Abstract
The Cockayne syndrome B (CSB) protein—defective in a majority of patients suffering from the rare autosomal disorder CS—is a member of the SWI2/SNF2 family with roles in DNA repair and transcription. We demonstrate herein that purified recombinant CSB and the major human apurinic/apyrimidinic (AP) endonuclease, APE1, physically and functionally interact. CSB stimulates the AP site incision activity of APE1 on normal (i.e. fully paired) and bubble AP–DNA substrates, with the latter being more pronounced (up to 6-fold). This activation is ATP-independent, and specific for the human CSB and full-length APE1 protein, as no CSB-dependent stimulation was observed with Escherichia coli endonuclease IV or an N-terminal truncated APE1 fragment. CSB and APE1 were also found in a common protein complex in human cell extracts, and recombinant CSB, when added back to CSB-deficient whole cell extracts, resulted in increased total AP site incision capacity. Moreover, human fibroblasts defective in CSB were found to be hypersensitive to both methyl methanesulfonate (MMS) and 5-hydroxymethyl-2′-deoxyuridine, agents that introduce base excision repair (BER) DNA substrates/intermediates.
INTRODUCTION
Cockayne syndrome (CS) is a rare autosomal recessive genetic disorder, classified as a segmental premature-aging syndrome (1–3). The clinical features of this disease include poor growth (‘cachectic dwarfism’), neurological abnormalities and cutaneous photosensitivity. However, in contrast to xeroderma pigmentosum (XP) patients—who also exhibit increased sensitivity to ultraviolet (UV) irradiation—individuals with CS do not display elevated cancer risk. CS is divided into two complementation groups: CSA (mutation in CKN1) and CSB (mutation in ERCC6). Of the patients suffering from CS, ∼80% have mutations in the CSB gene (1).
The CSB protein is composed of 1493 amino acids, and based on sequence homology, has been placed into the SWI2/SNF2 family of proteins that harbor seven helicase-like ATPase motifs (4,5). Although no helicase activity has been ascribed to CSB (6,7), the protein possesses a DNA-dependent ATPase activity (6–8). Moreover, since purified CSB (i) promotes alterations in the DNA conformation upon binding to the double-helix and (ii) alters the arrangement of nucleosome complexes (at the expense of ATP hydrolysis), the protein has been suggested to function as a chromatin remodeling factor (9). This function appears dependent on the ability of the protein to wrap and unwrap DNA molecules (10). More recently, CSB was found to possess homologous DNA strand pairing activity (11).
Numerous studies indicate that CSB participates in transcription-coupled nucleotide excision repair (TC-NER), as well as in global genome DNA repair and general transcription (1,12). In particular, CSB mutant cells exhibit hypersensitivity to a number of DNA-damaging agents, including UV light (4), 4-nitroquinoline-1-oxide (4-NQO) (13), and N-acetoxy-2-acetylaminofluorene (NA-AAF) (14,15). In the case of UV, the hypersensitivity has been attributed to a specific defect in the ability of CSB mutant cells to repair DNA damage within actively transcribed genes (16,17). A link of CSB to TC-NER is supported by the observations that (i) CSB-defective cells are unable to recover RNA synthesis following DNA-damaging agent treatment (4) and (ii) CSB binds RNA polymerase II when arrested at a template lesion in vitro and promotes recruitment of TFIIH, a factor involved in transcription and NER (18–20). Results also indicate that CSB plays a more general role in DNA repair, promoting changes in the chromatin structure to facilitate damage processing, particularly within active genes (21), and assists RNA polymerase I- or II-directed transcription (18,22–25).
Accumulating evidence suggests a role for CSB in base excision repair (BER) (1). BER is responsible for correcting most spontaneous, oxidative, or alkylation forms of DNA base or sugar damage. The observation that CSB−/− cells, at least certain cell types, display hypersensitivity to agents that generate reactive oxygen species (ROS), such as IR, paraquat and hydrogen peroxide, supports a role for the encoded protein in the repair of oxidative lesions (26–28). Moreover, biochemical assays using extracts from mutant cells indicate that CSB is responsible for promoting incision at 8-oxo-dG, a frequent oxidative base lesion and a marker of oxidative damage (26,29). In fact, global genome as well as mitochondrial DNA repair of 8-oxo-dG requires a functional CSB gene product in vivo (30–32). CSB mutant cells also exhibit a defect in the global repair of 8-hydroxyadenine, another oxidative base modification (33). Work from Flohr et al. (34) suggests that the efficient repair of oxidative base lesions through poly(ADP-ribose) polymerase 1 (PARP-1), a key DNA damage response protein, is likewise dependent on CSB. In addition, Thorslund et al. (35) identified a physical and functional interaction between CSB and PARP-1, and reported that poly(ADP)-ribosylation of CSB inhibits its DNA-dependent ATPase activity. Spivak et al. (36) have shown that reactivation of a plasmid containing a thymine glycol is defective in CSB mutant cells, further supporting a role for this protein in oxidative DNA damage repair. Although evidence exists for a function of CSB specifically in TC-BER, such a role has recently become unclear (37). It has been postulated that a defect in gene-specific and/or global genome repair of endogenous DNA damage gives rise to the premature-aging symptoms and increased neurological deficiencies associated with CS patients (38).
In mammalian cells, APE1 is the major apurinic/apyrimidinic (AP) endonuclease, operating to initiate repair of mutagenic and cytotoxic AP sites by incising the DNA backbone immediately adjacent to the lesion (39). AP sites are formed by spontaneous base loss (at a frequency of ∼10 000 events per mammalian genome per day), as well as by increased base release due to chemical modification (e.g. alkylation or oxidation) or through the action of DNA repair glycosylases that excise specific base damages. Following APE1 incision, subsequent participants of the BER pathway remove the 5′-terminal abasic fragment, replace the damaged nucleotide and seal the final nick to restore genome integrity. We describe herein data indicating a role for CSB in the repair of BER substrates/intermediates, and a specific functional interaction of CSB with the predominant AP site repair enzyme, APE1.
MATERIALS AND METHODS
Dot-blot interaction screen
The DiscoverLight Protein Array Kit (Pierce, Rockford, IL, USA) was used to screen for interactions between purified CSB and select proteins. As described elsewhere (40,41), various protein amounts were spotted on the membranes, which were then dried and blocked in PBS-T (1 × PBS, 0.1% Tween 20) containing 5% bovine serum albumin (BSA) for 1 h at room temperature. The membranes were then incubated with CSB (10 ng/ml) for 1 h at room temperature. A duplicate membrane was incubated with blocking buffer alone as a control for possible cross-reactivity with the CSB antibody. After washing with PBS-T, the membranes were probed with monoclonal mouse anti-CSB antibody (1 : 2000, kindly provided by Dr Jean Marc-Egly, Institut de Génétique et de Biologie Moléculaire et Cellulaire) overnight. After washing, the membranes were incubated with secondary antibody (1 : 10 000, anti-mouse IgG-horseradish peroxidase (HRP); Vector, Burlingame, CA, USA) for 1 h at room temperature, washed again and then developed with the SuperSignal West Pico chemiluminescent kit (Pierce).
Direct and indirect ELISA
Full-length, recombinant CSB and APE1 proteins were expressed and purified as previously detailed (8,42). Enzyme-linked immunosorbent assays (ELISAs) were performed essentially as described (43). Briefly, in the direct ELISAs, either BSA or APE1 (30 fmol) was coated onto a 96-well plate and then incubated with increasing amounts of CSB for 2 h at 37°C. After several washes, bound CSB was detected with anti-CSB antibody, followed by HRP-conjugated secondary antibody. For the indirect ELISA, either CSB (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or APE1 (Trevigen, Gaithersburg, MD, USA) polyclonal antibody was coated overnight at 4°C on a 96-well plate, and then incubated with either CSB (6 fmol = 1 ng) or APE1 (30 fmol = 1 ng) protein, respectively. After incubation with increasing amounts of the partner protein (i.e. APE1 or CSB) for 2 h at 37°C, bound protein was detected with either CSB or APE1 (Trevigen) monoclonal antibodies, followed by the secondary antibody steps described above. Where indicated, ethidium bromide (Invitrogen, Carlsbad, CA, USA) or DNase I (Sigma–Aldrich, St Louis, MO, USA) was added at 10 µg/ml or 5 µg/ml, respectively. The protein interaction was detected using OPD substrate (Sigma–Aldrich). Reactions were terminated after 3 min incubation with 3 N H2SO4 and plates were read at 490 nm using a Bio-Rad Benchmark Plus microplate spectrophotometer and Microplate Manager 5.2 software (Bio-Rad Laboratories, Hercules, CA, USA).
Immunoprecipitation
ECFP-CSB-CS1AN cells, which were created as described (8), were lysed in radioimmunoprecipitation assay (RIPA) buffer [50 mM Tris–HCl (pH 7.4), 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, 1 mM NaF, 10 U/ml DNase I and 1 tablet Complete protease inhibitor cocktail (Roche, Basel, Switzerland) per 50 ml] by sonication. After cell disruption, the suspension was centrifuged at 14 000 g for 15 min. The supernatant represented the whole cell extract, and the protein concentration was determined using the Bio-Rad Protein Assay (Bio-Rad Laboratories). For immunoprecipitation, ECFP–CSB whole cell extracts were pre-cleared with Protein G-Agarose beads (Invitrogen). The pre-cleared extracts (4 mg each) were then immunoprecipitated with either negative control rabbit IgG antibody (Santa Cruz Biotechnology), living colors full-length A.v. (i.e. anti-ECFP; 1 : 100) polyclonal rabbit antibody (BD biosciences, San Jose, CA, USA), or mouse monoclonal APE1 antibody (Novus, Littleton, CO; 1 : 50) for overnight at 4°C. Samples were next incubated with Protein G-Agarose beads (30 μl) at 4°C for 1 h, followed by multiple washes. Bound proteins were eluted by boiling in SDS sample buffer and were analyzed by SDS-PAGE and western blotting using mouse anti-CSB (1 : 1000; Dr Egly), or mouse anti-APE1 (1 : 1000; Novus) antibodies, followed by chemiluminescent analysis (Pierce).
AP endonuclease assay
The AP site-containing (i.e. the tetrahydrofuran, F, analog) 42-mer oligonucleotides used were: CCGCTGAATTGCACCCTCGAFCTAGGTCGATGATCCTAAGCA (42F-11), TGCTTAGGATCATCGACCTAGGTCGAGGGTGCAATTCAGCGG (42F comp) and TGCTTAGGATCATCGAGGATCGAGCTCGGTGCAATTCAGCGG (42F bubble comp). Following the generation of normal (42F-11:42 comp) or bubble (42F-11:42F bubble comp) 32P-end labeled duplex substrates, incision assays were performed at 37°C for 10 min with APE1 (10 or 30 fmol) and analyzed on denaturing polyacrylamide gels as previously described (44). Reactions consisted of 50 mM HEPES-KOH, pH 7.4, 50 mM KCl, 5% glycerol, 10 mM MgCl2, 100 µg/ml BSA, 0.05% Triton X-100 and 200 fmol of DNA. Where indicated, increasing amounts of CSB were added. Escherichia coli endonuclease IV and truncated Δ29APE1 were purified as described (42). Whole cell extract total AP endonuclease activity was determined for CSB-V and CSB-WT cells according to previously published methods (45).
Immunodepleted CSB supernatant fractions were obtained according to the procedure of (8). In brief, 15 µg of recombinant HA/His-tagged CSB was mixed with protein A magnetic beads (New England Biolabs, Ipswich, MA, USA) that had been pre-incubated with rabbit polyclonal anti-HA antibody (Santa Cruz Biotechnology). After an overnight incubation, the supernatant was separated from the beads by centrifugation, and kept as the immunodepleted fraction (ID CSB). Beads were washed and CSB was eluted with 0.5 mg/ml of HA peptide in buffer A (8) containing 0.5 M KCl to generate the immunoprecipitated (or immunopurified) CSB protein (IP CSB).
AP site measurements
The number of AP sites in isolated chromosomal DNA was determined using an aldehyde reactive probe reagent (N′-aminooxymethyl-carbonyl-hydrazino-d-biotin) and the DNA damage quantification kit of Dojindo Molecular Technology (Gaithersburg, MD, USA) as described (46).
Cellular sensitivity assays
CS1AN.S3.G2 SV40-transformed human skin fibroblast cell lines (4,47)—stably transfected with either the mammalian expression vector pcDNA3.1 (CSB-V), pcDNA3.1 containing the wild-type human CSB gene (CSB-WT), or pcDNA3.1 containing the E646Q ATPase domain II mutant CSB (CSB-E646Q) (48)—were cultured in Minimal Essential Medium plus 15% fetal bovine serum, antibiotics and 400 µg/ml of geneticin (Invitrogen). To investigate methyl methanesulfonate (MMS) and 5-hydroxymethyl-2′-deoxyuridine (HmdU) sensitivity, colony forming assays were performed essentially as described (49). Briefly, 5 × 102 cells were plated onto a 60-mm dish, incubated for 16 h, and then treated with different concentrations of MMS for 1 h. For HmdU treatment, cells were plated in complete medium at a density of 5 × 104 cells per 35-mm dish. After overnight incubation, HmdU was added, and the cells were incubated for an additional 24 h to permit incorporation. After agent treatment, cells were re-plated in complete media at a density of 1 × 102 per 60-mm dish. Colonies were stained and counted after 10 days of growth.
RNA synthesis recovery
CSB-V and CSB-WT cells were grown in the presence of [14C]thymidine (0.02 µCi/ml, Amersham or GE healthcare) for 3 days to label chromosomal DNA. Cells were then washed with PBS prior to a 1 h treatment with 1 mM MMS. Fresh media was subsequently added to allow cells to recover prior to measuring RNA synthesis at the indicated times. To measure transcription, cells were pulse-labeled with 5 µCi/ml [3H]uridine for 1 h at 37°C, washed twice with PBS, and lysed in 10 mM Tris (pH 8.0), 150 mM NaCl, 1 mM EDTA buffer containing 0.5% SDS and 100 µg/ml proteinase K for 1 h at 37°C. Trichloroacetic acid (TCA) (20%) was added to the cell lysate and the samples were spotted onto glass fiber discs (Whatman, Maidstone, UK). The filters were sequentially washed in 5% TCA, 70% ethanol and acetone. The TCA-precipitable radioactivity was scintillation-counted.
RESULTS
APE1 and CSB physically interact
Evidence suggests a role for CSB in facilitating global genome, and possibly transcription-coupled BER (1). As a means of elucidating the molecular involvement of CSB in BER, we examined for physical interactions of CSB with proteins operating in this and related DNA repair pathways. Specifically, initial studies employed a dot blot technique, where select proteins were (i) spotted and fixed to a capture membrane, (ii) incubated with purified CSB in solution and (iii) probed for binding of CSB using CSB-specific antibodies. Potential interactors identified using this technique were the strand break sensor protein PARP-1 (data not shown), the structure-specific endonuclease FEN1, the tyrosine kinase c-Abl and the major human abasic endonuclease APE1 (Supplementary Figure 1A: See online supplementary material for a color version of this figure). No significant interaction was seen with the tumor suppressor p53, the replication processivity factor PCNA, the end-joining binding complex Ku70/80, the Werner helicase, the protein defective in Nijmegen Breakage Syndrome NBS1, the telomere repeat binding protein TRF1, the recombination protein Rad51 and the single-stranded DNA binding protein RPA (Supplemental Figure 1A and data not shown). Detailed studies describing the interaction of CSB with PARP-1 have been reported elsewhere (35), and experiments with other putative binding partners (e.g. c-Abl and FEN1) are ongoing.
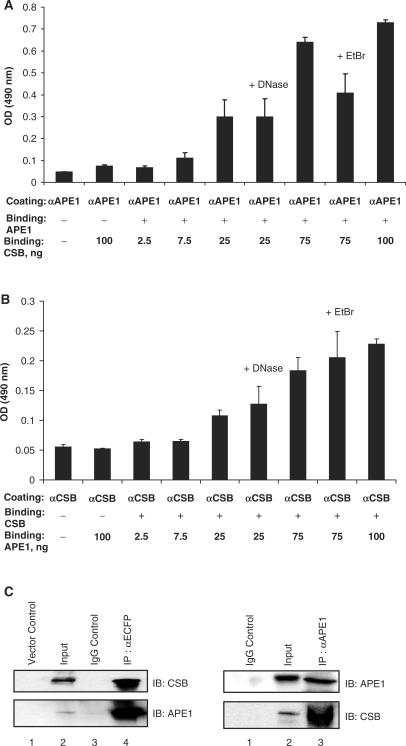
Figure 1.
CSB associates with APE1. Interaction between CSB and APE1 in indirect ELISA. Mouse anti-APE1 (panel A) or anti-CSB (panel B) was coated to the well. Purified APE1 (30 fmol, 1 ng) or CSB (6 fmol, 1 ng) was then bound to the appropriate antibody. These complexes were next incubated with different concentrations of CSB (panel A) or APE1 (panel B) as indicated. Ethidium bromide (EtBr) or DNase I was added where indicated. Values are means ± standard errors (n = 3). (C) CSB and Ape1 are in a common complex in human cell extracts. Lysates from CS1AN cells transfected with the ECFP-CSB plasmid were immunoprecipitated (IP) with either ECFP (left) or APE1 (right) antibodies. The IPs were then analyzed by immunoblotting (IB) with either anti-CSB or anti-APE1 antibodies. Lysates of 10% of the amount used for IP were loaded as input comparisons. As controls, lysates IPed with rabbit IgG only were loaded (IgG Control), and IPs with anti-ECFP were performed on extracts from ECFP-only expressing cells (Vector Control).
To interrogate the CSB–APE1 interaction further, we performed both direct and indirect ELISAs. Direct ELISAs, where APE1 was bound to the microtiter dish surface, supported that CSB physically interacts with APE1, as revealed by a concentration-dependent increase in the CSB protein signal that is not observed with the negative control protein BSA (Supplementary Figure 1B: See online Supplementary Data for a color version of this figure).Reciprocal experiments, however, where CSB was fixed to the surface, did not reveal an interaction (data not shown).
Thus, as an alternative method, an indirect ELISA approach, where either an APE1- (Figure 1A) or CSB-specific (Figure 1B) antibody was bound to the dish first and then incubated with the appropriate purified protein, was employed. In these studies, a reciprocal interaction was observed (albeit weaker when using APE1 as the binding probe; Figure 1B), and the increased signal in both cases was generally unaffected by the presence of ethidium bromide (EtBr) or DNase I (Figure 1A and B). The APE1 and CSB antibodies also did not cross-react non-specifically in the indirect ELISA experiments. These findings indicate that the APE1–CSB binding is the product of a direct, physical association and is independent of DNA. Analysis of the indirect ELISA experiments revealed an apparent binding affinity (i.e. the concentration at which approximately half maximal binding was observed) for CSB and APE1 of ≥4.5 (when APE1 was coated) to ≥20 nM (when CSB was coated).
APE1 and CSB are in a common complex within human cell extracts
We next examined whether APE1 and CSB existed in a common protein complex in human cell extracts using immunoprecipitation techniques. CSB mutant human cells (i.e. CS1AN.S3.G2) transfected with the pECFP–CSB expression vector were employed [ECFP-CSB-CS1AN; (8)], largely because antibodies directed against human CSB worked inconsistently in our hands in immunoprecipitation experiments. As shown in Figure 1C, immunoprecipitation with antibodies specific for either ECFP or APE1 pulled down both CSB and APE1 simultaneously from the ECFP-CSB-complemented CS1AN extracts. In contrast, neither the IgG antibody alone (negative control) nor the anti-ECFP antibody used against extracts containing only the ECFP fusion portion (vector control) precipitated either repair protein (Figure 1C). Semi-quantitative densitometry analysis of the western blot signals indicated that, depending on the experiment, between roughly 5% and 50% of either APE1 or CSB was co-immunoprecipitated with the other protein. The reason for the variability is presently unknown, but may reflect differences in cell culture conditions, cell cycle status or experimental handling.
CSB stimulates APE1 endonuclease activity
As APE1 and CSB were found to physically interact and co-immunoprecipitate from human cell extracts (Supplementary Figure 1 and Figure 1), we next explored whether this association had a functional effect on either APE1 or CSB enzymatic activities. Using a 42F normal (i.e. fully base-paired) duplex or a 42F 11-nt bubble substrate [where F is an abasic site analog (50)], we measured the effect of CSB on APE1 AP site incision activity. As shown in Figure 2A, APE1 cleavage of normal double-stranded AP–DNA was stimulated significantly in a CSB concentration-dependent manner, with roughly a 2-fold activation being observed at a molar ratio of APE1 to CSB of 1 : 3, and a 4-fold activation being detected at a 1 : 10 APE1 to CSB ratio. As expected, CSB itself exhibited no AP endonuclease activity. Moreover, no activation was observed with the CSB storage buffer (Figure 2A), suggesting that the stimulation was dependent on the CSB protein itself.
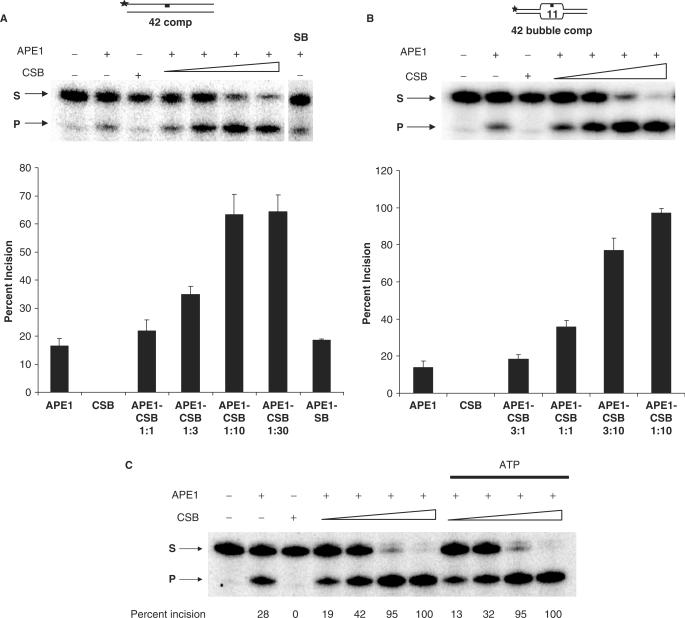
Figure 2.
CSB stimulates APE1 endonuclease activity, independent of ATP. (A) APE1 incision of 42F fully paired duplex substrates with or without CSB. Reactions contained 10 fmol of APE1 and 200 fmol of 42F-11: 42 comp duplex DNA, either alone or with increasing amounts of CSB (10, 30, 100 and 300 fmol). CSB alone (300 fmol) was also assayed. (B) APE1 incision of a 42-nt bubble substrate with or without CSB. Reactions were carried out as above, except with 30 fmol of APE1 and the 42F-11:42F bubble comp duplex substrate. AP endonuclease activity is the amount of radiolabeled F-DNA substrate (S) converted to the shorter incised DNA product (P) in 10 min. In panels A and B, values in graphs represent the average and standard deviation of a least three independent data points. The molar ratio of APE1 to CSB is indicated. (C) CSB stimulation of APE1 activity is not affected by ATP. Reactions were performed as above in panel B. ATP (0.5 mM) was added in the reactions indicated. No enzyme, as well as APE1 and CSB (300 fmol) alone, reactions are shown. Percentage of S converted to P is denoted under each lane.
With the 11-nt AP site-containing bubble substrate, which mimics a DNA transcription intermediate (44), CSB protein was again found to stimulate APE1 endonuclease activity in a concentration-dependent manner (Figure 2B). Significantly, CSB activation at either an equal molar ratio (2-fold) or at a higher ratio (up to 6-fold) was more profound with the bubble substrate than with the fully paired AP duplex (up to 4-fold). This observation suggests some DNA specificity in the activation, and is consistent with the higher affinity of CSB for transcription bubble structures than for normal double-stranded DNA (8)—an observation that supports that the stimulatory effect on APE1 is CSB-dependent. As above, CSB did not itself display AP endonuclease activity on the bubble substrate. Notably, addition of ATP did not affect the CSB stimulation of APE1 (Figure 2C). Last, APE1 had no effect on CSB ATPase activity in the presence of a 90-mer (35 nt)-bubble substrate (data not shown) or the 42F(11)-bubble substrate (Supplementary Figure 2: See online Supplementary Data for a color version of this figure).
To determine whether the activation of APE1 was indeed dependent on the CSB protein, we performed incision assays using immunodepleted and immunoprecipitated CSB protein fractions (see Materials and Methods section). As revealed by silver staining, immunodepletion of CSB with anti-HA antibodies reduced CSB protein levels in the recovered supernatant by ∼90% (data not shown). Thus, as expected, and supportive of the stimulation being CSB-dependent, the immunodepleted supernatant (ID CSB) had a ∼10-fold reduced ability to stimulate APE1 activity; that is, roughly equal stimulation was observed at a 10-fold higher amount of the immunodepleted CSB extract, relative to the purified CSB protein or the immunoprecipitated (i.e. immunopurified, IP CSB) CSB protein (Figure 3A).
Figure 3.
CSB specifically activates full-length human APE1. (A) APE1 stimulation is dependent on CSB. Reactions were carried out as in Figure 2. Immunodepleted supernatant (ID) or immunoprecipitated (IP) CSB fractions were prepared as described in Materials and Methods section. Percentage of intact substrate (S) converted to incised product (P) is denoted under each lane, and compared with the effects of purified, recombinant CSB protein. (B) CSB stimulation is specific for human APE1. Reactions were performed with E. coli AP endonuclease IV (0.1 fmol; ∼3 pg) and increasing amounts of CSB (0.01, 0.03, 0.1, 0.3, 1, 3, 10, 30, 100 and 300 fmol). (C) Incision activity of truncated APE1 protein is not activated by CSB. Truncated APE1 lacking the first 29 amino acids (5 fmol) was incubated with increasing concentrations of CSB (1, 5, 20, 50 and 100 fmol) as in Figure 2. Representative gel images are shown in panels A, B and C. In panels B and C, a full-length APE1 (30 fmol) and CSB (300 fmol) control reaction was simultaneously performed. Percentage of S converted to P is denoted.
As another means of evaluating the specificity of the CSB stimulation, we performed abasic site incision assays with purified bacterial AP endonuclease IV. Endonuclease IV is an E. coli protein with no sequence or structural homology to the human APE1 protein, although both enzymes exhibit many of the same biochemical repair functions (51). As shown in Figure 3B, CSB had no effect on endonuclease IV incision activity of the 11-nt bubble substrate, even when present at a molar ratio of 100:1 (i.e. at 10 fmol CSB); some inhibition was observed at the higher molar ratios. This result indicates that the AP endonuclease activation is specific to the human protein combination (i.e. CSB and APE1). As an aside, the findings here indicate that the ability to incise at AP sites in transcription bubble configurations is a general property of abasic endonucleases, both (unrelated) prokaryotic and eukaryotic.
We next examined whether CSB could stimulate the endonuclease activity of a truncated APE1 protein lacking the first 29 amino acid residues (i.e. Δ29APE1). As shown in Figure 3C, purified CSB did not activate Δ29APE1 incision activity of the 11 nt AP site-containing 42-mer bubble substrate, even at a molar ratio of 20 : 1. This result indicates that CSB physically interacts with the unique, unstructured N-terminal (REF-1) domain of APE1 (52) and that the protein contact is critical for the observed activation.
Employing a previously established electrophoretic mobility shift assay, which omits the divalent cation Mg2+ essential for APE1 catalysis (53), CSB was found not to obviously promote APE1 binding to fully paired or bubble-containing 42-mer AP–DNAs (Supplementary Figure 3: See online Supplementary Data for a color version of this figure). This finding argues that the endonuclease stimulation is mediated through a direct physical association with the APE1 protein (perhaps by promoting a favorable conformational change in APE1 through the N-terminal interaction) and/or the introduction of a topological alteration in the DNA substrate that permits a more efficient incision reaction. Interestingly, APE1 induces a kink in DNA to facilitate both recognition and strand cleavage of AP sites (52). Finally, some apparent binding of DNA was observed by CSB, particularly at the higher protein concentrations (Supplementary Figure 3).
The role of CSB in global genome AP site repair
As a means of evaluating the function of CSB in in vivo AP site repair, we measured the steady-state level of abasic lesions in total chromosomal DNA isolated from both CSB-V and CSB-WT fibroblasts using an established aldehyde reactive probe method (54). These studies found that both human cell lines maintain similar AP site levels (Figure 4A). Consistent with this, both CSB-WT and CSB-V whole cell extracts exhibited comparable total AP site incision capacities on fully paired 34-mer duplex substrates (Figure 4B and C). In addition, as with the whole cell extracts, we did not detect any difference in incision efficiency of normal duplex or bubble substrate AP site-containing DNAs using nuclear extracts prepared from CSB-WT or CSB-V cells (data not shown). These results suggest that CSB does not obviously modulate global genome repair of abasic lesions. However, the data does not exclude a more specialized role for the CSB–APE1 interaction in the repair of a subset of APE1 substrates, perhaps in regions of complex DNA structure. Significantly, addition of recombinant CSB protein to whole cell extracts prepared from CSB-V cells resulted in a concentration-dependent stimulation of AP site incision efficiency (Figure 4D), supportive of the idea that situations of high-localized concentrations of APE1 and CSB may indeed foster AP site repair. We note that semi-quantitative western blot analysis using CSB-WT whole cell extracts indicated an overall ratio of APE1:CSB of ≥100:1 (data not shown).
Figure 4.
Global AP site repair in CSB-V and CSB-WT cells. (A) Steady-state AP site levels in total chromosomal DNA from CSB-V and CSB-WT cells. AP sites measured per 106 nucleotides (nt) are shown. Values represent the average and standard deviation of three independent measurements. (B) Total AP endonuclease activity of CSB-V and CSB-WT whole cell extracts. Shown is a representative gel of reactions performed at the indicated concentration of CSB-V or CSB-WT whole cell extract (μg/10 μl) with 34F duplex substrates (44). NE = no enzyme control. Percentage of intact substrate (S) converted to incised product (P) is denoted under each lane. (C) AP site incision kinetics.Three microgram/10 μl of the indicated extract was incubated for the time specified, and the percentage of S converted to P was determined. Plotted is the average and standard deviation of at least three data points. (D) Recombinant CSB activates AP site incision in whole cell extracts. Extracts (0.2 μg) were prepared from CSB-V cells and assayed for AP site incision efficiency, without or with supplemented recombinant CSB protein (10, 30, 100, 300 fmol).
CSB contributes to MMS and HmdU resistance
As an additional means of evaluating the cellular contribution of CSB to the repair of BER substrates/intermediates, we determined the sensitivity of the SV40-transformed CSB mutant fibroblast cell line CS1AN.S3.G2 complemented with either a control vector (CSB-V) or a recombinant plasmid expressing wild-type human CSB (CSB-WT) (48) to the monofunctional alkylating agent MMS and the thymidine analog HmdU. For MMS, the most critical biological lesion is presumed to be the N-methylation base products, which frequently give rise to AP sites via enhanced hydrolysis of the N-glycosylic bond or DNA glycosylase-mediated base release (55). HmdU is an oxidative base product formed by attack of intracellular ROS as well as by way of deamination of HmdC, another oxidative base lesion. Repair of HmdU in chromosomal DNA proceeds through a SMUG1-initiated BER response (56). Prior studies have established that exposure to exogenous HmdU in the culture medium induces cell killing in a manner that is dependent on the formation of BER intermediates in DNA, and not the base itself (57). As shown in Figure 5, CSB-V cells exhibit an ∼3-fold increase in sensitivity to both MMS (panel A, 0.48 mM) and HmdU (panel B, 5.4 μM), as determined by the LD37 doses relative to the CSB-WT control (MMS, 1.3 mM; HmdU, 17.1 μM). These findings provide novel evidence for a direct role of CSB in the repair of BER substrates/intermediates, possibly AP sites or single-strand breaks (SSBs), which are common to the two agents. As seen previously (48), we did not detect an increased sensitivity of the CSB-V cells (studied here) to hydrogen peroxide (data not shown).
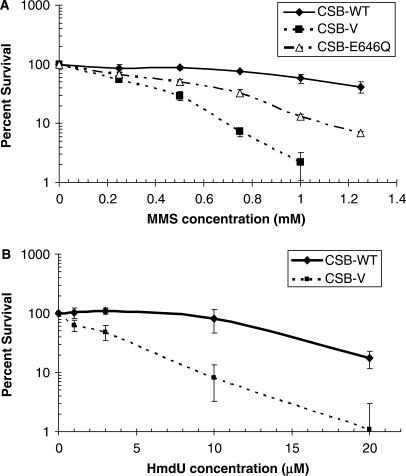
Figure 5.
CSB deficient cells exhibit hypersensitivity to MMS and HmdU. (A) MMS sensitivity. CSB-V, CSB-E646Q, and CSB-WT cells were exposed to MMS at different concentrations (indicated) for 1 h, and subsequently incubated for 10 days in drug-free medium. The surviving fraction of cells (% survival) was calculated by dividing the number of colonies in treated dishes by those counted in the untreated control. (B) HmdU sensitivity. The indicated cell lines were exposed to HmdU at different concentrations (denoted) for 24 h and plated in complete media. The colony formation ability was determined after 10 days. Values shown in both panels represent the mean and standard deviation of six independent data points.
As ATP was not critical to the APE1 activation (Figure 2C), we determined the contribution of the ATPase function of CSB to MMS resistance. Our studies reveal that CSB-deficient CS1AN cells expressing an ATPase domain II mutant CSB protein (i.e. CSB-E646Q) display intermediate sensitivity to MMS challenges (Figure 5A). This finding indicates both a structural and an enzymatic role for CSB in the MMS response.
DISCUSSION
We report herein that CSB and the major human AP endonuclease, APE1, directly interact, as well as exist in a common protein complex in human cells (Supplementary Figure 1 and Figure 1). As seen upon UV exposure (58), the composition of the CSB protein complex likely changes depending on the cellular environment, perhaps explaining the variability seen in the co-IP studies reported within (see Results section). Investigations to further analyze the nature of the CSB complex during different cell cycle stages or upon challenges with disparate DNA-damaging agents (e.g. oxidizing or alkylating compounds) are warranted. We also found that CSB stimulates APE1 AP site incision activity on abasic site-containing DNA substrates—in an ATP-independent, species-specific manner (no activation of E. coli endonuclease IV was observed by CSB)—likely via a direct interaction with the N-terminal ‘REF-1 region’ of APE1. This stimulation was more pronounced on a transcription-mimic bubble structure in comparison with a fully base-paired, ‘classical’ abasic site BER substrate (Figure 2). For instance, we detected a 2-fold enhanced incision activity of APE1 on the bubble duplex at a 1 : 1 CSB:APE1 molar ratio [keep in mind that CSB likely operates as a functional dimer (59)], whereas no stimulation was observed at this ratio on the fully paired double-stranded AP-DNA. Thus, in addition to coordination with DNA glycosylases in endogenous base damage repair and PARP-1 in DNA damage responses (see Introduction section), the interaction with APE1 suggests a more general function of CSB in modulating BER processes.
Despite the strong biochemical stimulation noted above, we found no clear evidence for a role of CSB in the global genome repair of endogenous, natural (hydrolytic) abasic sites (Figure 4). However, in light of the more pronounced activation seen with the bubble substrate, we favor a model whereby the CSB–APE1 interaction is most critical to regions of the genome where complex DNA structures are formed, such as during transcription or replication, or at sites of recombination (R-DNA or telomeres), where the local, relative concentrations of these proteins may also be higher. Moreover, a more specialized role for this interaction in the repair of a specific subset of APE1-targeted damages could exist. For instance, CSB may promote excision of oxidized AP sites or certain base modifications (e.g. 5,6-dihydro-2′-deoxyuridine, 5,6-dihydrothymidine, 5-hydroxy-2′-deoxyuridine, α-2′-deoxyadenosine and α-thymidine adducts), which have all been shown to be substrates for APE1 repair activity (60,61). We note that we did not see any obvious activation of the APE1 3′ to 5′ exonuclease activity on a 3′-recessed primer-template DNA duplex (unpublished data), supporting a specificity to the CSB–APE1 interaction.
While the idea of TCR of BER substrates is controversial, it is possible that CSB facilitates APE1 repair within actively transcribed genes. Interestingly, our preliminary data indicate a delayed RNA synthesis recovery in CSB mutant cells following MMS exposure (Supplementary Figure 4: See online Supplementary Data for a color version of this figure), suggesting that certain BER substrates/intermediates may indeed be handled in a gene-specific or potentially TCR-specific manner. Prior reports in fact indicate that hydrolytic or oxidized abasic lesions create a pause or arrest site for elongating RNA polymerases (62–68), and a stalled RNA polymerase is critical for the initiation of TCR and the recruitment of CSB (20,69). SSBs are also blocks to RNA polymerase progression (70), whereas alkylation base damage does not appear to be corrected in a TCR mechanism (71). Innovative methodologies to accurately and specifically evaluate genome (region)-specific repair and TCR of BER lesions are needed.
To our knowledge, we provide the first evidence that CSB mutant cells are defective in the repair of MMS-induced DNA damage, as well as DNA products generated after HmdU incorporation. Since both of these agents induce cell killing through the production of BER substrates/intermediates, the colony survival assays herein (Figure 5) provide compelling evidence that CSB mutant cells are defective in the efficient or productive removal of cytotoxic BER products, likely AP sites and/or DNA SSBs, which are common to these two agents. Consistent with this interpretation, prior studies have documented a clear and reproducible hypersensitivity of cells defective in a central BER participant to MMS and HmdU challenges. In particular, APE1, POLβ, XRCC1 and PARP1 mutant cells exhibit extreme sensitivity to both MMS (72–76) and HmdU [(77–79) and unpublished data]. Thus, combined with the reduced base damage removal and the increased sensitivity to oxidizing agents displayed by CSB mutant cells (see Introduction section), evidence is mounting that supports a role for CSB in facilitating a BER-related response.
The lack of an effect of ATP on the CSB-dependent activation of APE1, and the observation that the ATPase mutant protein (E646Q) only partially corrects the MMS sensitivity of CSB mutant cells (Figure 5A), suggests that this SNF2/SWI2 family member can function in BER in a chromatin remodeling-independent manner. This conclusion is in line with previous findings that uncovered normal hydrogen peroxide and γ irradiation sensitivity, as well as 8-oxo-dG incision activity, for CS1AN.S3.G2 cells transfected with the E646Q mutant (26,48). These results imply that CSB modulates BER primarily through protein complex associations (direct or indirect), or via a more subtle (ATP-independent) modification of the DNA structure. Notably, a functional ATPase activity, while necessary for chromatin remodeling, is not required for the induction of topological change in naked DNA (6). Thus, a story is emerging that suggests a distinct role for CSB in facilitating BER in comparison with NER, where the ATPase function of the protein (and energy) is more critical to the repair of UV-induced DNA damage (13,48). With that said, the partial complementation of E646Q (Figure 5A) does indicate a role for the ATPase function (and presumably the remodeling activity) of CSB in the MMS response as well. Further studies to more thoroughly define the precise biochemical contributions of CSB to the different DNA damage-response pathways (i.e. the contribution of protein–protein interactions, the ATP-dependent remodeling activity, or the ability to manipulate naked DNA) are a priority.
In summary, we report a physical and functional interaction between CSB and APE1, where the CSB-dependent stimulatory effect on APE1 incision activity, particularly on a bubble structure, is quite pronounced relative to other APE1 interactions (80). Moreover, we demonstrate that CSB mutant cells exhibit a profound hypersensitivity to DNA-damaging agents that create BER-type substrates/intermediates, namely MMS and HmdU, and display an impaired RNA synthesis recovery following MMS exposure. These findings provide further evidence for a role of CSB in the processing of BER-specific DNA substrates, potentially in regions of the genome that take on complex structures.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Mr Daniel R. McNeill (NIA) for assistance with a number of the experiments within and Mr Alfred May (NIA) for technical advice on this project. We thank Dr Robert Brosh and Dr. Mohammad Hedayati (NIA) for critical reading of this manuscript. This research and funding to pay the Open Access publication charges for this article were supported by the Intramural Research Program of the National Institute on Aging, NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Licht CL, Stevnsner T, Bohr VA. Cockayne syndrome group B cellular and biochemical functions. Am. J. Hum. Genet. 2003;73:1217–1239. doi: 10.1086/380399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehmann AR. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003;85:1101–1111. doi: 10.1016/j.biochi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer. 2005;5:564–573. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- 4.Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers JH. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 5.Durr H, Flaus A, Owen-Hughes T, Hopfner KP. Snf2 family ATPases and DExx box helicases: differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 2006;34:4160–4167. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Citterio E, Rademakers S, van der Horst GT, van Gool AJ, Hoeijmakers JH, Vermeulen W. Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J. Biol. Chem. 1998;273:11844–11851. doi: 10.1074/jbc.273.19.11844. [DOI] [PubMed] [Google Scholar]
- 7.Selby CP, Sancar A. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J. Biol. Chem. 1997;272:1885–1890. doi: 10.1074/jbc.272.3.1885. [DOI] [PubMed] [Google Scholar]
- 8.Christiansen M, Stevnsner T, Modin C, Martensen PM, Brosh RM, Jr, Bohr VA. Functional consequences of mutations in the conserved SF2 motifs and post-translational phosphorylation of the CSB protein. Nucleic Acids Res. 2003;31:963–973. doi: 10.1093/nar/gkg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Citterio E, van den Boom V, Schnitzler G, Kanaar R, Bonte E, Kingston RE, Hoeijmakers JH, Vermeulen W. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beerens N, Hoeijmakers JH, Kanaar R, Vermeulen W, Wyman C. The CSB protein actively wraps DNA. J. Biol. Chem. 2005;280:4722–4729. doi: 10.1074/jbc.M409147200. [DOI] [PubMed] [Google Scholar]
- 11.Muftuoglu M, Sharma S, Thorslund T, Stevnsner T, Soerensen MM, Brosh RM, Jr, Bohr VA. Cockayne syndrome group B protein has novel strand annealing and exchange activities. Nucleic Acids Res. 2006;34:295–304. doi: 10.1093/nar/gkj410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Hoffen A, Balajee AS, van Zeeland AA, Mullenders LH. Nucleotide excision repair and its interplay with transcription. Toxicology. 2003;193:79–90. doi: 10.1016/j.tox.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Brosh RM, Jr, Balajee AS, Selzer RR, Sunesen M, Proietti DS, Bohr VA. The ATPase domain but not the acidic region of Cockayne syndrome group B gene product is essential for DNA repair. Mol. Biol. Cell. 1999;10:3583–3594. doi: 10.1091/mbc.10.11.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Oosterwijk MF, Versteeg A, Filon R, van Zeeland AA, Mullenders LH. The sensitivity of Cockayne's syndrome cells to DNA-damaging agents is not due to defective transcription-coupled repair of active genes. Mol. Cell. Biol. 1996;16:4436–4444. doi: 10.1128/mcb.16.8.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sunesen M, Selzer RR, Brosh RM, Jr, Balajee AS, Stevnsner T, Bohr VA. Molecular characterization of an acidic region deletion mutant of Cockayne syndrome group B protein. Nucleic Acids Res. 2000;28:3151–3159. doi: 10.1093/nar/28.16.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayne LV, Lehmann AR. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne's syndrome and xeroderma pigmentosum. Cancer Res. 1982;42:1473–1478. [PubMed] [Google Scholar]
- 17.Laine JP, Egly JM. When transcription and repair meet: a complex system. Trends Genet. 2006;22:430–436. doi: 10.1016/j.tig.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Tantin D, Kansal A, Carey M. Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol. Cell. Biol. 1997;17:6803–6814. doi: 10.1128/mcb.17.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Boom V, Citterio E, Hoogstraten D, Zotter A, Egly JM, van Cappellen WA, Hoeijmakers JH, Houtsmuller AB, Vermeulen W. DNA damage stabilizes interaction of CSB with the transcription elongation machinery. J. Cell. Biol. 2004;166:27–36. doi: 10.1083/jcb.200401056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laine JP, Egly JM. Initiation of DNA repair mediated by a stalled RNA polymerase IIO. EMBO J. 2006;25:387–397. doi: 10.1038/sj.emboj.7600933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fousteri M, van HA, Vargova H, Mullenders LH. Repair of DNA lesions in chromosomal DNA impact of chromatin structure and Cockayne syndrome proteins. DNA Repair. 2005;4:919–925. doi: 10.1016/j.dnarep.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 22.Bradsher J, Auriol J, Proietti De SL, Iben S, Vonesch JL, Grummt I, Egly JM. CSB is a component of RNA pol I transcription. Mol. Cell. 2002;10:819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- 23.Balajee AS, May A, Dianov GL, Friedberg EC, Bohr VA. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc. Natl Acad. Sci. USA. 1997;94:4306–4311. doi: 10.1073/pnas.94.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tantin D. RNA polymerase II elongation complexes containing the Cockayne syndrome group B protein interact with a molecular complex containing the transcription factor IIH components xeroderma pigmentosum B and p62. J. Biol. Chem. 1998;273:27794–27799. doi: 10.1074/jbc.273.43.27794. [DOI] [PubMed] [Google Scholar]
- 25.van Gool AJ, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly JM, Bootsma D, Hoeijmakers JH. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tuo J, Muftuoglu M, Chen C, Jaruga P, Selzer RR, Brosh RM, Jr, Rodriguez H, Dizdaroglu M, Bohr VA. The Cockayne Syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J. Biol. Chem. 2001;276:45772–45779. doi: 10.1074/jbc.M107888200. [DOI] [PubMed] [Google Scholar]
- 27.de Waard H, de Wit J, Gorgels TG, van den AG, Andressoo JO, Vermeij M, van Steeg H, Hoeijmakers JH, van der Horst GT. Cell type-specific hypersensitivity to oxidative damage in CSB and XPA mice. DNA Repair. 2003;2:13–25. doi: 10.1016/s1568-7864(02)00188-x. [DOI] [PubMed] [Google Scholar]
- 28.de Waard H, de Wit J, Andressoo JO, van Oostrom CT, Riis B, Weimann A, Poulsen HE, van Steeg H, Hoeijmakers JH, et al. Different effects of CSA and CSB deficiency on sensitivity to oxidative DNA damage. Mol. Cell. Biol. 2004;24:7941–7948. doi: 10.1128/MCB.24.18.7941-7948.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osterod M, Larsen E, Le Page F, Hengstler JG, van der Horst GT, Boiteux S, Klungland A, Epe B. A global DNA repair mechanism involving the Cockayne syndrome B (CSB) gene product can prevent the in vivo accumulation of endogenous oxidative DNA base damage. Oncogene. 2002;21:8232–8239. doi: 10.1038/sj.onc.1206027. [DOI] [PubMed] [Google Scholar]
- 30.Sunesen M, Stevnsner T, Brosh RM, Jr, Dianov GL, Bohr VA. Global genome repair of 8-oxoG in hamster cells requires a functional CSB gene product. Oncogene. 2002;21:3571–3578. doi: 10.1038/sj.onc.1205443. [DOI] [PubMed] [Google Scholar]
- 31.Stevnsner T, Nyaga S, Souza-Pinto NC, van der Horst GT, Gorgels TG, Hogue BA, Thorslund T, Bohr VA. Mitochondrial repair of 8-oxoguanine is deficient in Cockayne syndrome group B. Oncogene. 2002;21:8675–8682. doi: 10.1038/sj.onc.1205994. [DOI] [PubMed] [Google Scholar]
- 32.Trapp C, Reite K, Klungland A, Epe B. Deficiency of the Cockayne syndrome B (CSB) gene aggravates the genomic instability caused by endogenous oxidative DNA base damage in mice. Oncogene. doi: 10.1038/sj.onc.1210167. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 33.Tuo J, Jaruga P, Rodriguez H, Bohr VA, Dizdaroglu M. Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. FASEB J. 2003;17:668–674. doi: 10.1096/fj.02-0851com. [DOI] [PubMed] [Google Scholar]
- 34.Flohr C, Burkle A, Radicella JP, Epe B. Poly(ADP-ribosyl)ation accelerates DNA repair in a pathway dependent on Cockayne syndrome B protein. Nucleic Acids Res. 2003;31:5332–5337. doi: 10.1093/nar/gkg715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thorslund T, von Kobbe C, Harrigan JA, Indig FE, Christiansen M, Stevnsner T, Bohr VA. Cooperation of the Cockayne syndrome group B protein and poly(ADP-ribose) polymerase 1 in the response to oxidative stress. Mol. Cell. Biol. 2005;25:7625–7636. doi: 10.1128/MCB.25.17.7625-7636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spivak G, Hanawalt PC. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA Repair. 2006;5:13–22. doi: 10.1016/j.dnarep.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 37.Check E. Retracted papers damage work on DNA repair. Nature. 2005;435:1015. doi: 10.1038/4351015a. [DOI] [PubMed] [Google Scholar]
- 38.Andressoo JO, Hoeijmakers JH. Transcription-coupled repair and premature ageing. Mutat. Res. 2005;577:179–194. doi: 10.1016/j.mrfmmm.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 39.Wilson DM, III, Barsky D. The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat. Res. 2001;485:283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- 40.Hu J, Imam SZ, Hashiguchi K, de Souza-Pinto NC, Bohr VA. Phosphorylation of human oxoguanine DNA glycosylase (alpha-OGG1) modulates its function. Nucleic Acids Res. 2005;33:3271–3282. doi: 10.1093/nar/gki636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muftuoglu M, Wong HK, Imam SZ, Wilson DM, III, Bohr VA, Opresko PL. Telomere Repeat Binding Factor 2 Interacts with Base Excision Repair Proteins and Stimulates DNA Synthesis by DNA Polymerase {beta} Cancer Res. 2006;66:113–124. doi: 10.1158/0008-5472.CAN-05-2742. [DOI] [PubMed] [Google Scholar]
- 42.Erzberger JP, Barsky D, Scharer OD, Colvin ME, Wilson DM., III Elements in abasic site recognition by the major human and Escherichia coli apurinic/apyrimidinic endonucleases. Nucleic Acids Res. 1998;26:2771–2778. doi: 10.1093/nar/26.11.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng WH, von KC, Opresko PL, Fields KM, Ren J, Kufe D, Bohr VA. Werner syndrome protein phosphorylation by abl tyrosine kinase regulates its activity and distribution. Mol. Cell. Biol. 2003;23:6385–6395. doi: 10.1128/MCB.23.18.6385-6395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson DM., III Ape1 abasic endonuclease activity is regulated by magnesium and potassium concentrations and is robust on alternative DNA structures. J. Mol. Biol. 2005;345:1003–1014. doi: 10.1016/j.jmb.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 45.McNeill DR, Narayana A, Wong HK, Wilson DM., III Inhibition of Ape1 nuclease activity by lead, iron, and cadmium. Environ. Health Perspect. 2004;112:799–804. doi: 10.1289/ehp.7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong HK, Kim D, Hogue BA, McNeill DR, Wilson DM., III DNA damage levels and biochemical repair capacities associated with XRCC1 deficiency. Biochemistry. 2005;44:14335–14343. doi: 10.1021/bi051161o. [DOI] [PubMed] [Google Scholar]
- 47.Mayne LV, Priestley A, James MR, Burke JF. Efficient immortalization and morphological transformation of human fibroblasts by transfection with SV40 DNA linked to a dominant marker. Exp. Cell Res. 1986;162:530–538. doi: 10.1016/0014-4827(86)90356-3. [DOI] [PubMed] [Google Scholar]
- 48.Selzer RR, Nyaga S, Tuo J, May A, Muftuoglu M, Christiansen M, Citterio E, Brosh RM, Jr, Bohr VA. Differential requirement for the ATPase domain of the Cockayne syndrome group B gene in the processing of UV-induced DNA damage and 8-oxoguanine lesions in human cells. Nucleic Acids Res. 2002;30:782–793. doi: 10.1093/nar/30.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong HK, Wilson DM., III XRCC1 and DNA polymerase beta interaction contributes to cellular alkylating-agent resistance and single-strand break repair. J. Cell. Biochem. 2005;95:794–804. doi: 10.1002/jcb.20448. [DOI] [PubMed] [Google Scholar]
- 50.Wilson DM, III, Takeshita M, Grollman AP, Demple B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J. Biol. Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 51.Mol CD, Hosfield DJ, Tainer JA. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3′ ends justify the means. Mutat. Res. 2000;460:211–229. doi: 10.1016/s0921-8777(00)00028-8. [DOI] [PubMed] [Google Scholar]
- 52.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 53.Wilson DM, III, Takeshita M, Demple B. Abasic site binding by the human apurinic endonuclease, Ape, and determination of the DNA contact sites. Nucleic Acids Res. 1997;25:933–939. doi: 10.1093/nar/25.5.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakamura J, Walker VE, Upton PB, Chiang SY, Kow YW, Swenberg JA. Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer Res. 1998;58:222–225. [PubMed] [Google Scholar]
- 55.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chem. Res. Toxicol. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boorstein RJ, Cummings A, Jr, Marenstein DR, Chan MK, Ma Y, Neubert TA, Brown SM, Teebor GW. Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J. Biol. Chem. 2001;276:41991–41997. doi: 10.1074/jbc.M106953200. [DOI] [PubMed] [Google Scholar]
- 57.Boorstein RJ, Chiu LN, Teebor GW. A mammalian cell line deficient in activity of the DNA repair enzyme 5-hydroxymethyluracil-DNA glycosylase is resistant to the toxic effects of the thymidine analog 5-hydroxymethyl-2'-deoxyuridine. Mol. Cell. Biol. 1992;12:5536–5540. doi: 10.1128/mcb.12.12.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 59.Christiansen M, Thorslund T, Jochimsen B, Bohr VA, Stevnsner T. The Cockayne syndrome group B protein is a functional dimer. FEBS J. 2005;272:4306–4314. doi: 10.1111/j.1742-4658.2005.04844.x. [DOI] [PubMed] [Google Scholar]
- 60.Demple B, DeMott MS. Dynamics and diversions in base excision DNA repair of oxidized abasic lesions. Oncogene. 2002;21:8926–8934. doi: 10.1038/sj.onc.1206178. [DOI] [PubMed] [Google Scholar]
- 61.Gros L, Ishchenko AA, Ide H, Elder RH, Saparbaev MK. The major human AP endonuclease (Ape1) is involved in the nucleotide incision repair pathway. Nucleic Acids Res. 2004;32:73–81. doi: 10.1093/nar/gkh165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verly WG, Tuy M, Brakier L, James M. Interaction between T7 coliphage denatured DNA and Sarcina lutea RNA. Biochim. Biophys. Acta. 1970;217:192–195. doi: 10.1016/0005-2787(70)90136-x. [DOI] [PubMed] [Google Scholar]
- 63.Boule-Charest L, Mamet-Bratley MD. Initiation by RNA polymerase on alkylated T7 DNA. Biochim. Biophys. Acta. 1972;277:276–279. doi: 10.1016/0005-2787(72)90408-x. [DOI] [PubMed] [Google Scholar]
- 64.Flamee PA, Verly WG. Action of intact AP (apurinic/apyrimidinic) sites and AP sites associated with breaks on the transcription of T7 coliphage DNA by Escherichia coli RNA polymerase. Biochem. J. 1985;229:173–181. doi: 10.1042/bj2290173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou W, Doetsch PW. Effects of abasic sites and DNA single-strand breaks on prokaryotic RNA polymerases. Proc. Natl Acad. Sci. USA. 1993;90:6601–6605. doi: 10.1073/pnas.90.14.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanchez G, Racine JF, Mamet-Bratley MD. Effect of abasic sites on bacteriophage T7 protein synthesis. Mutat. Res. 1994;325:39–45. doi: 10.1016/0165-7992(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 67.Sanchez G, Mamet-Bratley MD. Transcription by T7 RNA polymerase of DNA containing abasic sites. Environ. Mol. Mutagen. 1994;23:32–36. doi: 10.1002/em.2850230106. [DOI] [PubMed] [Google Scholar]
- 68.Wang Y, Sheppard TL, Tornaletti S, Maeda LS, Hanawalt PC. Transcriptional inhibition by an oxidized abasic site in DNA. Chem. Res. Toxicol. 2006;19:234–241. doi: 10.1021/tx050292n. [DOI] [PubMed] [Google Scholar]
- 69.Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, Campeau E, Tainer JA, Nogales E, et al. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne Syndrome. Mol. Cell. 2005;20:187–198. doi: 10.1016/j.molcel.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 70.Kathe SD, Shen GP, Wallace SS. Single-stranded breaks in DNA but not oxidative DNA base damages block transcriptional elongation by RNA polymerase II in HeLa cell nuclear extracts. J. Biol. Chem. 2004;279:18511–18520. doi: 10.1074/jbc.M313598200. [DOI] [PubMed] [Google Scholar]
- 71.Plosky B, Samson L, Engelward BP, Gold B, Schlaen B, Millas T, Magnotti M, Schor J, Scicchitano DA. Base excision repair and nucleotide excision repair contribute to the removal of N-methylpurines from active genes. DNA Repair. 2002;1:683–696. doi: 10.1016/s1568-7864(02)00075-7. [DOI] [PubMed] [Google Scholar]
- 72.Luo M, Kelley MR. Inhibition of the human apurinic/apyrimidinic endonuclease (APE1) repair activity and sensitization of breast cancer cells to DNA alkylating agents with lucanthone. Anticancer Res. 2004;24:2127–2134. [PubMed] [Google Scholar]
- 73.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 74.Thompson LH, Brookman KW, Jones NJ, Allen SA, Carrano AV. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol. Cell. Biol. 1990;10:6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ding R, Smulson M. Depletion of nuclear poly(ADP-ribose) polymerase by antisense RNA expression: influences on genomic stability, chromatin organization, and carcinogen cytotoxicity. Cancer Res. 1994;54:4627–4634. [PubMed] [Google Scholar]
- 76.Trucco C, Oliver FJ, de MG, Menissier-de MJ. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998;26:2644–2649. doi: 10.1093/nar/26.11.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Madhusudan S, Smart F, Shrimpton P, Parsons JL, Gardiner L, Houlbrook S, Talbot DC, Hammonds T, Freemont PA, et al. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 2005;33:4711–4724. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Horton JK, Joyce-Gray DF, Pachkowski BF, Swenberg JA, Wilson SH. Hypersensitivity of DNA polymerase beta null mouse fibroblasts reflects accumulation of cytotoxic repair intermediates from site-specific alkyl DNA lesions. DNA Repair. 2003;2:27–48. doi: 10.1016/s1568-7864(02)00184-2. [DOI] [PubMed] [Google Scholar]
- 79.Horton JK, Stefanick DF, Naron JM, Kedar PS, Wilson SH. Poly(ADP-ribose) polymerase activity prevents signaling pathways for cell cycle arrest after DNA methylating agent exposure. J. Biol. Chem. 2005;280:15773–15785. doi: 10.1074/jbc.M413841200. [DOI] [PubMed] [Google Scholar]
- 80.Fan J, Wilson DM., III Protein-protein interactions and posttranslational modifications in mammalian base excision repair. Free Radic. Biol. Med. 2005;38:1121–1138. doi: 10.1016/j.freeradbiomed.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 81.Muftuoglu M, Selzer R, Tuo J, Brosh RM, Jr, Bohr VA. Phenotypic consequences of mutations in the conserved motifs of the putative helicase domain of the human Cockayne syndrome group B gene. Gene. 2002;283:27–40. doi: 10.1016/s0378-1119(01)00870-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.