Abstract
We have applied correspondence analysis to electron micrographs of 2-D rafts of F-actin cross-linked with α-actinin on a lipid monolayer to investigate α-actinin: F-actin binding and cross-linking. More than 8,000 actin crossover repeats, each with 1–5 α-actinins bound, were selected, aligned, and grouped to produce class averages of α-actinin cross-links with ~9-fold improvement in the stochastic signal-to-noise ratio. Measurements and comparative molecular models show variation in the distance separating actin binding domains and the angle of the α-actinin cross-links. Rafts of F-actin and α-actinin formed predominantly polar 2-D arrays of actin filaments, with occasional insertion of filaments of opposite polarity. Unique to this study are the numbers of α-actinin molecules bound to successive crossovers on the same actin filament. These “monofilament”-bound α-actinins may reflect a new mode of interaction for α-actinin, particularly in protein dense actin-membrane attachments in focal adhesions. These results suggest that α-actinin is not simply a rigid spacer between actin filaments, but rather a flexible cross-linking, scaffolding, and anchoring protein. We suggest these properties of α-actinin may contribute to tension sensing in actin bundles.
Keywords: actin cytoskeleton, electron microscopy, focal adhesion, image processing, α-actinin
INTRODUCTION
α-Actinin is a modular protein belonging to the spectrin superfamliy that cross-links and bundles actin filaments in both muscle and non-muscle cells 1. There is no high resolution structure of the entire molecule, but atomic structures exist for most of its individual domains. α-Actinin has an N-terminal actin-binding domain (ABD) consisting of a tandem pair of non-equivalent calponin homology domains (CH1 and CH2) 2. Its structure has recently been solved by x-ray crystallography 3; 4. The placement of the ABD fragment on the actin filament has also been determined 5. In both of these structures, actin-bound and free, the ABD has a compact, closed arrangement of CH1 and CH2. In 2-D crystals, on the other hand, the ABD of intact α-actinin can adopt either an open or a closed conformation 6. The ABD is linked to the rest of the molecule by a 25-30 residue protease-sensitive flexible linker 7 whose structure is unknown. The linker is followed by a rod-like domain of four triple-helical, coiled-coil repeats (R1–R4). The R1–R4 domain lends the molecule an overall 90° left-handed twist 6; 8 that may contribute to its role as a protein docking platform 9. The C-terminus contains a calmodulin-like (CaM) domain consisting of a pair of structurally, but not necessarily functionally, conserved EF-hand motifs that bind Ca2+ in some isoforms (human, mouse ACTN1 & 4) while having evolutionarily lost this Ca2+-binding ability in other isoforms (human, mouse ACTN2 & 3) 1; 10; 11. α-Actinin forms antiparallel dimers through strong ~10 pM affinity associations between R1–R4 domains 12; 13. This arrangement places the CaM domain in close proximity to the ABD and is hypothesized to influence the ABD conformation 14; 15. These existing domain structures have been combined to generate a model of the dimer to fit 3-D images obtained by cryo-EM 6.
Previous studies on arrays of negatively-stained actin filaments have shown that α-actinin can cross-link in any orientation. Bundles formed in solution using chicken smooth muscle α-actinin favored an antiparallel orientation (9 of 11 filaments assayed) 16, while in other studies using the same isoform, 2-D bundles (rafts) formed on a lipid monolayer overwhelmingly preferred parallel cross-links 17; 18. Meyer and Aebi 16 suggested that the bundle characteristics were determined solely by the α-actinin molecular length and Taylor et al. 18 hypothesized that extrinsic factors were required to influence specificity of cross-linking orientation.
α-Actinin is localized to a variety of cellular structures requiring organized actin filament polarity. In Z-disks of striated muscle 19, cytoplasmic dense bodies of smooth muscle 20, and stress fibers of migrating cells 21, α-actinin cross-links oppositely oriented actin filaments to form bipolar assemblies. In focal adhesion plaques at cell membranes α-actinin is thought to cross-link similarly oriented actin filaments into polar bundles and also link them specifically to integrins 22; 23; 24. α-Actinin has been localized to these protein dense regions by GFP-tagged protein expression but its actin cross-linking function there is inferred.
α-Actinin also has numerous binding partners 25; 26; 27; 28. Through its interaction with the β-integrin cytoplasmic domains 24; 29; 30 α-actinin is thought to play a role in the formation and stabilization of focal adhesions in migrating cells 22. Interactions between α-actinin and other focal adhesion and stress fiber proteins include vinculin, zyxin, CRP, paxillin, MEKKI, PIP2, and FAK (reviewed by Otey and Carpen 28). Many of these interacting proteins are involved in cell signaling and regulation of transcription. One such protein, zyxin, has been demonstrated to mobilize from focal adhesions to stress fibers in response to cyclic stretch 31. In addition to being a focal adhesion component, zyxin is also a mechanosensitive transcription factor, translocating away from the cytoskeleton and into the nucleus 32.
The Z-disk of striated muscle is also described as a mechanosensory signaling interface 33. Here, α-actinin cross-links opposing actin filaments to form the Z-disk lattice while also interacting with titin Z-repeats 34; 35. Recent work has suggested a role for a titin/Tcap/MLP/α-actinin complex as a stretch sensor that is linked to cardiac hypertrophy. Mutations of these proteins lead to a lack of response to overstretching resulting in hypertrophic cardiomyopathy and muscular dystrophies 36. These observations suggest that α-actinin is more than a simple actin cross-linker but is also a scaffolding protein on which many additional factors bind and interact to specify α-actinin’s function more precisely.
The cytoskeleton is emerging as an integral component of cell tension sensing and signal transduction machinery. In order to further the understanding of α-actinin’s cross-linking function it is necessary to observe the intact molecule within an appropriate context. While actin bundles formed in solution are too thick to visualize with any molecular detail, 2-D arrays of actin filaments cross-linked by α-actinin can be readily formed on a lipid monolayer to make a specimen ideally suited for high resolution imaging 17; 18; 37. These arrays were not amenable to previously existing methods of spatial or helical averaging due to the highly variable nature of the cross-links and thus molecular detail was not obtainable. Now we are able to examine the range and flexibility of α-actinin binding and cross-linking actin filaments by employing correlation averaging, correspondence analysis, and classification schemes 38 to increase the signal-to-noise ratio (SNR) of the averages while retaining the variability of the α-actinin cross-links. We can distinguish the individual domains within the dimer in these averages and can quantify the angular distribution and lengths of the cross-links. We observe the occasional incorporation of antiparallel filaments into otherwise parallel arrays and determine that the antiparallel cross-links are no different in angular distribution or length from the parallel cross-links. We also report the intriguing and frequent occurrence of both α-actinin ABDs binding to a single actin filament. Most notable is the range of lengths measured for this molecule which suggests that α-actinin combines angular flexibility with linear elasticity in actin bundle formation.
RESULTS
The typical micrograph had four or more actin filaments within the raft and from one to five α-actinins bound to each crossover repeat. Although cross-links are clearly visible in the micrographs, their molecular details are not. We observed initially that the α-actinin cross-links all originate from the wide part of the actin crossover repeat with virtually no binding to the narrow portions. The cross-links are likely constrained to this periodicity by the planar form of the arrays. The symmetric sites located at ±90° are blocked by the lipid on one side of the filaments while the other side faces bulk solvent, where there may be too few actin filaments free in solution to be “grabbed” by the unbound end. Both actin and α-actinin are acidic proteins and strongly partition to the basic monolayer so that the interactions are constrained to occur in the 2-D phase. This is exactly the intent of the monolayer system as by keeping the specimen planar it imitates the effect of a membrane and keeps the specimen in an ideal form for electron microscopic examination. The polarity of the filaments cannot be determined in the original micrographs and there is no obvious change in the cross-linking pattern that would suggest a change in actin filament orientation. To obtain this level of information requires computation of averages with a higher SNR.
Alignment and Correspondence Analysis
The straightforward approach would involve alignment and classification of the crossover repeats to produce class averages. We refer to this as the simultaneous left-right approach because all cross-links in the repeat contribute to the classification. To eliminate or reduce actin filament misalignment as one source of variability all crossover repeats were aligned on a reference actin filament and subjected to several cycles of classification and multireference alignment. During this process, it became clear that some class averages, even from repeats within a single micrograph, were oppositely oriented compared to the majority. Since the linkage between repeats and their original micrograph was always maintained, it was easily determined that these oppositely oriented repeats came from a small subset of actin filaments (Fig. 1). Thus, arrays previously judged to have actin filaments with a single orientation were actually a mixture of parallel and antiparallel filaments. Note that there is no discernable difference in the cross-linking pattern between antiparallel actin filaments and parallel actin filaments in the original micrographs.
Figure 1.
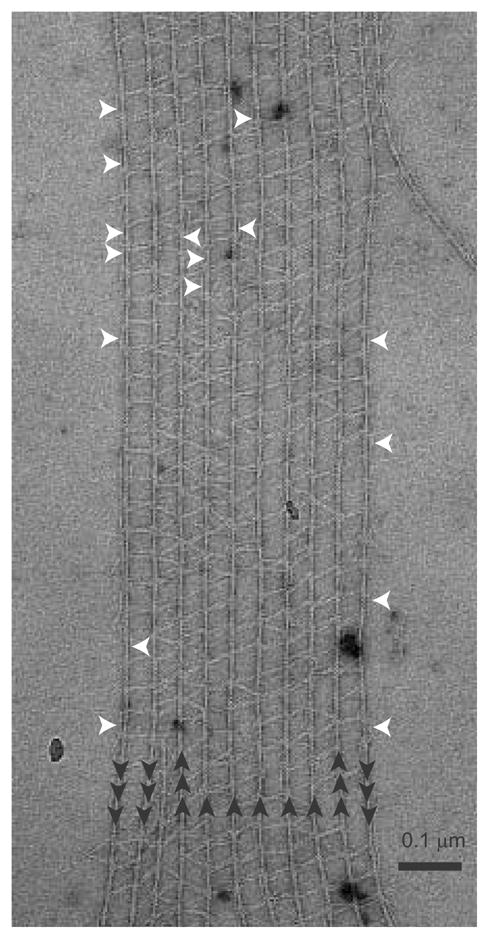
α-Actinin-F-actin raft. Polarity of the vertical actin filaments is indicated by the black arrows. White arrows point to mono-filament-bound α-actinin cross-links. Note the formation of girder-like struts.
The mixed orientation of the actin filaments in these rafts contradicts earlier studies that determined these same rafts contained actin filaments with a single orientation 17. That study utilized optical diffraction, changes in cross-links at inserted actin filaments within the rafts as well as the presence of actin filament spirals with α-actinin cross-links to argue bundle polarity. While cross-linking polarity in the spirals is indisputable, the other two approaches are subject to uncertainty. For example, small numbers of oppositely oriented actin filaments would not contribute significantly to the optical diffraction intensity which would report the dominant structure. The antiparallel cross-links account for ~15% of the ~8,000 total in this study. This would affect the diffraction intensity by 0.152 = .02, or 2% of the intensity. In addition, as shown in this analysis, inserted actin filaments (one of which is seen at the left of Fig. 1) are a weak discriminator of actin polarity in α-actinin bundles since the cross-linking pattern is unchanged by changes in filament orientation (see below). Correlation averaging as used here is much more powerful for establishing actin filament polarity.
Before applying correspondence analysis to the cross-links, all repeats were first rotated to the same actin orientation and aligned on the actin filament. A global average as well as eight class averages was then computed using a classification mask that included only a single crossover repeat centered on the wide part of the actin filament (Fig. 2). All of the averages show great detail in the actin filament, although the actin density becomes blurred at the ends indicating departures from strict linearity. Even given the detail in the actin filament, the averages do not show any α-actinin even in the wide part of the filament where the original micrographs show it to exist. The only hint of its presence is the appearance of distinct densities on either side of the actin crossover that we attribute to the ABDs. This blurring of the cross-links in the average emphasized their extreme variability. The variability is the primary reason that this data is not amenable to simple spatial averaging or helical averaging methods. Although there are some differences between the classes, the axial position of the actin monomers is identical. This suggested that the differences were primarily in the amount of filament bending at the ends of the repeats and the azimuthal orientation of the actin filament within the plane of the raft. In the global average the resolution by the Fourier Ring Correlation 39 was 1.6 nm indicating that the actin filament alignment is very accurate.
Figure 2.
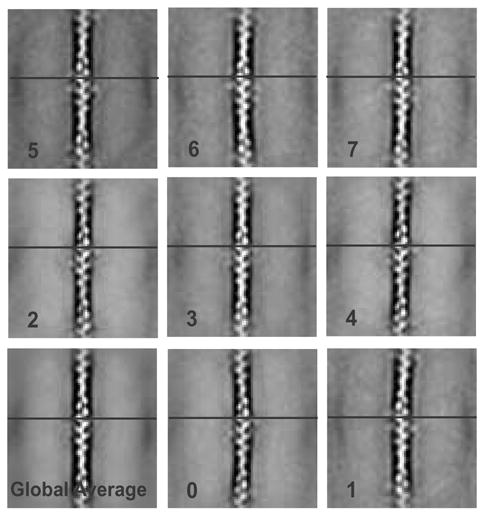
A global average of 7,903 actin crossover repeats and eight class averages based on the variance. Some curving of the filaments is present and can be seen in the averages. The horizontal line is consistent across each panel to demonstrate 2-D alignment accuracy.
Theoretically, α-actinin can pivot a full 180° about its actin-bound ABD, thus creating a semicircle of probability centered about the crossover repeat on each side of the actin filament. This range of flexibility produces considerable variation especially when both sides of the actin filament are considered together. A battery of different classification masks that excluded the pre-aligned actin filament were applied to the aligned repeats. Of all masks tried, none worked better, as judged by detail recovered in the averages, than a simple circle with a radius long enough to contain the length of α-actinin.
Two classification schemes were used. The initial classifications used a circular mask that included cross-links on both sides of the actin filament but excluded the pre-aligned actin filament. These classifications were not very satisfactory (Fig. 3) in that many α-actinins easily identified in raw repeats were not represented in the averages. Simply increasing the number of class averages only served to diminish the improvement in the SNR obtained by averaging.
Figure 3.
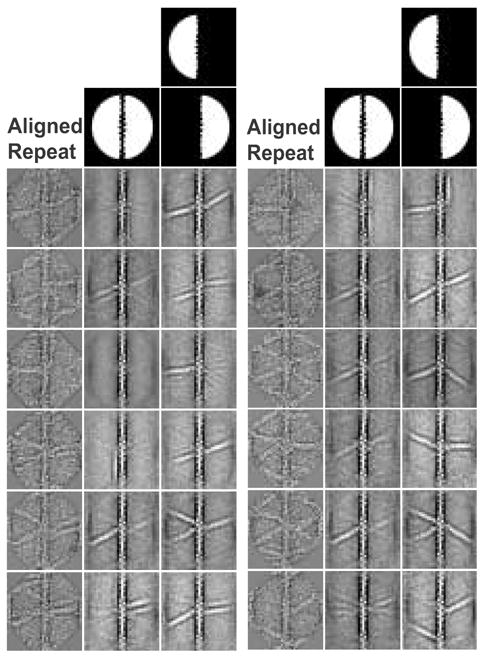
Comparison of whole image classification to left/right classification. The aligned, raw repeat class member in the left-most column is presented next to the results of both classification methods. The center column produced using a classification mask covering both sides of the filament, but excluding the filament itself. Areas in white are included in the classification. The method in the right column classifies on the left and right sides independently and then reassembles the averages to recreate the original repeat. Overall, the results of the independent left-right method compare more favorably to the raw repeat than simultaneous classification of both sides.
To improve on the number of cross-links retained in the class averages, but without sacrificing SNR improvement, we computed independent class averages of the two sides of the actin filament using semicircular left and right masks. This was based on the lack of any obvious correlation between cross-links on the left and right sides of the actin filament. Afterwards the class averages could be cut in half down the center of the aligned actin filament and “pasted” back together to regenerate the original repeat. This approach is capable of computing a “class average” for every original repeat using an optimal number of classes.
The left-right reassembled classes appeared to recover cross-links in the averages with better accuracy when compared to original images than the simultaneous classification and did so without sacrificing SNR improvement. It should be noted however, that some of the α-actinins visible in the original images were not picked up by classification even with this scheme. This is likely a consequence of the extreme amount of variation present. Patterns of cross-links that are statistically too infrequent to form a cluster of their own will be grouped instead with repeats that may lack one or more of its cross-links. Averaging will then eliminate the odd cross-links.
To evaluate the accuracy of the independent left-right method compared with the simultaneous left-right method, 100 original repeats containing 264 cross-links and the class averages to which they contributed were chosen at random and the number of visible cross-links in the original and two types of averages were scored (Table I). Independent evaluations by the three authors scored the number of cross-links that correctly appeared in the averages, the number of false negative cross-links (present in the original repeat but not picked up by classification), and the number of false positive cross-links (present in averages but not found in the original repeat). Cross-links in class averages generated by simultaneous left-right classification were correct ~30% of the time. The class averages reassembled from independent left and right classes were correct ~60% of the time. The numbers of false positive cross-links were relatively consistent between the two methods. Thus, the independent classification was twice as effective at retaining the cross-links in the reassembled averages.
Table I.
Assessment of Classification Accuracy
| Simultaneous Classification | Independent Classification | ||||||
|---|---|---|---|---|---|---|---|
| Total Cross-links | Correct | False Negative | False Positive | Correct | False Negative | False Positive | |
| 1 | 264 | 72 | 192 | 17 | 154 | 110 | 19 |
| 2 | 264 | 88 | 176 | 33 | 154 | 110 | 30 |
| 3 | 264 | 68 | 196 | 15 | 143 | 121 | 22 |
| Avg | 76 | 188 | 22 | 150 | 114 | 24 | |
| % | 29% | 71% | 8% | 57% | 43% | 9% | |
False negatives in this analysis represent cross-links that were present in some of the members of the class but with low frequency compared with other cross-links. These cross-links are blurred out when averaged. False positives are cross-links that exist in the data as they would otherwise disappear in the average. Their presence in the class average means that they were present in most members of the class, but not in the particular original repeat that was being scored.
Distribution of cross-link angles and lengths
Angles were measured from left and right class averages from the ABD on the actin filament outward. Numbers of crossover repeats included in each average were tallied for each angle in 10° increments. Histograms of the numbers of cross-links falling into a 180° range of possible angles show that the majority fall into 0°, 60°, 90°, 120°, and 180° bins (Fig. 4A). The 60°/120° cross-links are strut-like, while 90° cross-links appear as ladder rungs. Very often an actin filament crossover would have multiple cross-links of different angles which would suggest variations in length as well. Interestingly, the angular distribution of antiparallel cross-links is not very different from the parallel cross-links (Fig. 4B). This is contrary to the prediction 18 that the angular range should be narrower in bipolar cross-links because the helical nature of the actin filament would place alternating cross-links on opposite sides of the interfilament axis, i.e. both ends of cross-links between any pair of antiparallel actin filaments would be on the same side of the interfilament axis, either adjacent to the monolayer or in the bulk aqueous phase. For parallel cross-links, one end would be close to the monolayer and the other end closer to the bulk phase.
Figure 4.
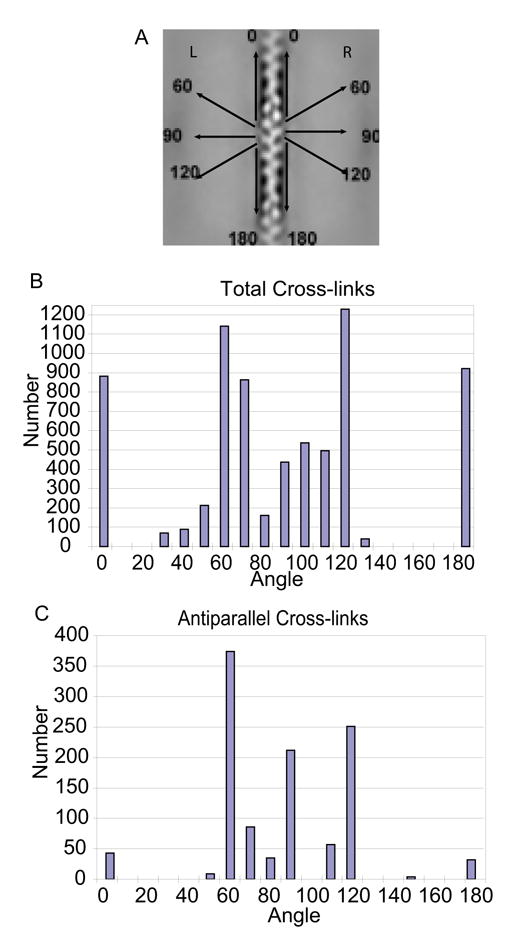
Angular distributions of total and antiparallel cross-links. A, Illustrates the angle convention used. B, Cross-link angles measured from left and right class averages of all repeats. Distribution is nearly symmetric because left and right angles are complementary. The departure from symmetry occurs because of cases where the repeat containing the opposite end of the cross-link was not extracted. C, Cross-link angles measured from left and right class averages of antiparallel repeats. The distribution is lopsided due to twice as many filaments having right-side antiparallel cross-links. Preferred orientations overall were the ~60°/120° “struts”. Total cross-links also strongly preferred 0°/180° “monofilament” binding.
The accuracy with which cross-links are recovered in the classes affects the accuracy of the histogram (Table I), but probably not its overall shape, nor the conclusions that can be drawn from it. False positives occur where cross-links were observed and overestimate the values in each bin by 9%. False negatives, which were 5× more numerous, are partially offset by the false positive. False negatives occur over the entire angular range with a roughly similar distribution as shown in Figure 4, though we made insufficient measurements to determine an accurate statistical distribution. Thus, correction for false positives and negatives would not affect our conclusion which is that cross-link angle is highly variable but preferentially oriented at 0°, 60°, 90°, 120°, and 180°.
To obtain an accurate measurement of the length of α-actinin as a cross-linker, we selected all molecules oriented in a particular angle, 60°, and aligned each of the repeats on the α-actinin itself. However, when class averages were computed, we discovered that the distal ends of the molecules were too variable for accurate measurement (Fig. 5). The central actin filament is usually aligned, but the second, outer actin filament is quite blurred due to translational/rotational variations. This suggests that the α-actinin length is variable and that the angle of the cross-link with respect to actin is not symmetrical with respect to the α-actinin.
Figure 5.
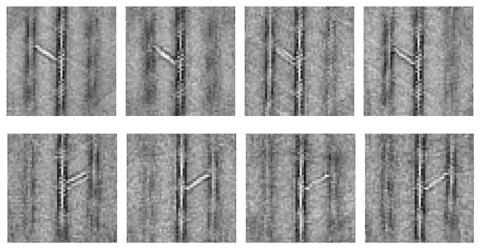
Examples of 60° cross-links realigned and classified on α-actinin. Note the blurring of distal actin filaments.
The difficulty posed by the high variability of even this highly populated class of cross-links was surmountable by “reassembling” cross-links from the left and right class averages (see Methods). In these correctly reassembled images, we were able to detect the density belonging to each ABD as well as the R1–R4 repeats because of the improved SNR. These “mapped-back” images (Fig. 6) reveal the α-actinin molecule from ABD to ABD, which is not possible in the original images. Traditionally these measurements would have been taken from the center of the actin filaments, which is the only possible measure in the original micrographs and each value would have experimental uncertainty because of the low SNR in the raw images. Each reassembled cross-link originates from two different class-averages. However, because the classification and averaging is accurate only ~60% of the time, reassembled images appear to correctly line up only about 35% of the time. Cross-links measured from ABD to ABD centers averaged 33.9 ± 2.5 nm. This compares favorably to the 34.5 nm α-actinin length estimated from 2-D arrays 15; 40. In many cases the ABD projection density appears to contribute only half as much intensity as might be expected from two superimposed CH domains placed on the actin filament. Our data are limited to projections at the moment so it is not possible to speculate on whether or not CH2 is actually bound to actin or to CH1 in the closed conformation.
Figure 6.
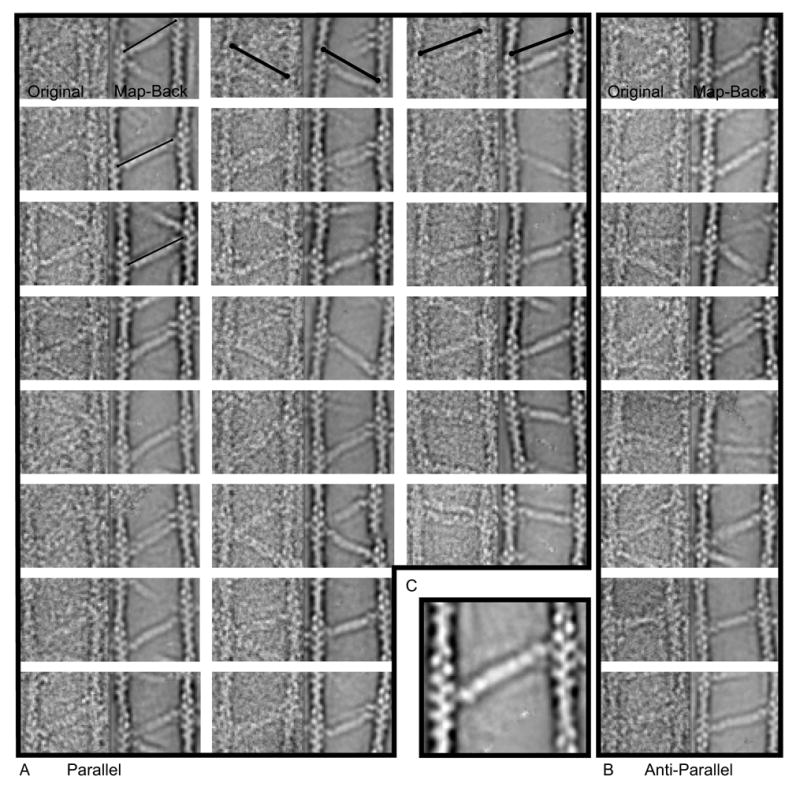
Mapped-back images. A and B represent direct comparisons of the raw image cross-link on the left and the re-assembled cross-link placed into the original coordinates for both (A) parallel and (B) antiparallel cross-links. Each half of the cross-link is contributed by a different class average. Each panel is 157 × 78.5 nm. The thin lines in the first column are all the same fixed length and illustrate the variation in total length of the α-actinin cross-link. The heavy lines in the first row are the same length in each panel and illustrate that the reassembled averages have identical positioning to the original motif which validates our use of these averages for measurement. C, Enlarged view. One can almost discern the four spectrin repeats in the rod domain, the CaM domain density, and a tiny nub of density nestled between the actin monomers attributable to the ABD.
α-Actinin binds to a single actin filament
An unexpected observation from these data is the frequency with which α-actinin binds with both ABDs onto single actin filaments. The α-actinins at first seemed to be aligning parallel to the actin filament as a consequence of drying in negative stain. However, a suggestion that the interaction is specific can be seen in the original micrographs (Fig. 1) where α-actinin ends are clearly placed on the wide part of the actin filament crossover repeat. “Cross-links” at 0° and 180° occur with close to the same frequency as the 60° and 120° struts (Fig. 4). With the repeats centered on the crossover only one end of the molecule was clearly visible in the averages and although a length could have been obtained by combining successive crossovers, for better accuracy we re-extracted the repeats with a larger box size to facilitate alignment centering on the narrow part of the filament between successive crossovers. Class averages could then be computed that contained both molecular ends of α-actinin. These averages show clear association of the ABD with the actin filament as a density comparable in size to that seen in a true cross-link and with both ends of the molecule placed on actin in a regular manner consistent with a specific interaction (Fig. 7). The R1–R4 domain is clearly separated from the actin filament suggesting that it does not contribute to the binding.
Figure 7.
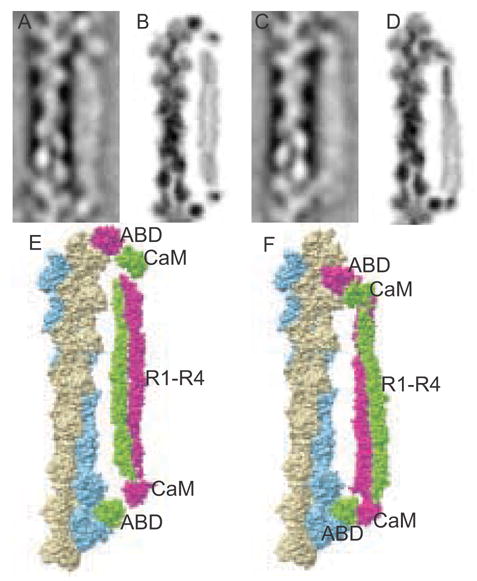
Models of α-actinin cross-linking a single actin filament. A and C, class averages representing long and short α-actinin binding to actin. B and D, long and short projection images in reverse contrast generated from PDB files modeled in O. E and F, images created in Chimera.
The two α-actinin ABDs span successive crossovers on the same filament so that the separating distance is quantized in length increments of 5.5 nm, the distance of one actin monomer along the filament (Fig. 7A, C). To help explain how α-actinin can accommodate this variation in length we constructed atomic models (Fig. 7E, F). When filtered to a similar resolution and projected these models compare favorably with the class averages (Fig. 7B, D). Of 4754 total repeats showing monofilament binding, 44.3%, fell into classes with a length of 35.9 nm, corresponding to 6.5 actin monomers along the filament, while 31.1% fell into classes with a length of 30.4 nm, equivalent to a length of 5.5 actin monomers. A total of 24.6% fell into classes whose lengths were ambiguous. This proportion of poorly defined images is not unusual in single particle reconstruction.
In building the long monofilament model, we extended the ABD-R1 linker to its maximum length consistent with a β-strand conformation. However, this may not be the only option. Less extension would be required if the CH2 domain of the ABD adopted an open conformation, rather than the closed one modeled here. CH2 domain movement could then contribute to the separation between CH1 domains and the rod in the long monofilament model. CH2 is not necessary for actin binding and does not bind actin by itself 41; 42. For the short model, the ABD-R1 linker would have to be folded and there are multiple ways to accomplish this, too many to model effectively.
DISCUSSION
α-Actinin was originally discovered as an actin cross-linking protein 19 but has since been shown to bind many cytoplasmic and membrane proteins. However, actin binding remains its most studied property. In vivo, α-actinin cross-links actin filaments into bipolar structures at the Z-disk, cytoplasmic dense bodies, and stress fibers. In the well ordered Z-disk cross-links of a length consistent with α-actinin are observed 43, while the angles of the cross-links have been estimated to be ~35° with respect to the actin filament 44.
The cytoplasmic dense bodies of smooth muscle, which are known to be the anchor points of oppositely oriented actin filaments 20, are much less well organized and clear cross-links due to α-actinin are not as easily seen as in Z-disks. Likewise, the cross-links in stress fibers are not unambiguously identifiable as α-actinin. α-Actinin’s presence in these structures is nonetheless undisputed.
The role of α-actinin at membranes, when actin filaments are oriented in the same direction is much less clear. Unlike with Z-disks, the structure of F-actin-membrane contacts is very obscure. Often, the identification of α-actinin at these actin-membrane contacts is done at the light microscopic level. It has been demonstrated that protein within the focal adhesion is relatively inaccessible by antibodies 45 and hence localization requires the expression and correct targeting of GFP-tagged α-actinin 46. At structures such as focal adhesions, α-actinin could have two roles, (1) anchoring actin filaments to membrane receptors such as integrins 24 and (2) cross-linking parallel actin filaments at the ends of stress fibers. The present study is relevant to both polar and bipolar actin filament bundles, but is perhaps more germane to polar bundles of actin filaments at cell membranes.
α-Actinin-Membrane Interactions
Our monolayer system was developed to simulate interactions between cytoplasmic proteins and membrane receptors on the inner leaflet of the cell membrane. The fluidity of the lipids combined with protein-lipid interactions facilitates sorting of the proteins into energetically favorable arrangements in a 2-D phase. The system has been used to identify the β1-integrin binding site on α-actinin 47 in a two component system as well as the structure of a ternary complex between α-actinin, the β1-integrin cytoplasmic domain and the first 258 residues of the vinculin head 48. These studies did not involve any linkage to the actin filament. The present study was undertaken to identify a plausible image processing scheme to enhance the SNR without loosing the structural variability of negatively-stained, macromolecular complexes involving F-actin which would then be applicable to future low contrast, frozen hydrated specimens.
The detail obtainable from the original micrographs is limited by the stochastic noise and the highly variable molecular arrangement. What has been lacking is a suitable analysis scheme to bin the highly variable cross-links for averaging. Our strategy of focused, independent left-right classification followed by reassembly recovers enough cross-link variability that reassembled class averages can be mapped-back to the original image to create large numbers of cross-links with enough detail to make measurements in a way that has not been possible before now. The accuracy of cross-link recovery could be improved by computing more class averages, but at the expense of the SNR improvement in the averages.
The 2-D rafts of F-actin and α-actinin show highly heterogeneous orientations and lengths for the cross-links, but their binding sites on actin are highly regular. The present result also shows that α-actinin can “cross-link actin crossovers,” an observation that broadens the range of possible α-actinin functions especially at the cell membrane.
Variability of bipolar cross-links
The present study demonstrates a promiscuous polarity preference for α-actinin: F-actin binding. Although 3-D images of the Z-disk reveal a highly regular and bi-polar arrangement of actin filaments cross-linked by α-actinin, the actual lengths of α-actinin linkers are inferred from inter-filament spacing which varies from 17–24 nm 43; 44. This difference is related to differences between the small square and the basket-weave lattice form, a function of thin filament activation and tension generation 44. The number of actin crossover repeats and α-actinin linkers in the Z-disk also vary across fiber types, and it was suggested that titin isoforms of varying numbers of Z-repeats may be a determinant 49. This suggests that there may be a number of extrinsic factors which govern the angle and polarity of α-actinin binding in cellular structures. These factors could be binding partners and/or mechanical effects. PIP2 binding to the CH2 domain of α-actinin induces a conformation that facilitates binding to titin, the Z-disk ruler, via two distinct interactions that alleviate a “pseudoligand” interaction between the CaM domain and the ABD-R1 flexible linker domain 35. Similarly, Actinin-associated LIM Protein, ALP, has been shown to strengthen the binding of α-actinin to actin 50; 51; 52. This protein binds α-actinin at both the CaM domain and the R1–R4 domain, crossing the short flexible linker domain 53. These authors postulate that the binding of ALP across this region may stabilize a conformation of α-actinin within the Z-disk that specifically limits it to antiparallel cross-linking.
Significance of “monofilament binding”
Our data suggest the ability of α actinin to bind from crossover to crossover on a single actin filament. As far as we know this is the first demonstration of this phenomenon, and raises the question of whether the length of α-actinin, which is close to the distance between actin filament crossovers, has evolved to maintain this monofilament binding capability rather than or in addition to an ability to cross-link actin filaments. Our work shows that variability in cross-linking angle is inherent to α-actinin; the separation between actin filaments in an α-actinin-actin bundle is therefore not an inherent property of α-actinin alone. Only the “struts” seem to maintain regular actin filament spacing.
Other than simple coincidence, what biological role could α-actinin monofilament binding perform? One possibility may be to sterically block interaction with other proteins. That is, regions of actin with α-actinin bound alongside would be excluded from interactions with other actin-binding proteins that induce branching, severing, or depolymerization. Another possibility could be to anchor individual actin filaments to membrane-associated adhesion proteins or channels. Many of these interactions, such as the α-actinin integrin interaction, have been characterized and involve the R1–R4 domain. Binding of α-actinin alongside, in fact, between the actin filament and the membrane, would present the entire length of the R1–R4 domain and hence two binding sites to the membrane surface thereby facilitating binding to adhesion proteins such as integrins.
Computer modeling of the appearance of α-actinin-vinculin-F-actin cross-links on a lipid monolayer surface suggested that simultaneous integrin binding and F-actin cross-linking was most likely to occur when the actin filaments are oriented away from the membrane whereas actin filament cross-linking alone was possible when the actin filaments run tangential to the membrane 48. Monofilament-bound α-actinin might circumvent this problem by facilitating linkage of tangential actin filaments to transmembrane elements such as integrins without forming cross-links.
Comparative modeling of the monofilament-bound α-actinins and the variation in total length of the cross-links raises the question of how this variation can be accommodated structurally. When building our comparative models for monofilament binding it was necessary to straighten this linker to accommodate the distance between the ABDs in the longer model. Although the α-actinin spectrin repeats can be unfolded with forces up to 40 pN 54, we do not expect that the molecules are experiencing such pulling forces at the monolayer surface in our specimens and there is no discernable change in length or morphology of the R1–R4 domain density that would suggest that this is occurring. One likely candidate then is the 25 amino acid linker region between the ABD and R1. At the moment there is no structural data on this linker. It has been demonstrated that the CaM domain can bind to this linker and its binding is relieved by PIP2 binding to the ABD, freeing the CaM domain for interaction with the titin Z-repeats 35. The length increase might also be accommodated if the ABD adopted an open conformation freeing CH2 to contribute to the separation. The CH2 domain is not required for actin binding as it only has one actin binding sequence and cannot bind actin on its own 41. The α-actinin ABD alone binds actin in a closed conformation; however, 2-D crystal structures suggest that the ABD can in fact assume an open conformation. Our data, which show a smaller density on actin than would be suggested by a combined CH1-CH2 ABD, suggest that CH2 may not be involved in actin binding in the intact molecule.
Significance of variable length of cross-links
α-Actinin has long been considered to be a fixed-length cross-linker based on its appearance in the Z-disk of striated muscle 19. The present study shows that α-actinin length when bound to actin can vary by more than 5.5 nm. We think that the length variation could facilitate a role for α-actinin in sensing tension within actin filament assemblies such as Z-disks and focal adhesions.
Although α-actinin is best known as an actin cross-linking protein, it has many other binding partners, some of which are known to be involved in tension sensing at the Z-disk 36 and at integrin-linked focal adhesions 55. We think that α-actinin itself is not likely to be the tension sensor but is more likely to act as a scaffold for tension-sensing signaling proteins. The present results suggest that α-actinin has the ability to respond to a change in tension by lengthening and could do so within limits without dissociating from actin. Hence it can maintain the integrity of the structure while responding to the stress. α-Actinin binding partners can then respond by dissociating. This would be facilitated if the binding partner bound α-actinin weakly but at more than one site, for instance the ABD and R1–R4, or bound both α-actinin and F-actin weakly. Then a stretch would cause dissociation from one site, destroying the cooperative interaction, facilitating dissociation from the other site followed by diffusion to a distant effector. As long as tension keeps the α-actinin extended, the tension sensor would be unlikely to reassociate with the structure. An example of a tentative tension-sensing complex at the Z-disk is the titin/Tcap/MLP/α-actinin complex 36. Mutations in MLP LIM domains that diminish its binding with α-actinin are linked to cardiac hypertrophy 56; 57. An example of a tension-sensing event at focal adhesions is the relocation of zyxin, another LIM-domain protein, from focal adhesions to stress fibers in response to cyclic stress 31. The specific interaction of zyxin with α-actinin has been well documented 58; 59; 60. Seemingly, the PDZ and LIM domain proteins that bind α-actinin are associated with signaling cell response to cytoskeletal changes.
Note that tension sensing in the focal adhesions may be different from tension sensing in Z-disks. In Z-disks α-actinin is stretched by opposing tension applied to actin filaments across the structure. In the focal adhesion, α-actinin acting as a cross-linker of parallel actin filaments would most likely not respond directly to the stretch. The stretch sensing would have to occur by pulling on α-actinins that are bound to another structure, such as the integrins. Tension sensing might then involve only a subset of the α-actinins in the structure.
Here we have applied a hybridization of techniques such as 2-D paracrystalline arrays to produce a favorable specimen orientation, correlation averaging methods, and a form of focused classification to overcome the disorder in our samples that prevented the use of conventional averaging techniques. Our scheme of alignment to a central element, left/right independent classification, reassembly, and mapping-back to the original coordinates effectively captures the variability of the α-actinin cross-link while improving the SNR in the average. This same technique can similarly be applied to tomograms of the same samples plus other adhesion-related proteins in vitreous ice to achieve new detail in our understanding of the interactions of adhesion proteins at the membrane.
MATERIALS AND METHODS
Protein Purification
Rabbit skeletal muscle α-actinin was purified from a myofibril preparation dissolved in 1.0 M NaCI, 25 mM Tris, 2 mM MgCl2, 0.02% β-mercaptoethanol, pH 8.0 followed by hydroxyapatite column purification 18. Actin was prepared from rabbit muscle acetone powder 61, followed by a Superose-12 column. Fresh G-actin was prepared by dialysis overnight against Buffer A: 2 mM Tris-Cl, 0.2 mM Na2ATP, 0.02% β-mercaptoethanol, 0.2 mM CaCl2, 0.01% NaN3, pH 8.0. The protein was clarified by high-speed centrifugation prior to sample set-up. Arrays were grown on positively charged lipid monolayers 17. A 3:7 volume ratio of DLPC: DDMA lipids (Avanti Polar Lipids) at 1 mg/mL in chloroform was layered over a solution containing 0.3 μM actin and α-actinin in phosphate-buffered polymerizing buffer. The actin was polymerized in the presence of α-actinin at 4 °C.
EM data collection
The 2-D arrays were recovered using 200–300 mesh copper grids with a reticulated carbon support film 62. Samples were stained with 1% uranyl acetate, dried, and stabilized by a thin layer of evaporated carbon. Data were collected on Philips EM300 and EM301 electron microscopes at 80 keV. Micrographs were scanned on a Z/I Imaging PhotoScan 2000 scanner (Carl Zeiss, Oberkochen, Germany) at either 7 or 14 μm.
Image Processing
Images were sampled and scaled to one another using the internal standard of the actin filament 5.9 nm layer line of the Fourier transform. This was achieved by windowing out single actin filaments from each micrograph and straightening using Phoelix 63. Filaments were then Fourier transformed and the number of pixels along the meridian to the 5.9 nm layer line was determined. All micrographs were resampled and scaled to 0.436 nm/pixel. The contrast transfer function (CTF) for each micrograph was corrected by phase flipping. Defocus and astigmatism angles were measured using the program ICE (Interactive Crystallographic Environment) 64. Images were normalized by setting the mean pixel value to 0 and the SD to 1. Repeat coordinates centered on each actin crossover from each micrograph were manually picked using the EMAN Boxer utility 65. Raw repeats were extracted as 180 × 80 pixel boxes and compiled into pseudo 3-D stacks. For classification of monofilament-binding α-actinins the repeats were re-extracted as 250 × 250 pixel boxes, a size large enough to contain the central actin filament and the two nearest neighbor actin filaments.
Classification
Repeats were aligned to the central actin filament using in-house software for alignment and multivariate statistical analysis. Classifications were done using 8, 16, 24, 32, and 64 factors. Eigenimages were evaluated by eye and by plotting the variance as a function of factor number and those seeming to contain only noise were left out of the classification. We also examined the resulting classes from each classification run for the ability to accurately recover cross-links in the averages and molecular detail until it was determined that 32 factors gave the best classification. In order to improve the amount of structure variance that can be recovered without sacrificing improvement in SNR, a left-right classification scheme was used. The CTF-corrected repeats were classified on the left side cross-links and the right-side cross-links separately. The repeats were then sorted into 88 classes left and 88 classes right; 88 being the square root of the total number of repeats, n. Because the SNR improves as the square root of the total number of images in each class average, the benefit of classifying each side separately followed by reassembly results in a SNR improvement of n¼ while capturing the variability as high as n. No images were discarded in the process as this leaves large gaps in the reassembled raft images. The resulting class averages from left and right were combined to reassemble the original repeat with an average 9.4-fold improvement in the SNR. Measurements of angular distribution were made from these averages.
These class averages, however, were somewhat blurred at the distal ends of the α-actinin molecules which prevented accurate length measurements. We first tried to align cross-links within a particular class, 60°, to the α-actinin itself. This resulted in a relatively well aligned central actin filament and α-actinin, but the distal filament was blurred due to translational differences and made the distal ABD hard to see. Instead, we reassembled these averaged repeats and mapped them back to the original micrograph. The inverse of the alignment shifts and rotation angle were applied to the repeats and each averaged repeat was placed back into the original image. First, we applied an oval mask with an apodized edge to each repeat. Then we applied the inverse of the shifts and rotations used in alignment and rescaled the repeat to its original parameters. Each repeat was then precisely inserted back onto the original micrograph from the original x and y coordinates. The reassembled image was then used to make length measurements.
Modeling
Models of cross-links were generated in O 66. The PDB coordinates of α-actinin from Liu et al. 6 were used to position the ABDs onto F-actin generated from the helical parameters of the averaged images which were determined by the ratio of the axial spacing of the 5.9 nm layer line to the first layer line. The MMTSB tool set 67 was used for manipulating the PDB files and positioning the spectrin-repeat rod domain. The EMAN utility pdb2mrc 65 was used to generate projections of the models which could be filtered and compared to the averaged repeats. Final images were prepared with Chimera 68.
Acknowledgments
This research was supported by NIH Grant GM64346 to the Cell Migration Consortium.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blanchard A, Ohanian V, Critchley D. The structure and function of alpha-actinin. J Muscle Res Cell Motil. 1989;10:280–9. doi: 10.1007/BF01758424. [DOI] [PubMed] [Google Scholar]
- 2.Mannherz HG, Gooch J, Way M, Weeds AG, McLaughlin PJ. Crystallization of the complex of actin with gelsolin segment 1. J Mol Biol. 1992;226:899–901. doi: 10.1016/0022-2836(92)90641-v. [DOI] [PubMed] [Google Scholar]
- 3.Franzot G, Sjoblom B, Gautel M, Djinovic Carugo K. The crystal structure of the actin binding domain from alpha-actinin in its closed conformation: structural insight into phospholipid regulation of alpha-actinin. J Mol Biol. 2005;348:151–65. doi: 10.1016/j.jmb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Borrego-Diaz E, Kerff F, Lee SH, Ferron F, Li Y, Dominguez R. Crystal structure of the actin-binding domain of alpha-actinin 1: Evaluating two competing actin-binding models. J Struct Biol. 2006;155:230–238. doi: 10.1016/j.jsb.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 5.McGough A, Way M, DeRosier D. Determination of the alpha-actinin-binding site on actin filaments by cryoelectron microscopy and image analysis. J Cell Biol. 1994;126:433–43. doi: 10.1083/jcb.126.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Taylor DW, Taylor KA. A 3-D reconstruction of smooth muscle alpha-actinin by CryoEM reveals two different conformations at the actin-binding region. J Mol Biol. 2004;338:115–25. doi: 10.1016/j.jmb.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 7.Imamura M, Endo T, Kuroda M, Tanaka T, Masaki T. Substructure and higher structure of chicken smooth muscle alpha-actinin molecule. J Biol Chem. 1988;263:7800–5. [PubMed] [Google Scholar]
- 8.Ylanne J, Scheffzek K, Young P, Saraste M. Crystal structure of the alpha-actinin rod reveals an extensive torsional twist. Structure (Camb) 2001;9:597–604. doi: 10.1016/s0969-2126(01)00619-0. [DOI] [PubMed] [Google Scholar]
- 9.Djinovic-Carugo K, Gautel M, Ylanne J, Young P. The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Lett. 2002;513:119–23. doi: 10.1016/s0014-5793(01)03304-x. [DOI] [PubMed] [Google Scholar]
- 10.Burridge K, Feramisco JR. Non-muscle alpha actinins are calcium-sensitive actin-binding proteins. Nature. 1981;294:565–7. doi: 10.1038/294565a0. [DOI] [PubMed] [Google Scholar]
- 11.Witke W, Hofmann A, Koppel B, Schleicher M, Noegel AA. The Ca(2+)-binding domains in non-muscle type alpha-actinin: biochemical and genetic analysis. J Cell Biol. 1993;121:599–606. doi: 10.1083/jcb.121.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahana E, Gratzer WB. Properties of the spectrin-like structural element of smooth-muscle alpha-actinin. Cell Motil Cytoskeleton. 1991;20:242–8. doi: 10.1002/cm.970200307. [DOI] [PubMed] [Google Scholar]
- 13.Flood G, Rowe AJ, Critchley DR, Gratzer WB. Further analysis of the role of spectrin repeat motifs in alpha-actinin dimer formation. Eur Biophys J. 1997;25:431–5. doi: 10.1007/s002490050057. [DOI] [PubMed] [Google Scholar]
- 14.Noegel A, Witke W, Schleicher M. Calcium-sensitive non-muscle alpha-actinin contains EF-hand structures and highly conserved regions. FEBS Lett. 1987;221:391–6. doi: 10.1016/0014-5793(87)80962-6. [DOI] [PubMed] [Google Scholar]
- 15.Tang J, Taylor DW, Taylor KA. The three-dimensional structure of alpha-actinin obtained by cryoelectron microscopy suggests a model for Ca(2+)-dependent actin binding. J Mol Biol. 2001;310:845–58. doi: 10.1006/jmbi.2001.4789. [DOI] [PubMed] [Google Scholar]
- 16.Meyer RK, Aebi U. Bundling of actin filaments by alpha-actinin depends on its molecular length. J Cell Biol. 1990;110:2013–24. doi: 10.1083/jcb.110.6.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor KA, Taylor DW. Formation of two-dimensional complexes of F-actin and crosslinking proteins on lipid monolayers: demonstration of unipolar alpha-actinin-F-actin crosslinking. Biophys J. 1994;67:1976–83. doi: 10.1016/S0006-3495(94)80680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor KA, Taylor DW, Schachat F. Isoforms of alpha-actinin from cardiac, smooth, and skeletal muscle form polar arrays of actin filaments. J Cell Biol. 2000;149:635–46. doi: 10.1083/jcb.149.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masaki T, Endo M, Ebashi S. Localization of 6S component of a alpha-actinin at Z-band. J Biochem (Tokyo) 1967;62:630–2. doi: 10.1093/oxfordjournals.jbchem.a128717. [DOI] [PubMed] [Google Scholar]
- 20.Bond M, Somlyo AV. Dense bodies and actin polarity in vertebrate smooth muscle. J Cell Biol. 1982;95:403–13. doi: 10.1083/jcb.95.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazarides E, Burridge K. Alpha-actinin: immunofluorescent localization of a muscle structural protein in nonmuscle cells. Cell. 1975;6:289–98. doi: 10.1016/0092-8674(75)90180-4. [DOI] [PubMed] [Google Scholar]
- 22.Rajfur Z, Roy P, Otey C, Romer L, Jacobson K. Dissecting the link between stress fibres and focal adhesions by CALI with EGFP fusion proteins. Nat Cell Biol. 2002;4:286–93. doi: 10.1038/ncb772. [DOI] [PubMed] [Google Scholar]
- 23.Burridge K, Nuckolls G, Otey C, Pavalko F, Simon K, Turner C. Actin-membrane interaction in focal adhesions. Cell Differ Dev. 1990;32:337–42. doi: 10.1016/0922-3371(90)90048-2. [DOI] [PubMed] [Google Scholar]
- 24.Otey CA, Pavalko FM, Burridge K. An interaction between alpha-actinin and the beta 1 integrin subunit in vitro. J Cell Biol. 1990;111:721–9. doi: 10.1083/jcb.111.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hampton CM, Taylor KA. alpha-Actinin-2. AfCS-Nature Molecule Pages; 2006. DOI: 10.1038/mp.a000195.01. [Google Scholar]
- 26.Hampton CM, Taylor KA. alpha-Actinin-3. AfCS-Nature Molecule Pages; 2006. DOI: 10.1038/mp.a000196.01. [Google Scholar]
- 27.Hampton CM, Taylor KA. alpha-Actinin-4. AfCS-Nature Molecule Pages. 2006 DOI: 10.1038/mp.a000197.01.
- 28.Otey CA, Carpen O. Alpha-actinin revisited: a fresh look at an old player. Cell Motil Cytoskeleton. 2004;58:104–11. doi: 10.1002/cm.20007. [DOI] [PubMed] [Google Scholar]
- 29.Pavalko FM, Burridge K. Disruption of the actin cytoskeleton after microinjection of proteolytic fragments of alpha-actinin. J Cell Biol. 1991;114:481–91. doi: 10.1083/jcb.114.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otey CA, Vasquez GB, Burridge K, Erickson BW. Mapping of the alpha-actinin binding site within the beta 1 integrin cytoplasmic domain. J Biol Chem. 1993;268:21193–7. [PubMed] [Google Scholar]
- 31.Yoshigi M, Hoffman LM, Jensen CC, Yost HJ, Beckerle MC. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J Cell Biol. 2005;171:209–15. doi: 10.1083/jcb.200505018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cattaruzza M, Lattrich C, Hecker M. Focal adhesion protein zyxin is a mechanosensitive modulator of gene expression in vascular smooth muscle cells. Hypertension. 2004;43:726–30. doi: 10.1161/01.HYP.0000119189.82659.52. [DOI] [PubMed] [Google Scholar]
- 33.Frank D, Kuhn C, Katus HA, Frey N. The sarcomeric Z-disc: a nodal point in signalling and disease. J Mol Med. 2006:1–23. doi: 10.1007/s00109-005-0033-1. [DOI] [PubMed] [Google Scholar]
- 34.Sorimachi H, Freiburg A, Kolmerer B, Ishiura S, Stier G, Gregorio CC, Labeit D, Linke WA, Suzuki K, Labeit S. Tissue-specific expression and alpha-actinin binding properties of the Z-disc titin: implications for the nature of vertebrate Z-discs. J Mol Biol. 1997;270:688–95. doi: 10.1006/jmbi.1997.1145. [DOI] [PubMed] [Google Scholar]
- 35.Young P, Gautel M. The interaction of titin and alpha-actinin is controlled by a phospholipid-regulated intramolecular pseudoligand mechanism. EMBO J. 2000;19:6331–40. doi: 10.1093/emboj/19.23.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayashi T, Arimura T, Itoh-Satoh M, Ueda K, Hohda S, Inagaki N, Takahashi M, Hori H, Yasunami M, Nishi H, Koga Y, Nakamura H, Matsuzaki M, Choi BY, Bae SW, You CW, Han KH, Park JE, Knoll R, Hoshijima M, Chien KR, Kimura A. Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. J Am Coll Cardiol. 2004;44:2192–201. doi: 10.1016/j.jacc.2004.08.058. [DOI] [PubMed] [Google Scholar]
- 37.Taylor KA, Taylor DW. Structural studies of cytoskeletal protein arrays formed on lipid monolayers. J Struct Biol. 1999;128:75–81. doi: 10.1006/jsbi.1999.4167. [DOI] [PubMed] [Google Scholar]
- 38.Frank J. Three-Dimensional Elecron Microscopy of Macromolecular Assemblies. Academic Press; San Diego, CA: 1996. [Google Scholar]
- 39.van Heel M, Bretaudiere J-P, Frank J. Proceedings of the 10th International Congress on Electron Microscopy. Vol. 1. 1982. Classification and multireference alignment of images of macromolecules; pp. 563–564. [Google Scholar]
- 40.Taylor KA, Taylor DW. Projection image of smooth muscle alpha-actinin from two-dimensional crystals formed on positively charged lipid layers. J Mol Biol. 1993;230:196–205. doi: 10.1006/jmbi.1993.1136. [DOI] [PubMed] [Google Scholar]
- 41.Way M, Pope B, Weeds AG. Evidence for functional homology in the F-actin binding domains of gelsolin and alpha-actinin: implications for the requirements of severing and capping. J Cell Biol. 1992;119:835–42. doi: 10.1083/jcb.119.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gimona M, Winder SJ. Single calponin homology domains are not actin-binding domains. Curr Biol. 1998;8:R674–5. doi: 10.1016/s0960-9822(98)70432-4. [DOI] [PubMed] [Google Scholar]
- 43.Luther PK. Three-dimensional structure of a vertebrate muscle Z-band: implications for titin and alpha-actinin binding. J Struct Biol. 2000;129:1–16. doi: 10.1006/jsbi.1999.4207. [DOI] [PubMed] [Google Scholar]
- 44.Goldstein MA, Schroeter JP, Sass RL. The Z lattice in canine cardiac muscle. J Cell Biol. 1979;83:187–204. doi: 10.1083/jcb.83.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavalko FM, Schneider G, Burridge K, Lim SS. Immunodetection of alpha-actinin in focal adhesions is limited by antibody inaccessibility. Exp Cell Res. 1995;217:534–40. doi: 10.1006/excr.1995.1119. [DOI] [PubMed] [Google Scholar]
- 46.Dabiri GA, Turnacioglu KK, Sanger JM, Sanger JW. Myofibrillogenesis visualized in living embryonic cardiomyocytes. Proc Natl Acad Sci U S A. 1997;94:9493–8. doi: 10.1073/pnas.94.17.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelly DF, Taylor KA. Identification of the beta1-integrin binding site on alpha-actinin by cryoelectron microscopy. Journal of structural biology. 2005;149:290–302. doi: 10.1016/j.jsb.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 48.Kelly DF, Taylor DW, Bakolitsa C, Bobkov AA, Bankston L, Liddington RC, Taylor KA. Structure of the alpha-actinin-vinculin head domain complex determined by cryo-electron microscopy. Journal of molecular biology. 2006;357:562–73. doi: 10.1016/j.jmb.2005.12.076. [DOI] [PubMed] [Google Scholar]
- 49.Luther PK, Padron R, Ritter S, Craig R, Squire JM. Heterogeneity of Z-band structure within a single muscle sarcomere: implications for sarcomere assembly. J Mol Biol. 2003;332:161–9. doi: 10.1016/s0022-2836(03)00883-0. [DOI] [PubMed] [Google Scholar]
- 50.Xia H, Winokur ST, Kuo WL, Altherr MR, Bredt DS. Actinin-associated LIM protein: identification of a domain interaction between PDZ and spectrin-like repeat motifs. J Cell Biol. 1997;139:507–15. doi: 10.1083/jcb.139.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pomies P, Macalma T, Beckerle MC. Purification and characterization of an alpha-actinin-binding PDZ-LIM protein that is up-regulated during muscle differentiation. J Biol Chem. 1999;274:29242–50. doi: 10.1074/jbc.274.41.29242. [DOI] [PubMed] [Google Scholar]
- 52.Pashmforoush M, Pomies P, Peterson KL, Kubalak S, Ross J, Jr, Hefti A, Aebi U, Beckerle MC, Chien KR. Adult mice deficient in actinin-associated LIM-domain protein reveal a developmental pathway for right ventricular cardiomyopathy. Nat Med. 2001;7:591–7. doi: 10.1038/87920. [DOI] [PubMed] [Google Scholar]
- 53.Klaavuniemi T, Kelloniemi A, Ylanne J. The ZASP-like motif in actinin-associated LIM protein is required for interaction with the alpha-actinin rod and for targeting to the muscle Z-line. J Biol Chem. 2004;279:26402–10. doi: 10.1074/jbc.M401871200. [DOI] [PubMed] [Google Scholar]
- 54.Ortiz V, Nielsen SO, Klein ML, Discher DE. Unfolding a linker between helical repeats. J Mol Biol. 2005;349:638–47. doi: 10.1016/j.jmb.2005.03.086. [DOI] [PubMed] [Google Scholar]
- 55.Bershadsky AD, Balaban NQ, Geiger B. Adhesion-dependent cell mechanosensitivity. Annu Rev Cell Dev Biol. 2003;19:677–95. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- 56.Gehmlich K, Geier C, Osterziel KJ, Van der Ven PF, Furst DO. Decreased interactions of mutant muscle LIM protein (MLP) with N-RAP and alpha-actinin and their implication for hypertrophic cardiomyopathy. Cell Tissue Res. 2004;317:129–36. doi: 10.1007/s00441-004-0873-y. [DOI] [PubMed] [Google Scholar]
- 57.Geier C, Perrot A, Ozcelik C, Binner P, Counsell D, Hoffmann K, Pilz B, Martiniak Y, Gehmlich K, van der Ven PF, Furst DO, Vornwald A, von Hodenberg E, Nurnberg P, Scheffold T, Dietz R, Osterziel KJ. Mutations in the human muscle LIM protein gene in families with hypertrophic cardiomyopathy. Circulation. 2003;107:1390–5. doi: 10.1161/01.cir.0000056522.82563.5f. [DOI] [PubMed] [Google Scholar]
- 58.Crawford AW, Michelsen JW, Beckerle MC. An interaction between zyxin and alpha-actinin. J Cell Biol. 1992;116:1381–93. doi: 10.1083/jcb.116.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reinhard M, Zumbrunn J, Jaquemar D, Kuhn M, Walter U, Trueb B. An alpha-actinin binding site of zyxin is essential for subcellular zyxin localization and alpha-actinin recruitment. J Biol Chem. 1999;274:13410–8. doi: 10.1074/jbc.274.19.13410. [DOI] [PubMed] [Google Scholar]
- 60.Li B, Trueb B. Analysis of the alpha-actinin/zyxin interaction. J Biol Chem. 2001;276:33328–35. doi: 10.1074/jbc.M100789200. [DOI] [PubMed] [Google Scholar]
- 61.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85(Pt B):164–81. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 62.Kubalek EW, Kornberg RD, Darst SA. Improved transfer of two-dimensional crystals from the air/water interface to specimen support grids for high-resolution analysis by electron microscopy. Ultramicroscopy. 1991;35:295–304. doi: 10.1016/0304-3991(91)90082-h. [DOI] [PubMed] [Google Scholar]
- 63.Whittaker M, Carragher BO, Milligan RA. PHOELIX: a package for semi-automated helical reconstruction. Ultramicroscopy. 1995;58:245–59. doi: 10.1016/0304-3991(95)00057-8. [DOI] [PubMed] [Google Scholar]
- 64.Wild DL, Tucker PA, Choe S. A visual data flow environment for macromolecular crystallographic computing. J Mol Graph. 1995;13:291–8. 299–300. doi: 10.1016/0263-7855(95)00068-2. [DOI] [PubMed] [Google Scholar]
- 65.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 66.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47 ( Pt 2):110–9. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 67.Feig M, Karanicolas J, Brooks CL., 3rd MMTSB Tool Set: enhanced sampling and multiscale modeling methods for applications in structural biology. J Mol Graph Model. 2004;22:377–95. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 68.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]


