Abstract
The binding of oxidatively damaged red blood cells (OxRBCs) to resident mouse peritoneal macrophages correlates with an increase in phosphatidylserine on the external leaflet of the plasma membrane. Liposomes rich in phosphatidylserine can inhibit this binding and also the binding of certain apoptotic cells. We have shown previously that oxidized low density lipoproteins (OxLDL) also can inhibit the binding of OxRBCs to resident mouse peritoneal macrophages. The present studies show that microemulsions prepared from the lipids extracted from OxLDL are very effective in inhibiting the binding of OxRBCs and also, to a lesser extent, of apoptotic thymocytes to macrophages. OxRBC binding was also inhibited by cholesterol phospholipid liposomes containing oxidized 1-stearoyl-2-linoleoyl-phosphatidylcholine. The binding and uptake of 125I-labeled OxLDL were also strongly inhibited by microemulsions of the lipids extracted from OxLDL and by cholesterol phospholipid liposomes containing oxidized 1-palmitoyl-2-arachidonoyl-phosphatidylcholine. Earlier studies have shown that the delipidated protein moiety of OxLDL can competitively inhibit macrophage binding of intact OxLDL, implicating the protein moiety as an effective receptor-binding domain of OxLDL with respect to some macrophage scavenger receptors. The present studies suggest that the lipid moiety of OxLDL may also play a role.
We have shown previously that the binding of oxidatively damaged red blood cells (OxRBCs) to resident mouse peritoneal macrophages can be strongly inhibited by oxidized low density lipoprotein (OxLDL) (1, 2). The same was true for the binding of apoptotic thymocytes, although the extent of inhibition was not as great. The implication was that one or more of the receptors involved in the binding of OxLDL could also bind OxRBCs and apoptotic cells. Now, the binding of OxLDL to scavenger receptors is generally assumed to be mediated by the modified apoprotein B—modified by direct oxidative attack and by conjugation to oxidized lipid moieties, such as malondialdehyde and 4-hydroxynonenal. The fact that acetylation (and a number of other chemical modifications that block lysine ɛ-amino groups) under nonoxidative conditions also generates a modified LDL that can compete with OxLDL strongly supports that view. During LDL oxidation, lysine ɛ-amino groups are indeed progressively masked (3). Moreover, Parthasarathy et al. (4) showed that the delipidated protein moiety of OxLDL, resolubilized in octyl glucoside, can competitively inhibit uptake and degradation of intact OxLDL and vice versa. So there would seem to be little doubt that the modified apoprotein of OxLDL plays a major role in its binding and uptake via some macrophage receptors. It should be noted, however, that Parthasarathy et al. (4) did not directly test whether the lipids of OxLDL had any ability to compete with intact OxLDL.
On the other hand, the binding of OxRBCs and, under some conditions, the binding of apoptotic cells to macrophages correlate with an increase in the appearance of phosphatidylserine (PS) on the external leaflet of the plasma membrane of the damaged cells (2, 5–9) and pure lipid liposomes rich in PS are effective competitors for macrophage binding of OxRBCs and, under some conditions, apoptotic cells also. These findings strongly imply that in this case the macrophage is indeed recognizing and binding lipids on the cell surface of the damaged cell and that the protein of the damaged cell is not necessarily involved. How are we to rationalize this apparent discordance—recognition of lipids when an OxRBC is the ligand and recognition of protein when OxLDL is the ligand? And how do we account for the competition of OxLDL for OxRBC binding? The present studies were undertaken to explore the possibility that the lipid moiety of OxLDL may bind to some scavenger receptors and thus play a role in the ability of OxLDL to inhibit the binding of OxRBCs to mouse peritoneal macrophages.
MATERIALS AND METHODS
Materials.
CuSO4, dexamethasone, EDTA, ascorbic acid, and cholesterol were from Sigma; acetic anhydride and 2,6-di-tert-butyl-p-cresol were from Aldrich; all phospholipids were from Avanti Polar Lipids; 3,3′-dihexadecyloxacarbocyanine (DiO) were from Molecular Probes; DMEM was from Life Technologies; RPMI 1640 medium was from BioWhittaker; fetal bovine serum and penicillin-streptomycin were from Gemini Biological Products (Calabasas, CA); Hepes was from Irvine Scientific; 125I-labeled sodium iodide was from ICN; d-octyl glucoside was from Boehringer-Mannheim; 0.1-μm polycarbonate membranes were from Poretics; and 70-μm nylon cell strainers were from Falcon.
Cells.
Resident mouse peritoneal macrophages were isolated from 6- to 8-week-old female Swiss-Webster mice by peritoneal lavage with PBS. Thymocytes were isolated by disrupting murine thymus tissue and passing it through a 70-μm cell strainer. Thymocytes were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum and penicillin-streptomycin for 6 hr in the presence of 1 μM dexamethasone to induce apoptosis. Control thymocytes were kept in RPMI/10% fetal bovine serum at 4°C. Red blood cells (RBCs) were isolated from the blood of healthy male volunteers, washed three times with PBS, and stored at 20% hematocrit in PBS at 4°C. RBCs were used for binding and phagocytosis within 2 days. OxRBCs and opsonized RBCs were prepared as described (1).
Lipoproteins.
Human LDL (d = 1.019–1.063) was isolated from fresh plasma containing 100 μM EDTA by preparative ultracentrifugation (10). To protect native LDL from oxidation during isolation and handling, 30 μM 2,6-di-tert-butyl-p-cresol was added. No 2,6-di-tert-butyl-p-cresol was added to LDL intended for oxidation; it was dialyzed, adjusted to 100 μg/ml, and incubated for 18 hr in PBS with 10 μM CuSO4. Acetylation was carried out with acetic anhydride by the method of Basu et al. (11). The degree of oxidation was measured by assessing electrophoretic mobility on a 1% agarose gel and by measuring fatty acid composition by gas liquid chromatography. Lipoprotein protein and cell protein were measured according to Lowry et al. (12).
Lipid Extraction and Liposome Preparation.
Lipids from native LDL, from OxLDL, and from acetylated LDL were extracted with chloroform/methanol. To 2 mg of lipoprotein in a total volume of 1.8 ml of PBS containing 0.01 M HCl, 2 ml of methanol and 2 ml of chloroform were added. After vortex mixing for 10 sec, the mixture was centrifuged at 1,800 × g for 10 min. The chloroform phase was removed and dried under a stream of N2. To the dried lipids, PBS/10 mM Hepes was added and vortex mixed until the lipids were in suspension, presumably as a microemulsion.
These microemulsions were then extruded 8–10 times through 0.1-μm polycarbonate membranes under N2 flow (13). During extrusion, the temperature was kept at 37°C for the natural phospholipid mixtures and also for lipids from OxLDL but at 45°C for saturated phospholipids (distearoylphosphatidylcholine and dimyristoylphosphatidylserine) and for oxidized 1-stearoyl-2-linoleoyl-phosphatidylcholine. Labeled dipalmitoylphosphatidylserine liposomes were prepared by incorporating the fluorescent dye DiO into the lipid mixture (1%). The size and heterogeneity of the microemulsions and liposomes were assessed by light scattering and by electron microscopy of negatively stained preparations. Liposomes prepared from partially purified natural phospholipids or from synthetic phospholipids [phospholipid/cholesterol, 2:1 (mol/mol)] were relatively uniform (diameters, 80–120 nm). Microemulsions prepared from the lipids extracted from either native LDL or oxidized LDL were more heterogeneous and larger (200–600 nm) because of some aggregation. Each particle was centrally electron dense and showed a multilamellar periphery consisting of three to six layers, presumably phospholipid. The protein content of these microemulsions was estimated by acid hydrolysis of the unfractionated preparation (2 M HCl, 50–56 min at 160°C, under vacuum or under argon) followed by quantitative amino acid analysis using ion-exchange chromatography.
Fatty Acid Composition.
Lipids from liposomes were extracted by a modification of the method of Folch et al. (14). The fatty acids were transmethylated (15) and analyzed in a Varian gas chromatograph model 3700, equipped with a column of 10% Silar 5 CP on 100/120 Gas Chrom Q2. Total lipid concentrations in the liposome samples used were estimated from the total fatty acids measured by gas chromatography and expressed as mg/ml. This underestimates the true value for OxLDL lipids to the extent that polyunsaturated fatty acids have been degraded.
Binding and Phagocytosis of RBCs and Thymocytes.
Resident mouse peritoneal macrophages (0.6 × 106 cells per ml) were plated in 48-well plates in DMEM/10% fetal calf serum/penicillin-streptomycin and allowed to adhere for 4 hr. Wells were washed four times with DMEM before incubation with RBCs or thymocytes. RBCs (final concentration, 0.1% hematocrit) or thymocytes (10 thymocytes per macrophage) were added in DMEM/10 mM Hepes and incubated at 37°C for 1 hr in the presence of competitors as indicated. Nonadherent cells were removed by washing five times with DMEM. The percentage of macrophages binding or phagocytosing one or more RBCs or thymocytes was counted.
Binding of Liposomes.
Macrophages (1 × 106 cells per well) were incubated with DiO-labeled liposomes for 2 hr at 4°C, washed, scraped gently from the bottoms of the wells, and resuspended in PBS/0.1% BSA/0.01% NaN3. Mean fluorescence intensity of 10,000 events per incubation was measured by flow cytometry (Becton Dickinson) and analyzed by using cellquest software.
Statistical Analysis.
All cell binding and phagocytosis assays were done in triplicate and at least 100 macrophages per well were counted. Results are expressed as the mean ± SD and P values were calculated with ANOVA.
RESULTS
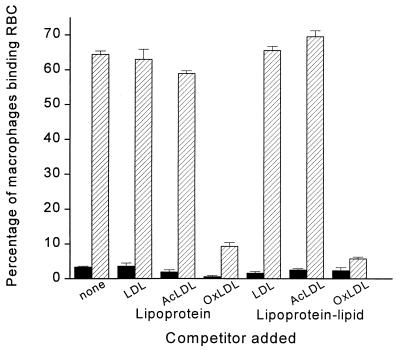
As shown in Fig. 1, when resident mouse peritoneal macrophages were incubated with native unopsonized RBCs, fewer than 5% of the macrophages showed any binding. In contrast, almost 65% of them bound one or more RBCs when incubated with OxRBCs. The addition of native LDL or of acetyl LDL had no significant effect on the binding of OxRBCs, but the addition of intact OxLDL almost completely blocked the binding, in agreement with the results of previous studies from this laboratory (1). We then extracted the lipids from native LDL, acetyl LDL, and OxLDL, prepared microemulsions, and tested the ability of these microemulsions to inhibit the binding of OxRBCs. As shown in Fig. 1, the microemulsions prepared from the lipids of native LDL or acetyl LDL had no effect, whereas those prepared from the lipids of OxLDL inhibited to about the same extent as did intact OxLDL. As shown in Fig. 1, binding of OxRBCs was inhibited by 87 ± 10% (n = 7). Phagocytosis of OxRBCs (data not shown) was inhibited to the same extent—86 ± 5% (n = 5). As a control for possible nonspecific effects of the oxidized lipids on RBC binding, we tested for inhibition of the Fc receptor-mediated binding of opsonized RBCs by using the same protocol and found none. In control wells, opsonized RBCs were bound by 45.9 ± 5.4% of the macrophages; in the presence of microemulsions prepared from the lipids of native LDL and of OxLDL, the values were 45.3 ± 9.4% and 56.9 ± 4.1%, respectively.
Figure 1.
Binding of native RBCs (solid bars) and OxRBCs (hatched bars) to resident mouse peritoneal macrophages in the presence of lipoproteins (100 μg of protein per ml) or lipoprotein–lipid microemulsions (25 μg/ml). Data are the mean ± SD of triplicate samples. The experiment was repeated twice with similar results.
When LDL is extensively oxidized, the apoprotein B breaks down to yield a mixture of smaller fragments, most more than 80 kDa but some as small as 14 kDa (16). It was therefore important to rule out carry-over of apoprotein B fragments into the liposomes prepared from OxLDL lipids. This was done by subjecting the microemulsion preparations to acid hydrolysis and carrying out an amino acid analysis. Microemulsions prepared from the lipids extracted from 1 mg of native LDL (expressed in terms of protein) contained 6.3 μg of total amino acids, presumably representing low molecular weight contaminants. Recovery of added BSA (20 μg) added as an internal standard was quantitative. Microemulsions prepared from the lipids extracted from OxLDL (1 mg of lipoprotein protein) contained 13 μg of total amino acids. This would correspond to a maximum of 26 pmol of intact apoprotein B. Because the concentrations of microemulsions needed to inhibit OxRBC binding were in the range of 25–50 μg of total fatty acids per ml, it seems unlikely that protein contamination can explain the observed results.
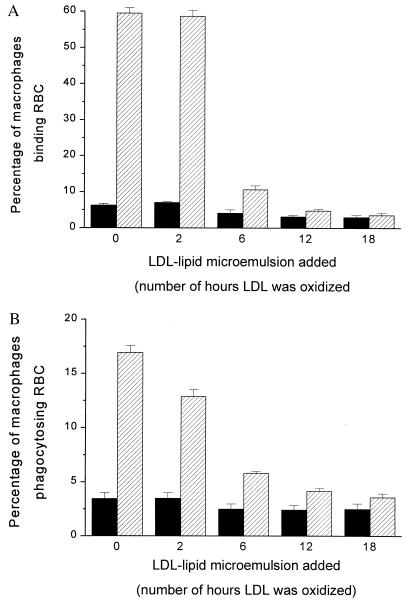
The OxLDL used in the experiment shown in Fig. 1 had been incubated 18 hr in PBS in the presence of 10 μM CuSO4. To determine the effects of different degrees of oxidation, we prepared OxLDL varying the incubation time from 2 to 18 hr. As shown in Fig. 2, microemulsions prepared from the lipids of LDL oxidized for only 2 hr had little or no effect on binding or phagocytosis of OxRBCs; microemulsions prepared from the lipids of LDL oxidized for 6 hr inhibited phagocytosis by about 60% (Fig. 2B); and a maximal effect was seen at 12 hr (80% inhibition). Incubation with Cu2+ for 2 hr approximates the degree of oxidation in minimally oxidized LDL as studied by Kim et al. (17). Minimally oxidized LDL is defined in part by the fact that it is still recognized by the native LDL receptor and that it is not a ligand for the scavenger receptors (18). Thus the failure of microemulsions made from the lipids of minimally oxidized LDL to inhibit OxRBC binding is consistent with the receptor binding properties of the intact lipoproteins.
Figure 2.
Binding (A) and phagocytosis (B) of native RBCs (solid bars) and OxRBCs (hatched bars) to resident mouse peritoneal macrophages in the presence of microemulsions (50 μg of fatty acids per ml) prepared from lipids of LDL oxidized to different degrees. The time of incubation of LDL with CuSO4 (10 μM) is indicated in hours. Lipids were extracted from LDL after oxidation. Data are the mean ± SD of three determinations in a single experiment.
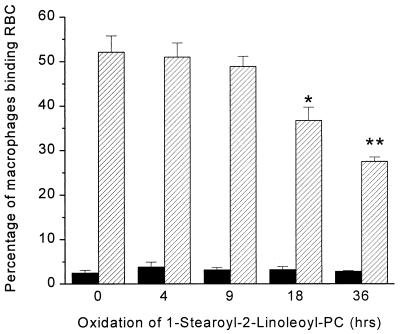
One way to explain the competition by OxLDL (or by its lipid components in a microemulsions) is that oxidation of phospholipids generates more polar moieties that possess some of the properties of PS-rich liposomes. Thus, intact OxLDL and PS-rich liposomes show reciprocal competition for binding to macrophages (2, 13). It is well established that the polyunsaturated fatty acids in the sn-2 position of phospholipids undergo oxidation to yield a variety of modified products, including hydroperoxides of the same carbon chain length as the original together with an array of fragmented fatty acid chains (aldehydes, ketones, carboxylic acids, and alkanes) that can remain attached via ester linkage to the glycerol backbone or, eventually, be completely lost, generating lysolecithin (19–22). We therefore tested the possibility that an oxidized phospholipid might yield a liposome that would compete with OxRBCs. Pure 1-stearoyl-2-linoleoyl-phosphatidylcholine, either in its native state or after oxidation to different degrees, was incorporated into liposomes along with free cholesterol (molar ratio, 2:1). As shown in Fig. 3, the liposomes containing highly oxidized 1-stearoyl-2-linoleoyl-phosphatidylcholine gave highly significant inhibition of the binding of OxRBCs, whereas the native phospholipid or the phospholipid oxidized less extensively gave little or no inhibition. Analysis of the fatty acid composition of the phospholipid after 36 hr of oxidation in air showed that 42% of the linoleic acid had been destroyed; after 18 hr 26% had been destroyed. At the shorter times, there was no significant change in the initial linoleic acid content (46.4% of total fatty acid) and no significant inhibition of OxRBC binding.
Figure 3.
Binding of native RBCs (solid bars) and OxRBCs (hatched bars) to resident mouse peritoneal macrophages in the presence of liposomes prepared from 1-stearoyl-2-linoleoyl-phosphatidylcholine and cholesterol (2:1 molar ratio) at 15 μg of fatty acid per ml. Data are the mean ± SD of three determinations. The experiment was done twice with similar results. ∗, P < 0.005; ∗∗, P < 0.0005.
Table 1 summarizes the effects of microemulsions on the binding of OxRBCs and of apoptotic thymocytes to macrophages. The results were very similar for OxRBCs and for apoptotic thymocytes, with the exception that inhibition of the latter by microemulsion prepared from the lipids of OxLDL and by liposomes containing oxidized 1-stearoyl-2-linoleoyl-phosphatidylcholine was less than the inhibition of binding of OxRBCs. Note that liposomes prepared from fully saturated PS were also inhibitory, showing that PS need not be oxidized to exert its inhibitory effect. However, it will be of interest to determine whether PS liposomes containing oxidized fatty acids in the sn-2 positions would be still more effective as inhibitors.
Table 1.
Effects of microemulsions on binding of OxRBCs and of apoptotic thymocytes by resident mouse peritoneal macrophages
| Ligand | Addition | Concentration, μg/ml* | % inhibition |
|---|---|---|---|
| OxRBCs | LDL–lipid microemulsions (n = 6)† | 20–30 | 2 ± 3 |
| OxLDL–lipid microemulsions (n = 7) | 20–30 | 87 ± 10 | |
| PS liposomes (n = 8) | 30–35 | 95 ± 4 | |
| Saturated-PS liposomes‡(n = 6) | 30–35 | 81 ± 8 | |
| PC liposomes (n = 5) | 30–35 | 3 ± 4 | |
| Oxidized SLPC liposomes§(n = 4) | 30–35 | 55 ± 10 | |
| Apoptotic thymocytes | LDL–lipid microemulsion (n = 3) | 20–30 | 2 ± 2 |
| OxLDL–lipid microemulsion (n = 2) | 20–30 | 47 ± 4 | |
| PS liposomes (n = 4) | 30–35 | 89 ± 3 | |
| Saturated-PS liposomes (n = 1) | 31 | 87 | |
| PC liposomes (n = 1) | 33 | 0 | |
| Oxidized SLPC liposomes (n = 1)§ | 33 | 33 |
Concentrations of liposome lipids added are expressed in terms of the total fatty acid content determined by gas liquid chromatography.
n indicates the number of experiments. Each experiment was done in triplicate.
Saturated PS liposomes were made from 1,2-dimyristoyl-phosphatidylserine or 1,2-dipalmitoyl-phosphatidylserine in a 1:1:1 molar ratio with unoxidized, fully saturated phosphatidylcholine and cholesterol.
1-stearoyl-2-linoleoyl-phosphatidylcholine (SLPC) that had been oxidized for 18–36 hr.
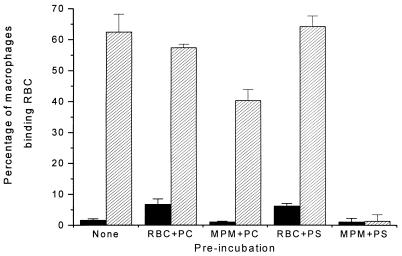
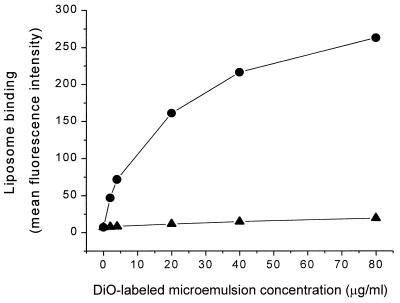
It was necessary to consider the possibility that the inhibition of OxRBC binding to the macrophage might be due to interaction of the liposome with the OxRBC rather than interaction with the surface of the macrophage. To test this possibility, we preincubated OxRBCs with phosphatidylcholine (PC) or PS liposomes for 30 min at 37°C, washed several times to remove unbound liposomes, and then added the washed OxRBCs to the macrophage culture for 1 hr. As shown in Fig. 4 this preincubation, whether with PC liposomes or PS liposomes, did not significantly alter the subsequent binding of OxRBCs to the macrophage. In contrast, preincubation of the macrophages with PS liposomes almost completely blocked the subsequent binding of OxRBCs. There was in this experiment some inhibition also by preincubation of the macrophages with PC liposomes but this was only about 35% compared with the 90% inhibition on preincubation with PS liposomes. In our experience PC preparations can undergo oxidation during handling and storage, in which case they can compete for OxRBC binding (data now shown). The results shown in Fig. 4 are compatible with the interpretation that PS liposomes and OxRBCs compete for one or more saturable receptor sites on the macrophage plasma membrane. That interpretation is also supported by the fact that binding of DiO-labeled microemulsions prepared from OxLDL lipids, shown in Fig. 5, was concentration-dependent and included a sizable saturable component (saturating at 20–30 μg of lipid per ml), whereas the labeled microemulsions prepared from native LDL lipids showed almost no binding.
Figure 4.
Comparison of the effects of preincubation of either the mouse peritoneal macrophages (MPM) or the OxRBCs with liposomes on the subsequent binding of OxRBCs to MPM (hatched bars). Binding of native RBC is shown for reference (solid bars). PC, liposomes prepared from egg PC/cholesterol (molar ratio 2:1); PS, liposomes prepared from brain PS/egg PC/cholesterol (molar ratio 1:1:1). Macrophages or RBCs were preincubated with liposomes (25 μg of fatty acids per ml) in DMEM for 30 min at 37°C before assaying RBC binding. Data are the mean ± SD of three determinations in a single experiment. Preincubations of MPM with PS liposomes inhibited subsequent OxRBC binding by more than 90% (P < 0.0001). Preincubation of MPM with PC liposomes inhibited by less than 35% (P < 0.01).
Figure 5.
Binding of fluorescence-labeled LDL-lipid (▴) or OxLDL-lipid (•) microemulsions to resident mouse peritoneal macrophages, measured by flow cytometry. Liposomes were labeled by incorporating 1% DiO (wt/wt). Approximately 1 × 106 macrophages were incubated with microemulsions for 2 hr at 4°C. The mean fluorescence intensity of 10,000 events was measured and plotted against the concentration of the microemulsion added (expressed as μg of fatty acids per ml).
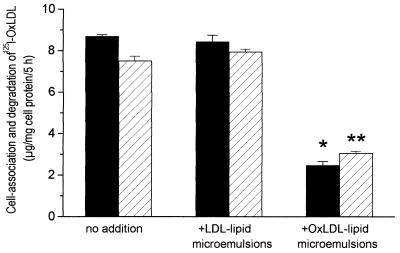
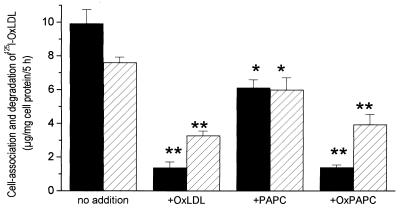
We have also shown that the cell association and degradation of 125I-labeled OxLDL is strongly inhibited by microemulsions prepared from the lipids of OxLDL but not by those prepared from the lipids of native LDL (Fig. 6). Finally, as shown in Fig. 7, cell association and degradation of 125I-labeled OxLDL was also inhibited by oxidized 1-palmitoyl-2-arachidonoyl-phosphatidylcholine presented in liposomes containing equimolar amounts of 1,2-distearoyl-phosphatidylcholine and free cholesterol. The lesser but significant inhibition by nonoxidized 1-palmitoyl-2-arachidonoyl-phosphatidylcholine may reflect oxidation during the 5-hr incubation.
Figure 6.
Inhibition of the cell association (solid bars) and degradation (hatched bars) of 125I-labeled OxLDL by OxLDL–lipid microemulsions. 125I-labeled OxLDL (2 μg of protein per ml) was incubated with resident mouse peritoneal macrophages for 5 hr. Degradation products in the medium and 125I remaining associated with the washed cells were assayed as described (25). ∗, P < 0.001; ∗∗, P < 0.002.
Figure 7.
Inhibition of the cell association (solid bar) and degradation (hatched bars) of 125I-labeled OxLDL by liposomes containing oxidized 1-palmitoyl-2-arachidonoyl-phosphatidylcholine. Methods are as described in Fig. 6. ∗, P < 0.05; ∗∗, P < 0.0001.
DISCUSSION
These studies show that microemulsions prepared from the unfractionated lipids of OxLDL share with PS-rich liposomes the ability to inhibit the binding of OxRBCs and of apoptotic thymocytes to mouse peritoneal macrophages. Additionally, liposomes prepared from oxidized PC (but not liposomes prepared from native PC) were similarly able to compete. Thus, liposomes prepared from 1-stearoyl-2-linoleoyl-phosphatidylcholine were inhibitory when 25–50% of the linoleic acid had been degraded by oxidation. The microemulsions prepared from OxLDL lipids were shown to contain less than 0.5% of the amount of apoprotein B that would normally be associated with the amount of LDL lipid in them. Of course the liposomes prepared from well-characterized PC and cholesterol contained no protein at all. Thus we can say that the inhibition observed is almost certainly protein-independent. This does not necessarily mean, however, that the apoprotein B in intact OxLDL plays no role at all in the competition between OxLDL and damaged cells. Indeed, earlier studies have shown that the resolubilized apoprotein B of OxLDL does inhibit the binding and uptake of intact OxLDL (4), and we confirmed that observation in the course of the present studies.
The ability of intact OxLDL to mimic the inhibitory effects of PS-rich liposomes on the binding of OxRBCs and of apoptotic cells implies that all three may share at least one common binding site on the macrophage surface (although each may of course have independent binding sites as well) (1, 2, 13). The present findings suggest that whatever the nature of the common binding sites or receptors, it is primarily lipid that is being recognized. For OxRBCs that has been accepted for some time; i.e., there is a considerable body of evidence that the binding of OxRBCs is attributable to an increase in the amount of PS on the outer leaflet of the RBC membrane (2, 5, 7–9). Macrophage binding of apoptotic cells is under some circumstances also believed to rest on an increased exposure of PS on the membrane (8, 9, 23). The inhibition of binding of damaged cells by protein-free liposomes containing PS, other anionic phospholipids, or as shown herein, lipids from OxLDL certainly focuses attention on the lipid moiety.
The simplest interpretation of our current results is that oxidized phospholipids—but not native phospholipids—may be responsible for a significant part of the binding of OxLDL to macrophage scavenger receptors. The presumption is that one or more of the polar phospholipid products in which the polyunsaturated fatty acid in the sn-2 position has been oxidatively attacked somehow mimic the physicochemical properties of PS. These could be hydroperoxide or hydroxy forms of the fatty acids or they could be degradation products in which the fatty acid carbon chain has been fragmented, generating aldehydes, ketones, or carboxylic acids. Watson et al. (22), using elegant chromatographic/mass spectrometric approaches, have identified two of the biologically active oxidized phospholipids in minimally oxidized LDL and verified their structure by synthesis: 1-palmitoyl-2(5-oxovaleryl)-sn-glyero-3-phosphocholine and 1-palmitoyl-2-glutaryl-sn-glycero-3-phosphocholine. Studies of that kind will be needed to determine whether these or other specific subfractions of oxidized lipids are responsible for the liposome effects observed in the present studies.
The nature of the binding site for OxRBCs and PS-rich liposomes on the macrophage remains unknown. Despite attempts in a number of laboratories, the elusive “PS receptor” remains elusive. Is it a protein receptor? Favoring this is the observation that previous treatment of the macrophage with trypsin or Pronase (but not collagenase) destroys its ability to bind OxRBCs and PS liposomes (whereas such prior treatment of the OxRBCs has no effect; unpublished observations). However, this in itself is inconclusive because protease treatment could affect binding indirectly, e.g., by weakening adhesion to ligands on the substratum. Whether protein in nature or not, there appears to be a finite saturable number of receptor sites, shown herein by the saturable binding of fluorescent-labeled liposomes prepared from OxLDL lipids (Fig. 5) and shown previously for a variety of other liposomes that bind to macrophages or monocytes (24).
The possibility that scavenger receptor A (SRA) might be a common receptor for OxLDL and OxRBCs was recently tested by comparing peritoneal macrophages of wild-type mice with macrophages from mice in which the SRA had been targeted (25). The uptake and degradation of acetyl LDL by the mutant cells was reduced 80% compared with that in the wild-type cells (25), in agreement with the findings of Suzuki et al. (26) and Lougheed et al. (27). The uptake and degradation of OxLDL was also significantly reduced, although to a lesser extent. However, there was no decrease at all in the binding of OxRBCs, suggesting that SRA is not a major participant in the recognition of OxRBCs by macrophages. Macrophages from SRA-deficient mice did show a significant reduction in the binding of apoptotic thymocytes, both to thymic macrophages (28) and to resident mouse peritoneal macrophages (25). Thus there is not a complete congruency between receptors that recognize OxRBCs, on the one hand, and apoptotic cells, on the other. Nishikawa et al. (13) presented evidence that resident mouse peritoneal macrophages bound PS-rich liposomes via the scavenger receptor but Lee et al. (29), using transfected cells expressing SRA, found no binding or uptake of PS-rich liposomes nor did they find uptake by smooth muscle cells treated to induce expression of SRA. The basis for the recognition of apoptotic cells by SRA remains to be established.
CD36 plays a role in the binding and uptake of OxLDL in mouse peritoneal macrophages (30) and in human monocyte/macrophages (31, 32). This receptor has been clearly established to play a role in the recognition and phagocytosis of apoptotic cells. Several lines of evidence support the interpretation that CD36 must interact with the vitronectin receptor (αvβ3 integrin) and with thrombospondin to bind apoptotic cells (33). The nature of the ligand on the apoptotic cell has not been characterized. On the other hand, Rigotti et al. (34) have shown that human CD36 expressed by transfected COS cells binds anionic phospholipid liposomes and that these liposomes compete for the binding of 125I-labeled acetyl LDL. A rat adipocyte protein (FAT) that is 85% homologous with human CD36 binds long-chain fatty acids (35) and so does CD36 (31). Nicholson et al. (31) found no binding of delipidated OxLDL to CD36-transfected cells and inferred that recognition might reside in the lipid moiety of OxLDL. However, the ability of the lipid moiety to bind was not directly tested. Most recently Ryeom et al. (36) presented convincing evidence that retinal pigment epithelial cells express CD36 and that the phagocytosis of photoreceptor outer segments by these epithelial cells is inhibited by liposomes containing PS or phosphatidylinositol and also by a mAb against CD36. Thus, CD36 is the first well-defined receptor shown to bind apoptotic cells through a PS-related mechanism. As shown by Fadok et al. (9, 37) and Rose et al. (38), the ability of macrophages to recognize PS-rich membranes is under complex regulation.
Additional OxLDL binding proteins in the macrophage include CD68 (39, 40) and SR-B1 (34, 41) and its human homologue CLA-1 (42). However, their role in recognition and uptake of apoptotic cells remains to be more fully explored.
It is still not clear whether the recognition of anionic phospholipid liposomes rests on some topologically specific array of negative charges or polar groups, on the one hand, or on specific recognition of phospholipid head groups, i.e., recognition of some narrowly defined microdomain(s). The ability of annexin V to bind specifically to artificial membranes rich in anionic phospholipids offers precedent that proteins can indeed recognize such lipid microdomains (or clusters of them) and bond tightly via calcium bridges (43). The recent studies by Hörkkö et al. (44), characterizing mAbs that recognize oxidized phospholipids but not native phospholipids, show again that specific protein recognition of microdomains (or clusters of them) is quite possible. The polar fatty acid degradation products generated as a result of oxidation of cell membrane lipids or LDL lipids (including ketones, aldehydes, and carboxylic acids) probably reorient themselves with the polar constituents presenting at the water–lipid interface. This could result in a surface configuration that in charge and polarity resembles that of a PS-rich liposome.
We have shown previously that the delipidated protein moiety of OxLDL competes with intact OxLDL for binding to resident peritoneal macrophages (4). The possibility that the lipid moiety might also play a role was not tested. It now appears that both may be involved. Further studies will be needed to determine which receptors recognize one or the other or, possibly, both.
The present findings raise the interesting possibility that oxidative damage to the membranes of cells may be an additional way to generate configurations recognized by scavenger receptors, (i.e., in addition to the loss of membrane phospholipid asymmetry with exposure of PS on the outer leaflet). There is evidence that apoptosis is associated with the generation of reactive oxygen species even if these are not essential for cell death (45). Thus there could be, in parallel, an increase in PS expression on the outer leaflet of the plasma membrane and an appearance of oxidized phospholipids in the membrane. Either or both could then be involved in the recognition of the damaged cells by scavenger receptors. This hypothesis is attractive because it could explain why macrophages are able to recognize and take up a wide variety of oxidatively damaged cells, namely, while they express very different plasma membrane proteins, their phospholipid composition is much less variable.
Acknowledgments
We thank Dr. George H. Rothblat, Dr. Michael C. Phillips, and Dr. Donald M. Small for reading and commenting on this manuscript. Dr. Jack Kyte, Mr. Steve Smith, and Mr. Robert S. Garrett provided important help in characterizing the liposomes used and the microemulsions reconstituted from LDL lipids. The mAb against glycophorin A was generously provided by Dr. Ann Rearden. This work was supported by the National Heart Lung and Blood Institute (HL-14197) and by the Sam and Rose Stein Institute for Research on Aging. V.T. was supported in part by the Dutch Heart Foundation.
ABBREVIATIONS
- RBC
red blood cell
- OxRBC
oxidatively damaged red blood cell
- OxLDL
oxidized low density lipoprotein
- PS
phosphatidylserine
- PC
phosphatidylcholine
- SRA
scavenger receptor A
- DiO
3,3′-hexadecyloxacarbocyanine
References
- 1.Sambrano G R, Parthasarathy S, Steinberg D. Proc Natl Acad Sci USA. 1994;91:3265–3269. doi: 10.1073/pnas.91.8.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sambrano G R, Steinberg D. Proc Natl Acad Sci USA. 1995;92:1396–1400. doi: 10.1073/pnas.92.5.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinbrecher U P, Witztum J L, Parthasarathy S, Steinberg D. Arteriosclerosis. 1987;7:135–143. doi: 10.1161/01.atv.7.2.135. [DOI] [PubMed] [Google Scholar]
- 4.Parthasarathy S, Fong L G, Otero D, Steinberg D. Proc Natl Acad Sci USA. 1987;84:537–540. doi: 10.1073/pnas.84.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schroit A J, Tanaka Y, Madsen J, Fidler I J. Biol Cell. 1984;51:227–238. doi: 10.1111/j.1768-322x.1984.tb00303.x. [DOI] [PubMed] [Google Scholar]
- 6.Schlegel R A, Prendergast T W, Williamson P. J Cell Physiol. 1985;123:215–218. doi: 10.1002/jcp.1041230210. [DOI] [PubMed] [Google Scholar]
- 7.Schlegel R A, Callahan M, Krahling S, Pradhan D, Williamson P. Mechanisms of Lymphocyte Activation and Immune Regulation. 1996. pp. 21–28. [DOI] [PubMed] [Google Scholar]
- 8.Fadok V A, Voelker D R, Campbell P A, Cohen J J, Bratton D L, Henson P M. J Immunol. 1992;148:2207–2216. [PubMed] [Google Scholar]
- 9.Fadok V A, Savill J S, Haslett C, Bratton D L, Doherty D E, Campbell P A, Henson P M. J Immunol. 1992;149:4029–4035. [PubMed] [Google Scholar]
- 10.Havel R J, Eder H A, Bragdon J H. J Clin Invest. 1955;34:1345–1353. doi: 10.1172/JCI103182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basu S K, Goldstein J L, Anderson G W, Brown M S. Proc Natl Acad Sci USA. 1976;73:3178–3182. doi: 10.1073/pnas.73.9.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowry O H, Rosebrough N J, Farr A L, Randall R J. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 13.Nishikawa K, Arai H, Inoue K. J Biol Chem. 1990;265:5226–5231. [PubMed] [Google Scholar]
- 14.Folch J, Lees M, Stanley H. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 15.Karmen A, Whyte M, Goodman D. J Lipid Res. 1963;4:312–321. [PubMed] [Google Scholar]
- 16.Fong L G, Parthasarathy S, Witztum J L, Steinberg D. J Lipid Res. 1987;28:1466–1477. [PubMed] [Google Scholar]
- 17.Kim J A, Territo M C, Wayner E, Carlos T M, Parhami F, Smith C W, Haberland M E, Fogelman A M, Berliner J A. Arterioscler Thromb. 1994;14:427–433. doi: 10.1161/01.atv.14.3.427. [DOI] [PubMed] [Google Scholar]
- 18.Navab M, Berliner J A, Watson A D, Hama S Y, Territo M C, Lusis A J, Shih D M, Van Lenten B J, Frank J S, Demer L L, Edwards P A, Fogelman A M. Arterioscler Thromb Vasc Biol. 1996;16:831–842. doi: 10.1161/01.atv.16.7.831. [DOI] [PubMed] [Google Scholar]
- 19.Esterbauer H, Jurgens G, Quehenberger O, Koller E. J Lipid Res. 1987;28:495–509. [PubMed] [Google Scholar]
- 20.Esterbauer H, Dieber-Rotheneder M, Waeg G, Striegl G, Jurgens G. Chem Res Toxicol. 1990;3:77–92. doi: 10.1021/tx00014a001. [DOI] [PubMed] [Google Scholar]
- 21.Steinbrecher U P, Parthasarathy S, Leake D S, Witztum J L, Steinberg D. Proc Natl Acad Sci USA. 1984;81:3883–3887. doi: 10.1073/pnas.81.12.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson A D, Leitinger N, Navab M, Faull K F, Horkko S, Witztum J L, Palinski W, Schwenke D. J Biol Chem. 1997;272:13597–13607. doi: 10.1074/jbc.272.21.13597. [DOI] [PubMed] [Google Scholar]
- 23.Savill J, Fadok V, Henson P, Haslett C. Immunol Today. 1993;14:131–136. doi: 10.1016/0167-5699(93)90215-7. [DOI] [PubMed] [Google Scholar]
- 24.Lee K D, Nir S, Papahadjopoulos D. Biochemistry. 1993;32:889–899. doi: 10.1021/bi00054a021. [DOI] [PubMed] [Google Scholar]
- 25.Terpstra V, Kondratenko N, Steinberg D. Proc Natl Acad Sci USA. 1997;94:8127–8131. doi: 10.1073/pnas.94.15.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki H, Kurihara Y, Takeya M, Kamada N, Kataoka M, Jishage K, Ueda O, Sakaguchi T, Higashi T, Suzuki T, et al. Nature (London) 1997;386:292–296. doi: 10.1038/386292a0. [DOI] [PubMed] [Google Scholar]
- 27.Lougheed M, Ming Lum C, Ling W, Suzuki H, Kodama T, Steinbrecher U P. J Biol Chem. 1997;272:12938–12944. doi: 10.1074/jbc.272.20.12938. [DOI] [PubMed] [Google Scholar]
- 28.Platt N, Suzuki H, Kurihara Y, Kodama T, Gordon S. Proc Natl Acad Sci USA. 1996;93:12456–12460. doi: 10.1073/pnas.93.22.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee K D, Pitas R E, Papahadjopoulos D. Biochim Biophys Acta. 1992;1111:1–6. doi: 10.1016/0005-2736(92)90267-p. [DOI] [PubMed] [Google Scholar]
- 30.Endemann G, Stanton L W, Madden K S, Bryant C M, White R T, Protter A A. J Biol Chem. 1993;268:11811–11816. [PubMed] [Google Scholar]
- 31.Nicholson A C, Frieda S, Pearce A, Silverstein R L. Arterioscler Thromb Vasc Biol. 1995;15:269–275. doi: 10.1161/01.atv.15.2.269. [DOI] [PubMed] [Google Scholar]
- 32.Nozaki S, Kashiwagi H, Yamashita S, Nakagawa T, Kostner B, Tomiyama Y, Nakata A, Ishigami M, Miyagawa J, Kameda-Takemura K. J Clin Invest. 1995;96:1859–1865. doi: 10.1172/JCI118231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savill J, Hogg N, Ren Y, Haslett C. J Clin Invest. 1992;90:1513–1522. doi: 10.1172/JCI116019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rigotti A, Acton S, Krieger M. J Biol Chem. 1995;270:16221–16224. doi: 10.1074/jbc.270.27.16221. [DOI] [PubMed] [Google Scholar]
- 35.Abumrad N, Raafat El-Maghrabi M, Amri E, Lopez E, Grimaldi P. J Biol Chem. 1993;268:17665–17668. [PubMed] [Google Scholar]
- 36.Ryeom S, Silverstein R, Scotto A, Sparrow J. J Biol Chem. 1996;271:20536–20539. doi: 10.1074/jbc.271.34.20536. [DOI] [PubMed] [Google Scholar]
- 37.Fadok V A, Laszlo D, Noble P, Weinstein L, Riches D, Henson P. J Immunol. 1993;151:4274–4285. [PubMed] [Google Scholar]
- 38.Foster D M, Barrett H R, Toffolo G, Beltz W F, Cobelli C. J Lipid Res. 1994;34:2193–2205. [PubMed] [Google Scholar]
- 39.Ramprasad M, Fischer W, Witztum J, Sambrano G, Quehenberger O, Steinberg D. Proc Natl Acad Sci USA. 1995;92:9580–9584. doi: 10.1073/pnas.92.21.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramprasad M, Terpstra V, Kondratenko N, Quehenberger O, Steinberg D. Proc Natl Acad Sci USA. 1996;93:14833–14838. doi: 10.1073/pnas.93.25.14833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukasawa M, Adachi H, Hirota K, Tsujimoto M, Arai H, Inoue K. Exp Cell Res. 1996;222:246–250. doi: 10.1006/excr.1996.0030. [DOI] [PubMed] [Google Scholar]
- 42.Murao K, Terpstra V, Green S, Kondratenko N, Steinberg D, Quehenberger O. J Biol Chem. 1997;272:17551–17557. doi: 10.1074/jbc.272.28.17551. [DOI] [PubMed] [Google Scholar]
- 43.Tait J, Gibson D. Arch Biochem Biophys. 1992;298:187–191. doi: 10.1016/0003-9861(92)90111-9. [DOI] [PubMed] [Google Scholar]
- 44.Horkko S, Miller E, Dudl E, Reaven P, Palinski W, Zvaifler N, Terkeltaub R, Pierangeli S, Curtiss L, Branch W, Witztum J. J Clin Invest. 1996;98:815–825. doi: 10.1172/JCI118854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacobson M. Trends Biochem Sci. 1996;21:83–86. [PubMed] [Google Scholar]