA.
13C MFAs in growth phase and two glutamate production phases
The 13C MFA in the growth phase was performed by a previously reported method [26], where 13C-labeled glucose was added at the beginning of fermentation, that is, the GC-MS data in the log growth phase was used for analysis (22 h). The 13C MFAs in the two glutamate production phases were performed using the GC-MS data mentioned in the previous section. The GC-MS data of C. glutamicum in the growth phase and two glutamate production phases are shown in Tables 2, 3, 4, respectively. The measurement of each sample was performed five times, and all data errors (standard deviation divided by mean values) were below 2%. The 13C MFA results in the growth phase and two glutamate production phases are shown in Fig. 3. For the result in the growth phase, cross validation was performed using the NMR data shown in Table 5. For the 13C MFA results in the two production phases, the results based on the 13C labeling patterns of proteinogenic amino acids were verified using the GC-MS data of intermediate metabolites (pyruvate, succinate, and fumarate) because of difficulty in cross validation using the NMR data due to a small number of cells in the production phase, as shown in Tables 6 and 7.
Table 2.
Mass isotopomer distributions of amino acids in growth phase of C. glutamicum.
| m0 | m1 | m2 | m3 | m4 | m5 | ||
| Ala-57 | measured | 0.677 | 0.128 | 0.040 | 0.155 | ||
| estimated | 0.665 | 0.138 | 0.041 | 0.156 | |||
| Gly-57 | measured | 0.753 | 0.076 | 0.171 | |||
| estimated | 0.754 | 0.076 | 0.170 | ||||
| Val-57 | measured | 0.472 | 0.162 | 0.158 | 0.138 | 0.035 | 0.034 |
| estimated | 0.465 | 0.178 | 0.164 | 0.139 | 0.026 | 0.028 | |
| Ser-57 | measured | 0.654 | 0.154 | 0.047 | 0.146 | ||
| estimated | 0.669 | 0.136 | 0.047 | 0.147 | |||
| Phe-57 | measured | 0.310 | 0.165 | 0.135 | 0.151 | 0.103 | 0.061 |
| estimated | 0.308 | 0.174 | 0.141 | 0.153 | 0.106 | 0.057 | |
| Asp-57 | measured | 0.495 | 0.237 | 0.128 | 0.106 | 0.033 | |
| estimated | 0.506 | 0.227 | 0.155 | 0.099 | 0.013 | ||
| Glu-57 | measured | 0.389 | 0.226 | 0.221 | 0.111 | 0.039 | 0.013 |
| estimated | 0.386 | 0.243 | 0.230 | 0.099 | 0.032 | 0.009 | |
| His-57 | measured | 0.411 | 0.209 | 0.130 | 0.103 | 0.058 | 0.058 |
| estimated | 0.422 | 0.243 | 0.123 | 0.097 | 0.053 | 0.054 | |
| Val-159 | measured | 0.496 | 0.156 | 0.258 | 0.054 | 0.036 | |
| estimated | 0.489 | 0.170 | 0.265 | 0.044 | 0.032 | ||
| Leu-159 | measured | 0.361 | 0.243 | 0.239 | 0.105 | 0.040 | 0.012 |
| estimated | 0.357 | 0.256 | 0.240 | 0.103 | 0.035 | 0.009 | |
| Ile-159 | measured | 0.398 | 0.224 | 0.229 | 0.099 | 0.036 | 0.013 |
| estimated | 0.386 | 0.243 | 0.230 | 0.099 | 0.032 | 0.009 | |
| Ser-159 | measured | 0.691 | 0.131 | 0.178 | |||
| estimated | 0.702 | 0.131 | 0.166 | ||||
| Phe-159 | measured | 0.350 | 0.157 | 0.194 | 0.109 | 0.093 | 0.046 |
| estimated | 0.323 | 0.170 | 0.212 | 0.103 | 0.103 | 0.042 |
The underlined values are those used for 13C MFA based on the threshold (value > 0.06, error < 3%). -57 and -159 in amino acids indicate [M-57] and [M-159] groups of GC-MS analysis, whose ion fragments have high intensities [26, 42, 43].
Table 3.
Mass isotopomer distributions of amino acids in glutamate production phase of C. glutamicum, where glutamate flux was 20 with respect to glucose uptake of 100.
| m0 | m1 | m2 | m3 | m4 | m5 | ||
| Ala-57 | measured | 0.702 | 0.129 | 0.047 | 0.122 | ||
| estimated | 0.706 | 0.135 | 0.041 | 0.117 | |||
| Gly-57 | measured | 0.782 | 0.087 | 0.131 | |||
| estimated | 0.790 | 0.083 | 0.127 | ||||
| Val-57 | measured | 0.529 | 0.166 | 0.139 | 0.111 | 0.029 | 0.026 |
| estimated | 0.527 | 0.178 | 0.148 | 0.111 | 0.019 | 0.017 | |
| Ser-57 | measured | 0.701 | 0.139 | 0.058 | 0.102 | ||
| estimated | 0.708 | 0.139 | 0.058 | 0.095 | |||
| Phe-57 | measured | 0.404 | 0.167 | 0.123 | 0.121 | 0.082 | 0.048 |
| estimated | 0.381 | 0.181 | 0.135 | 0.135 | 0.088 | 0.041 | |
| Asp-57 | measured | 0.572 | 0.215 | 0.117 | 0.077 | 0.019 | |
| estimated | 0.575 | 0.211 | 0.130 | 0.075 | 0.008 | ||
| Glu-57 | measured | 0.466 | 0.226 | 0.196 | 0.077 | 0.028 | 0.007 |
| estimated | 0.462 | 0.236 | 0.202 | 0.073 | 0.021 | 0.005 | |
| His-57 | measured | 0.487 | 0.199 | 0.117 | 0.100 | 0.048 | 0.036 |
| estimated | 0.492 | 0.228 | 0.112 | 0.082 | 0.035 | 0.045 | |
| Val-159 | measured | 0.550 | 0.164 | 0.210 | 0.050 | 0.026 | |
| estimated | 0.557 | 0.163 | 0.227 | 0.032 | 0.021 | ||
| Leu-159 | measured | 0.452 | 0.231 | 0.198 | 0.083 | 0.028 | 0.008 |
| estimated | 0.430 | 0.253 | 0.213 | 0.076 | 0.023 | 0.005 | |
| Ile-159 | measured | 0.471 | 0.225 | 0.194 | 0.076 | 0.026 | 0.008 |
| estimated | 0.462 | 0.236 | 0.202 | 0.073 | 0.021 | 0.005 | |
| Ser-159 | measured | 0.721 | 0.158 | 0.121 | |||
| estimated | 0.747 | 0.138 | 0.115 | ||||
| Phe-159 | measured | 0.418 | 0.169 | 0.167 | 0.092 | 0.077 | 0.037 |
| estimated | 0.402 | 0.170 | 0.200 | 0.085 | 0.085 | 0.029 |
The underlined values are those used for 13C MFA based on the threshold (value > 0.06, error < 3%).-57 and -159 in amino acids indicate [M-57] and [M-159] groups of GC-MS analysis, whose ion fragments have high intensities [26, 42, 43].
Table 4.
Mass isotopomer distributions of amino acids in glutamate production phase of C. glutamicum, where glutamate flux was 68 with respect to glucose uptake of 100.
| m0 | M1 | m2 | m3 | m4 | m5 | ||
| Ala-57 | measured | 0.676 | 0.145 | 0.025 | 0.154 | ||
| estimated | 0.670 | 0.140 | 0.042 | 0.148 | |||
| Gly-57 | measured | 0.768 | 0.073 | 0.158 | |||
| estimated | 0.769 | 0.072 | 0.159 | ||||
| Val-57 | measured | 0.463 | 0.189 | 0.164 | 0.133 | 0.021 | 0.030 |
| estimated | 0.472 | 0.179 | 0.164 | 0.134 | 0.025 | 0.026 | |
| Ser-57 | measured | 0.651 | 0.170 | 0.082 | 0.097 | ||
| estimated | 0.657 | 0.163 | 0.087 | 0.093 | |||
| Phe-57 | measured | 0.324 | 0.160 | 0.150 | 0.150 | 0.104 | 0.051 |
| estimated | 0.334 | 0.169 | 0.139 | 0.141 | 0.104 | 0.055 | |
| Asp-57 | measured | 0.586 | 0.185 | 0.102 | 0.116 | 0.010 | |
| estimated | 0.582 | 0.183 | 0.104 | 0.117 | 0.014 | ||
| Glu-57 | measured | 0.435 | 0.211 | 0.227 | 0.091 | 0.028 | 0.009 |
| estimated | 0.436 | 0.207 | 0.229 | 0.088 | 0.031 | 0.010 | |
| His-57 | measured | 0.477 | 0.223 | 0.099 | 0.073 | 0.032 | 0.080 |
| estimated | 0.466 | 0.232 | 0.099 | 0.072 | 0.047 | 0.071 | |
| Val-159 | measured | 0.484 | 0.162 | 0.272 | 0.050 | 0.031 | |
| estimated | 0.495 | 0.168 | 0.263 | 0.042 | 0.031 | ||
| Leu-159 | measured | 0.346 | 0.256 | 0.246 | 0.110 | 0.032 | 0.010 |
| estimated | 0.359 | 0.258 | 0.237 | 0.103 | 0.034 | 0.009 | |
| Ile-159 | measured | 0.446 | 0.201 | 0.228 | 0.086 | 0.033 | 0.007 |
| estimated | 0.436 | 0.207 | 0.229 | 0.088 | 0.031 | 0.010 | |
| Ser-159 | measured | 0.693 | 0.206 | 0.101 | |||
| estimated | 0.689 | 0.203 | 0.108 | ||||
| Phe-159 | measured | 0.351 | 0.164 | 0.212 | 0.084 | 0.105 | 0.040 |
| estimated | 0.350 | 0.162 | 0.214 | 0.084 | 0.105 | 0.037 |
The underlined values are those used for 13C MFA based on the threshold (value > 0.06, error < 3%).-57 and -159 in amino acids indicate [M-57] and [M-159] groups of GC-MS analysis, whose ion fragments have high intensities [26, 42, 43].
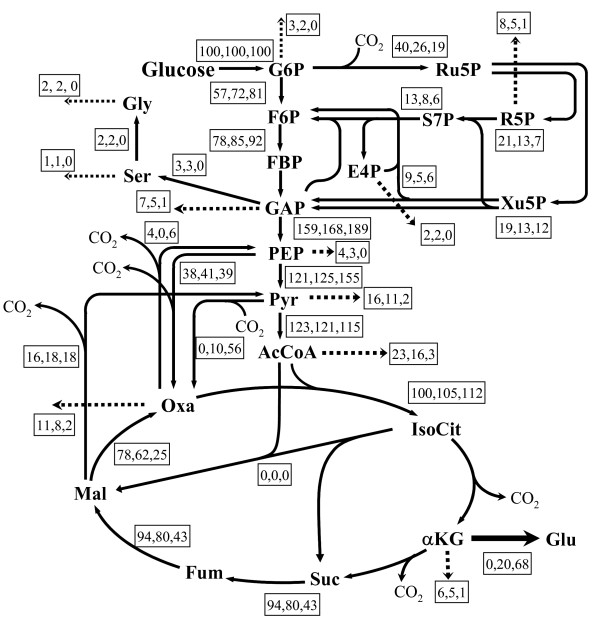
Figure 3.
Metabolic fluxes in C. glutamicum in growth and production phases of two different glutamate production activities. Dotted arrows indicate fluxes for biomass. Left, middle, and right values in boxes indicate fluxes in the growth and production phases, where glutamate fluxes were 20 and 68, respectively. In this study, the fluxes with backward (exchange) reactions, i.e., those in glycolysis, the pentose phosphate pathway, the latter steps of the TCA cycle (succinate → oxaloacetate), and C1 metabolisms, are shown as net values [22]. Abbreviations: Gly, glycine; Ser, serine; Glu, glutamate; G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; FBP, fructose-1,6-bisphosphate; GAP, glyceraldehyde-3-phosphate; PEP, phosphoenolpyruvate; Pyr, pyruvate; Ru5P, ribulose-5-phosphate; R5P, ribose-5-phosphate; Xu5P, xylulose-5-phosphate; S7P, sedoheptulose-7-phosphate; E4P, erythrose-4-phosphate; AcCoA, acetyl-CoA; IsoCit, isocitrate; αKG, 2-oxoglutarate; Suc, succinate; Fum, fumarate; Mal, malate; Oxa, oxaloacetate.
Table 5.
Measured values of amino acids by NMR analysis in growth phase of C. glutamicum.
| Amino Acid | Pre. | Is | Id1 | Id2 | Idd | It | |
| Ser C3 | GAP C3 | measured | 0.37 | 0.63 | |||
| estimated | 0.35 | 0.65 | |||||
| Gly C2 | GAP C2 | measured | 0.17 | 0.83 | |||
| estimated | 0.18 | 0.82 | |||||
| Ala C2 | Pyr C2 | measured | 0.09 | 0.06 | 0.13 | 0.72 | |
| estimated | 0.11 | 0.06 | 0.11 | 0.72 | |||
| Ala C3 | Pyr C3 | measured | 0.33 | 0.67 | |||
| estimated | 0.34 | 0.66 | |||||
| Val C4 | Pyr C3 | measured | 0.33 | 0.67 | |||
| estimated | 0.34 | 0.66 | |||||
| Leu C5 | Pyr C3 | measured | 0.33 | 0.67 | |||
| estimated | 0.34 | 0.66 | |||||
| Asp C3 | Oxa C3 | measured | 0.31 | 0.20 | 0.28 | 0.21 | |
| estimated | 0.31 | 0.21 | 0.29 | 0.19 | |||
| Thr C3 | Oxa C3 | measured | 0.36 | 0.54 | 0.11 | ||
| estimated | 0.40 | 0.51 | 0.09 | ||||
| Thr C4 | Oxa C4 | measured | 0.42 | 0.58 | |||
| estimated | 0.38 | 0.62 | |||||
| Glu C3 | αKG C3 | measured | 0.43 | 0.47 | 0.10 | ||
| estimated | 0.44 | 0.45 | 0.11 | ||||
| Arg C5 | αKG C5 | measured | 0.18 | 0.82 | |||
| estimated | 0.17 | 0.83 |
Symbols: Is, Id1, Id2, Idd, and It indicate coupling pattern of NMR, that is, singlet, doublet1, doublet2, double-doublet, and triplet, respectively. Abbreviations: Pre., precursor of amino acid. For others, see Fig. 1.
Table 6.
Mass isotopomer distributions of intermediate metabolites in glutamate production phase 1, where glutamate flux was 20 with respect to glucose uptake of 100.
| m0 | m1 | m2 | m3 | m4 | ||
| Pyruvate | measured | 0.701 | 0.131 | 0.055 | 0.113 | |
| estimated | 0.707 | 0.135 | 0.041 | 0.117 | ||
| Succinate | measured | 0.581 | 0.214 | 0.122 | 0.074 | 0.008 |
| estimated | 0.570 | 0.214 | 0.134 | 0.073 | 0.009 | |
| Fumarate | measured | 0.561 | 0.220 | 0.144 | 0.065 | 0.009 |
| estimated | 0.572 | 0.213 | 0.133 | 0.074 | 0.009 |
Table 7.
Mass isotopomer distributions of intermediate metabolites in glutamate production phase 2, where glutamate flux was 68 with respect to glucose uptake of 100.
| m0 | m1 | m2 | m3 | m4 | ||
| Pyruvate | measured | 0.669 | 0.129 | 0.052 | 0.149 | |
| estimated | 0.670 | 0.140 | 0.042 | 0.148 | ||
| Succinate | measured | 0.551 | 0.194 | 0.103 | 0.124 | 0.029 |
| estimated | 0.562 | 0.189 | 0.127 | 0.106 | 0.016 | |
| Fumarate | measured | 0.576 | 0.180 | 0.132 | 0.089 | 0.022 |
| estimated | 0.568 | 0.187 | 0.120 | 0.109 | 0.015 |
As the glutamate production increased with the amount of Tween 40 added, fluxes in the central metabolic pathway changed. Particularly, the flux distributions between glycolysis and the pentose phosphate pathway around the glucose-6-phosphate (G6P) branch point obviously changed. As the glutamate production increased, the flux distribution in the pentose phosphate pathway decreased (from 40 to 26 and 19).
Correlation between glutamate production and each flux in anaplerosis
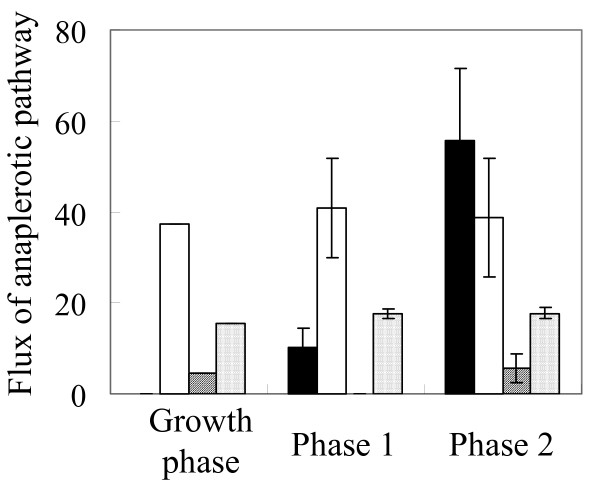
We determined which reaction in the anaplerotic pathways of C. glutamicum has the greatest correlation with the amount of glutamate produced by the quantitative analysis of the 13C MFA results. As shown in Fig. 4, reactions from PEP to oxaloacetate catalyzed by PEPc and from malate to pyruvate catalyzed by malic enzymes were mainly active in the growth phase. The flux catalyzed by PEPc, which showed a certain extent value (around 40) in the growth phase, was not significantly changed, even though the condition of fermentation was changed from cell growth to glutamate production by Tween 40 addition. The flux of the reaction from pyruvate to oxaloacetate catalyzed by Pc was almost zero in the growth phase. As glutamate production increased with the amount of Tween 40 added, the flux of this reaction catalyzed by Pc increased. The fluxes of the other anaplerotic pathways from oxaloacetate to PEP catalyzed by PEPck and glyoxylate shunt were almost zero in all cases.
Figure 4.
Comparison of fluxes in anaplerotic pathways in different phases. Black, white, zebra, and dotted bars indicate the reactions catalyzed by Pc, PEPc, PEPck, and malic enzymes, respectively. Phases 1 and 2 indicate the fermentations with the glutamate production fluxes of 20 and 68, respectively. Flux values of the anaplerosis in three phases (growth, Phase 1, and Phase 2) were as follows: 0, 10, 56 by Pc, 37, 41, 39 by PEPc, 4, 0, 6 by PEPck, and 13, 18, 18 by malic enzymes, respectively. Error bars indicate those with fluxes induced by the errors of GC-MS data, which were obtained from 20 sets of artificial GC-MS data with a variance due to the observed experimental errors (below 2% in this study), as previously described [25].
Discussion
In this study, different glutamate production activities dependent on the amount of Tween 40 added were obtained (Fig. 1). The GC-MS data of amino acids did not change 32–44 h after 13C-labeled glucose addition (48–60 h in the fermentation) (Fig. 2). This finding indicates that the condition of 13C in each amino acid in the cell reaches a plateau. The 13C labeling patterns of the proteinogenic amino acids, that is, the GC-MS data at the time point, indicate the information obtained only in the glutamate production phase. This is also confirmed by verifying the 13C MFA results in the two glutamate production phases (Tables 6 and 7). The GC-MS data of proteinogenic amino acids are good agreement with the estimated ones (Tables 3 and 4). Furthermore, the estimated ones are good agreement with the data of precursor ones which are considered to represent the conditions where sample was harvested and the extraction was performed (Tables 6 and 7). This strongly supports that the estimated fluxes using the GC-MS data of proteinogenic amino acids (Tables 3 and 4) represent the condition of the two glutamate production phase. Therefore, 13C MFAs in the glutamate production phases by obtaining the time courses of proteinogenic amino acids are considered to be successful.
As the glutamate production of C. glutamicum increased, the flux distribution in the pentose phosphate pathway around the glucose-6-phosphate branch point decreased from 40 to 26 and 19, as shown in Fig. 3. These results are similar to the previous study [10], and in contrast to those in the lysine production of C. glutamicum [24,25,27-30]. In lysine production, the energy metabolite NADPH, which is mainly produced via the pentose phosphate pathway, is necessary for not only cell growth but also lysine synthesis. Therefore, the flux distribution in the pentose phosphate pathway increases in the lysine production phase, compared with that in the growth phase. Also, in the glutamate production phase, NADPH is necessary for synthesizing glutamate from 2-oxoglutarate (α-KG) catalyzed by glutamate dehydrogenase (GDH). However, NADPH for glutamate synthesis was considered to be sufficiently supplied via a reaction from isocitrate to 2-oxoglutarate in the TCA cycle catalyzed by isocitrate dehydrogenase (ICDH) [31,32]. In lysine production, NADPH has to be supplied via the pentose phosphate pathway, since this reaction in the TCA cycle is related to lysine production. Therefore, as the cell growth decreased with the increase in the amount of Tween 40 added, the amount of NADPH necessary for the syntheses of the cell components decreased; that is, the flux distribution in the pentose phosphate pathway decreased. If glutamate production is more increased, and consequently cell growth was stopped completely as previous described [7,8], the flux on the pentose phosphate pathway would decrease more.
For glutamate production, anaplerotic pathways are essential, as mentioned in Background. In this study, the direct quantitative analysis of the flux changes in the anaplerotic pathways was performed by a 13C MFA method in the wild-type strain of C. glutamicum, which is different from the previously described method comparing the glutamate productions of the parental and recombinant strains [17]. The malic enzyme activities in three batch fermentations allow us to precisely and independently determine the fluxes of Pc and PEPc in anaplerosis, as previously described [16,21]. This was strongly supported by a 10% increase in the sum of absolute residual errors between the estimated and measured GC-MS data, when metabolic fluxes were re-determined by replacing the flux of Pc with that of PEPc. In the growth phase of C. glutamicum, the main active reaction was from PEP to oxaloacetate catalyzed by PEPc (Figs. 3 and 4). Although it might be claimed that this result is in contrast to the previous studies that strains deleted PEPc activity shows no growth defects on glucose [33,34]. In these reports, it is considered that Pc instead of PEPc is active due to the PEPc deletion It was also separately reported that not PEPc but Pc is mainly active in continuous cultures [16,35]. In this study, batch fermentations were performed, and thus, environmental condition around cell such as glucose and ammonia concentrations was quite different from those of continuous culture. In the growth phase in the batch cultures, PEPc is considered to be mainly active. However, in the glutamate production phase, this flux did not increase, even though a higher activity in anaplerosis was required than that in the growth phase. From the results of the quantitative 13C MFA in the glutamate production phase, it is considered that this reaction catalyzed by PEPc is not affected by environmental changes due to Tween 40 addition (Fig. 4). Peters-Wendisch and coworkers also found that PEPc is dispensable for glutamate production using a C. glutamicum WT (pMF1014) strain with a specific PEPc activity more than 10-fold higher than that of the wild-type strain [17,38]. It is found that the flux of Pc proportionally increases to glutamate production; in contrast, the flux of PEPc remains constant throughout fermentation (Fig. 4). This result directly indicates that the flux of Pc is the most important of those in the anaplerotic pathways in the glutamate overproduction induced by Tween 40 addition. It is considered that the flux of Pc would be more active as glutamate production increases, and simultaneously, cell growth is rarely induced using a larger amount of Tween 40 added than that in this study.
Furthermore, a backward flux from oxaloacetate catalyzed by PEPck, which was considered to be a negative factor for glutamate production, was inactive in both the growth and glutamate production phases of the batch fermentations. In the batch fermentations in this study, malic enzymes is active but PEPck is not active, owing to high ammonium ion concentration compared to that in continuous cultures [36], although this result is in contrast to that in continuous cultures as previously described [16,37]. Thus, the inhibition or attenuation of the PEPck activity of C. glutamicum by genetic modification might not be a necessary strategy for improving in glutamate production by Tween 40 addition. This would also be considered to be related to the cases involving malic enzymes and a glyoxylate shunt, because these fluxes did not change and were inactive in all the batch fermentations, as previously described [21,28].
Conclusion
We established experimental conditions under which both cell growth and glutamate production occurred simultaneously by Tween 40 addition. We quantitatively investigated how fluxes in the central metabolism of C. glutamicum change as glutamate production increases. The flux distribution in the pentose phosphate pathway around the glucose-6-phosphate branch point decreased as glutamate production increased. The flux of Pc proportionally increased to glutamate production despite the flux of PEPc remaining constant throughout batch fermentation. This result directly indicates that the flux of Pc is the most important of those in the anaplerotic pathways in the glutamate overproduction induced by Tween 40 addition, and is well consistent with that of the genetic evidence. The quantitative 13C MFA in the glutamate production phase in this study indicated which reaction in the anaplerotic pathways should be controlled; that is, what type of cell modification should be performed.
Methods
Bacterial strains and medium
C. glutamicum AJ-1511 (ATCC13869) was used. The same medium as that employed in our previous study was used for the preculture of the microorganism [26] (per liter deionized water): 40 g of glucose, 30 g of (NH4)2SO4, 3.0 g of Na2HPO4, 6.0 g of KH2PO4, 2.0 g of NaCl, 84 mg of CaCl2, 3.9 mg of FeCl3, 0.9 mg of ZnSO4·7H2O, 0.3 mg of CuCl2·H2O, 5.56 mg of MnSO4·5H2O, 0.1 mg of (NH4)6MO7O24·' m4H2O, 0.3 mg of Na2B4O7·10H2O, 0.4 g of MgSO4·7H2O, 40 mg of FeSO4·7H2O, 500 μg of vitamin B1·HCl, 0.1 g of EDTA, and 10 μg of biotin. The main culture medium composition was the same as the preculture medium composition except for the initial glucose concentration of 20 g/L. For the 13C MFA in the growth phase, the initial glucose (natural abundance of glucose) concentration was 15 g/L (see below and Fig. 1-a).
Culture conditions
The cultivation of C. glutamicum for 13C metabolic flux analysis (13C MFA) was carried out using a 250-mL jar fermenter with a liquid working volume of 80 mL (ABLE Co., Japan). The culture temperature was 31.5°C. The pH was maintained at 7.2 by adding 2.0 g of CaCO3 at the beginning of fermentation. The dissolved oxygen concentration was maintained above 3.0 mg/L by controlling the agitation speed in the range of 400–800 revolutions per minute (rpm). The aeration rate was maintained at 2 L of air volume per liter of liquid volume per minute (vvm). Two types of glutamate production were induced using two different amounts of Tween 40 added, that is, the final concentrations of 0.5 and 0.8 mg/mL. Subsequently, 1 mL of 400 g/L [1-13C]-labeled and 1 mL of [U-13C]-labeled glucose were added to the batch culture. For the 13C MFA in the growth phase, such components were added at the beginning of fermentation (see Fig. 1-a).
Off-line measurement
Cell concentration was measured at OD660. Glucose concentration was measured enzymatically using a glucose analyzer (Model-2700, YSI, USA). Glutamate concentration was measured by a colorimetric method using a glutamate kit (Roche Co., Germany), as previously described [8].
Sample preparation for GC-MS analysis
For 13C labeling experiment, cells were harvested in the log growth and glutamate production phases by centrifugation and then hydrolyzed in 6 M HCl at 105°C for 18 hours. Amino acids were purified and evaporated, as previously described [26,27]. For GC-MS, the amino acids were derivatized prior to their analysis. The dried hydrolysate was derivatized by N-(tert-butyldimethylsilyl (TBDMS))-N-methyl-trifluoroacetamide (MTBSTFA) [39,40], and 1 μL of the sample was injected to the GC-MS system.
For the GC-MS measurements of the 13C labeling patterns of the intermediate metabolites considered in this study (pyruvate, succinate, and fumarate), these metabolites were extracted from cells. In this study, the modified method previously described was used [41,42]. Cell broth (5 mL) was harvested at 60 and 69 h in two batch fermentations and immediately dropped in liquid nitrogen for cell quenching. Subsequently, cells were washed twice using 1.0% KCl at 4°C by centrifugation and added to 500 μL of methanol cooled at -20°C for 30 min. After centrifugation, the supernatant was used for GC-MS analysis after evaporation in dried nitrogen stream. The dried residue was redissolved and derivatized for 90 min at 30°C in 100 μL of 20 mg/mL methoxyamine hydrochloride in pyridine. Subsequently, trimethylsilylation (TMS derivatization) was performed for 30 min at 37°C and then for 2 hours at room temperature with 100 μL of N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA) [42,43]. The sample (1 μL) was injected to the GC-MS system.
Metabolic flux analysis (MFA)
The metabolic pathways for analyzing the flux distribution in glutamate production and the metabolic reaction model developed here were previously reported [26]. In the main part of the model, the central metabolic pathway including glycolysis, the tricarboxylic acid (TCA) cycle and the pentose phosphate pathway were considered for 13C MFA. Furthermore, the anaplerotic pathways (anaplerosis) essential for glutamate production and biosynthesis pathways for a cell were also considered in the model [26]. The fluxes for biomass synthesis of C. glutamicum were cited from Marx et al. [27], except for the biosynthesis fluxes of amino acids for proteins, which were calculated based on the content of each amino acid determined by GC-MS as previously described [26]. To evaluate precision of estimated fluxes, standard deviations of fluxes with respect to experimental errors was studied by investigating how much the estimated flux varied with such dispersion of artificial GC-MS data [26]. NMR analysis of the same amino acids was performed to check for consistency with the 13C labeling patterns estimated based on the MFA using only GC-MS.
GC-MS
GC-MS was carried out using a Shimadzu GC-17A gas chromatograph equipped with an InertCap1MS capillary column (25 m × 0.25 mm × 0.25 μm; GL Science, Tokyo) that was directly connected to a JMS-AMSUN200HS mass spectrometer (JEOL, Co., Japan), as previously described [26]. The settings of the measurements of TBDMS amino acid derivatives were used, as previously described [26].
For the measurements of TMS derivatives, an initial oven temperature of 70°C was maintained for 5 min, then increased to 300°C at 10°C/min and maintained for 8 min. The total running time was 35 min. The other settings were the same as those of the measurements of TBDMS derivatives (amino acids). In this study, the ion fragment of [M-15] was measured by GC-MS [43]. The GC-MS data of TMS derivatives were obtained by calculating the ratio of each intensity to the sum of all intensities in the [M-15] groups. For 13C MFA, correct mass data, where abundances of natural isotopes were removed from the raw GC-MS data, were used as previously described [44].
Authors' contributions
TS participated in the design of this study, performing fermentation experiments, the measurements of metabolites by GC-MS and the calculation of fluxes using computers. He analyzed the data and wrote the manuscript.
KF participated in the design of the study and measuring metabolites by GC-MS.
CF participated in the design of the study and reviewed the manuscript.
KN participated in the design of the study and reviewed the manuscript.
SS participated in the design of the study and reviewed the manuscript.
HS participated in the design of the study and reviewed the manuscript.
All authors have read and approved the manuscript.
Acknowledgments
Acknowledgements
This work was also supported in part by a grant of the 21st century Center of Excellence (COE) Program, a Grant-in-Aid for Scientific Research (B), and Special Coordination Funds for Promoting Science and Technology: Yuragi Project from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Contributor Information
Tomokazu Shirai, Email: shirai@bio.eng.osaka-u.ac.jp.
Koki Fujimura, Email: koki_fujimura@bio.eng.osaka-u.ac.jp.
Chikara Furusawa, Email: furusawa@ist.osaka-u.ac.jp.
Keisuke Nagahisa, Email: nagahisa@ist.osaka-u.ac.jp.
Suteaki Shioya, Email: shioya@bio.eng.osaka-u.ac.jp.
Hiroshi Shimizu, Email: shimizu@ist.osaka-u.ac.jp.
References
- Kinoshita S, Udaka S, Shimono M. Studies on the amino acid fermentation. J Gen Appl Microbiol Jpn. 1957;3:193–205. [PubMed] [Google Scholar]
- Shiio I, Otsuka S, Takahashi M. Effect of biotin on the bacterial formation of glutamic acid. I. Glutamate formation and cellular permeability of amino acids. J Biochem. 1962;51:56–62. doi: 10.1093/oxfordjournals.jbchem.a127500. [DOI] [PubMed] [Google Scholar]
- Takenami K, Yoshii H, Tsuri H, Okada H. Biochemical effect of fatty acid and its derivatives on glutamic acid fermentation. III. Biotin-tween 60 relationship in the accumulation of L-glutamic acid and the growth of Brevibacterium lactofermentum. Agric Biol Chem. 1965;29:351–359. [Google Scholar]
- Nunheimer TD, Birnbaum J, Ihnen ED, Demain AL. Product inhibition of the fermentative formation of glutamic acid. Appl Microbiol. 1970;20:215–217. doi: 10.1128/am.20.2.215-217.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay S, Gourdon P, Lapujade P, Mailly E, Oriol E, Engasser JM, Lindley ND, Goergen JL. An improved temperature-triggered process for glutamate production with Corynebacterium glutamicum. Enzyme Microbiol Technol. 1999;25:762–768. doi: 10.1016/S0141-0229(99)00120-9. [DOI] [Google Scholar]
- Kawahara Y, Takahashi-Fuke K, Shimizu E, Nakamatsu T, Nakamori S. Relationship between the glutamate production and the activity of 2-oxoglutarate dehydrogenase in Brevibacterium lactofermentum. Biosci Biotech Biochem. 1997;61:1109–1112. doi: 10.1271/bbb.61.1109. [DOI] [PubMed] [Google Scholar]
- Shimizu H, Tanaka H, Nakato A, Nagahisa K, Kimura E, Shioya S. Effects of the changes in enzyme activities on metabolic flux redistribution around the 2-oxoglutarate branch in glutamate production by Corynebacterium glutamicum. Bioprocess Biosyst Eng. 2003;25:291–298. doi: 10.1007/s00449-002-0307-8. [DOI] [PubMed] [Google Scholar]
- Shirai T, Nakato A, Izutani N, Nagahisa K, Shioya S, Kimura E, Kawarabayashi Y, Yamagishi A, Gojobori T, Shimizu H. Comparative study of flux redistribution of metabolic pathway in glutamate production by two coryneform bacteria. Metab Eng. 2005;7:59–69. doi: 10.1016/j.ymben.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Asakura Y, Kimura E, Usuda Y, Kawahara Y, Matsui K, Osumi T, Nakamatsu T. Altered metabolic flux deletion of odhA causes L-glutamate overproduction in Corynebacterium glutamicum. Appl Env Microb. 2007;73:1308–1319. doi: 10.1128/AEM.01867-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx A, Striegel T, de Graaf AA, Sahm H, Eggeling L. Response of the central metabolism of Corynebacterium glutamicum to different flux burdens. Biotech Bioeng. 1997;56:168–180. doi: 10.1002/(SICI)1097-0290(19971020)56:2<168::AID-BIT6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Ozaki H, Shiio I. Regulation of the TCA and glyoxylate cycles in Brevibacterium flavum. II. Regulation of phosphoenolpyruvate carboxylase and pyruvate kinase. J Biochem. 1969;66:297–311. doi: 10.1093/oxfordjournals.jbchem.a129148. [DOI] [PubMed] [Google Scholar]
- Mori M, Shiio I. Purification and some properties of phosphoenolpyruvate carboxylase from Brevibacterium flavum and its aspartate-overproducing mutant. J Biochem. 1985;97:1119–28. doi: 10.1093/oxfordjournals.jbchem.a135156. [DOI] [PubMed] [Google Scholar]
- Peters-Wendisch PG, Wendisch VF, Paul S, Eikmanns BJ, Sahm H. Pyruvate carboxylase as an anaplerotic enzyme in Corynebacterium glutamicum. Microbiology. 1997;143:1095–1103. doi: 10.1099/00221287-143-4-1095. [DOI] [PubMed] [Google Scholar]
- Jetten MSM, Sinskey AJ. Characterization of phosphoenolpyruvate carboxykinase from Corynebacterium glutamicum. FEMS Microbiol Lett. 1993;111:183–188. doi: 10.1111/j.1574-6968.1993.tb06383.x. [DOI] [Google Scholar]
- Cocaign-Bousquet M, Guyonvarch A, Lindley ND. Growth rate-dependent modulation of carbon flux through central metabolism and the kinetic consequences for glucose-limited chemostat cultures of Corynebacterium glutamicum. Appl Environ Microbiol. 1996;62:429–436. doi: 10.1128/aem.62.2.429-436.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen S, de Graaf AA, Eggeling L, Möllney M, Wiechert W, Sahm H. In vivo quantification of parallel and bidirectional fluxes in the anaplerosis of Corynebacterium glutamicum. J Biol Chem. 2000;275:35932–35941. doi: 10.1074/jbc.M908728199. [DOI] [PubMed] [Google Scholar]
- Peters-Wendisch PG, Schiel B, Wendisch VF, Katsoulidis E, Mökel B, Sahm H, Eikmanns BJ. Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J Mol Microbiol Biotechnol. 2001;3:295–300. [PubMed] [Google Scholar]
- Schmidt K, Nielsen J, Villadsen J. Quantitative analysis of metabolic fluxes in Escherichia coli, using two-dimensional NMR spectroscopy and complete isotopomer models. J Biotech. 1999;71:175–190. doi: 10.1016/S0168-1656(99)00021-8. [DOI] [PubMed] [Google Scholar]
- Yang C, Hua Q, Shimizu K. Quantitative analysis of intracellular metabolic fluxes using GC-MS and two-dimensional NMR spectroscopy. J Biosci Bioeng. 2002;93:78–87. [PubMed] [Google Scholar]
- Fischer E, Sauer U. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur J Biochem. 2003;270:880–891. doi: 10.1046/j.1432-1033.2003.03448.x. [DOI] [PubMed] [Google Scholar]
- Klapa MI, Aont JC, Stephanopoulos G. Systematic quantification of complex metabolic flux networks using stable isotopes and mass spectrometry. Eur J Biochem. 2003;270:3525–3542. doi: 10.1046/j.1432-1033.2003.03732.x. [DOI] [PubMed] [Google Scholar]
- Frick O, Wittmann C. Characterization of the metabolic shift between oxidative and fermentative growth in Saccharomyces cerevisiae by comparative 13C flux analysis. Microb Cell Fact. 2005;3:30. doi: 10.1186/1475-2859-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann C. Fluxome analysis using GC-MS. Microb Cell Fact. 2007 doi: 10.1186/1475-2859-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann C, Heinzle E. Modeling and experimental design for metabolic flux analysis of lysine-producing corynebacteria by mass spectrometry. Metab Eng. 2001;3:173–191. doi: 10.1006/mben.2000.0178. [DOI] [PubMed] [Google Scholar]
- Wittmann C, Heinzle E. Genealogy profiling through strain improvement by using metabolic network analysis: metabolic flux genealogy of several generations of lysine-producing corynebacteria. Appl Environ Microbiol. 2002;68:5843–5859. doi: 10.1128/AEM.68.12.5843-5859.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai T, Matsuzaki K, Kuzumoto M, Nagahisa K, Furusawa C, Shioya S, Shimizu H. Precise metabolic flux analysis of coryneform bacteria by gas chromatography-mass spectrometry and verification by nuclear magnetic resonance. J Biosci Bioeng. 2006;102:413–424. doi: 10.1263/jbb.102.413. [DOI] [PubMed] [Google Scholar]
- Marx A, de Graaf AA, Wiechert W, Eggeling L, Sahm H. Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing. Biotechnol Bioeng. 1996;49:111–129. doi: 10.1002/(SICI)1097-0290(19960120)49:2<111::AID-BIT1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Wendisch VF, de Graaf AA, Sahm H, Eikmanns BJ. Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J Bacteriol. 2002;182:3088–3096. doi: 10.1128/JB.182.11.3088-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz B, Striegel K, de Graaf AA, Sahm H. Changes of pentose phosphate pathway flux in vivo in Corynebacterium glutamicum during leucine-limited batch cultivation as determined from intracellular metabolite concentration measurements. Metab Eng. 2002;4:295–305. doi: 10.1006/mben.2002.0233. [DOI] [PubMed] [Google Scholar]
- Wittmann C, Kim HM, Heinzle E. Metabolic network analysis of lysine-producing Corynebacterium glutamicum at a miniaturized scale. Biotechnol Bioeng. 2004;87:1–6. doi: 10.1002/bit.20103. [DOI] [PubMed] [Google Scholar]
- Eikmanns BJ, Rittmann D, Sahm H. Cloning, sequence analysis, expression, and inactivation of the Corynebacterium glutamicum icd gene encoding isocitrate dehydrogenase and biochemical characterization of the enzyme. J Bacteriol. 1995;177:774–782. doi: 10.1128/jb.177.3.774-782.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RD, Gadal P. Structure, functions, and regulation of NAD and NADP dependent isocitrate dehydrogenases in higher plants and in other organisms. Plant Physiology Biochem. 1990;28:411–427. [Google Scholar]
- Peters-Wendisch PG, Eikmanns BJ, Thierbach G, Bachmann B, Sahm H. Phosphoenolpyruvate carboxylase in Corynebacterium glutamicum is dispensable for growth and lysine production. FEMS Microbiol Lett. 1993;112:269–274. doi: 10.1111/j.1574-6968.1993.tb06461.x. [DOI] [Google Scholar]
- Gubler M, Park SM, Jetten M, Stephanopoulos G, Sinskey AJ. Effects of phosphoenol pyruvate carboxylase deficiency on metabolism and lysine production in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 1994;40:857–863. doi: 10.1007/s002530050080. [DOI] [Google Scholar]
- Petersen S, Mack C, de Graaf AA, Riedel C, Eikmanns BJ, Sahm H. Metabolic consequences of altered phosphoenolpyruvate carboxykinase activity in Corynebaterium glutamicum reveal anaplerotic regulation mechanisms in vivo. Metab Eng. 2001;3:344–361. doi: 10.1006/mben.2001.0198. [DOI] [PubMed] [Google Scholar]
- Gourdon P, Baucher MF, Lindley ND, Guyonvarch A. Cloning of the malic enzyme gene from Corynebacterium glutamicum and role of the enzyme in lactate metabolism. Appl Environ Microbiol. 2000;66:2981–2987. doi: 10.1128/AEM.66.7.2981-2987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel C, Rittmann D, Dangel P, Mockel B, Petersen S, Sahm H, Eikmanns BJ. Characterization of the phosphoenolpyruvate carboxykinase gene from Corynebacterium glutamicum and significance of the enzyme for growth and amino acid production. J Mol Microbiol Biotechnol. 2001;3:573–583. [PubMed] [Google Scholar]
- Cremer J, Eggeling L, Sahm H. Control of the lysine biosynthetic sequence in Corynebacterium glutamicum as analyzed by overexpression of the individual corresponding genes. Appl Environ Microbiol. 1991;57:1746–1752. doi: 10.1128/aem.57.6.1746-1752.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawhinny TP, Robinett RSR, Atalay A, Madson MA. Analysis of amino acids as their tert-butyldimethylsilyl derivatives by gas-liquid chromatography and mass spectrometry. J Chromatogr. 1986;358:231–242. doi: 10.1016/S0021-9673(01)90333-4. [DOI] [PubMed] [Google Scholar]
- MacKenzie SL, Tenaschuk D, Fortier G. Analysis of amino acids by gas-liquid chromatography as tert-butyldimethylsilyl derivatives: preparation of derivatives in single reaction. J Chromatogr. 1987;387:241–253. doi: 10.1016/S0021-9673(01)94528-5. [DOI] [PubMed] [Google Scholar]
- Devantier R, Scheithauer B, Villas-Bôas SG, Pedersen S, Olsson L. Metabolite profiling for analysis of yeast stress response during very high gravity ethanol fermentations. Biotechnol Bioeng. 2005;90:703–714. doi: 10.1002/bit.20457. [DOI] [PubMed] [Google Scholar]
- Strelkov S, von Elstermann M, Schomburg D. Comprehensive analysis of metabolites in Corynebacterium glutamicum by gas chromatography/mass spectrometry. Biol Chem. 2004;385:853–861. doi: 10.1515/BC.2004.111. [DOI] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 2000;23:131–142. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- van Winden WA, Wittmann C, Heinzle E, Heijnen JJ. Correcting mass isotopomer distributions for naturally occurring isotopes. Biotechnol Bioeng. 2002;80:477–479. doi: 10.1002/bit.10393. [DOI] [PubMed] [Google Scholar]