Abstract
We have examined the folding, stability and kinetics of intramolecular quadruplexes formed by DNA sequences containing four G3 tracts separated by either single T or T4 loops. All these sequences fold to form intramolecular quadruplexes and 1D-NMR spectra suggest that they each adopt unique structures (with the exception of the sequence with all three loops containing T4, which is polymorphic). The stability increases with the number of single T loops, though the arrangement of different length loops has little effect. In the presence of potassium ions, the oligonucleotides that contain at least one single T loop exhibit similar CD spectra, which are indicative of a parallel topology. In contrast, when all three loops are substituted with T4 the CD spectrum is typical of an antiparallel arrangement. In the presence of sodium ions, the sequences with two and three single T loops also adopt a parallel folded structure. Kinetic studies on the complexes with one or two T4 loops in the presence of potassium ions reveal that sequences with longer loops display slower folding rates.
INTRODUCTION
DNA sequences that contain four or more closely spaced G-tracts can fold to form intramolecular quadruplexes, which consist of stacked G-quartets that are linked by three loops between the four G-strands (1–4). These structures are stabilized by monovalent cations (especially potassium) (5,6) and can adopt a variety of different folding patterns dependent on the relative orientation of the strands and the position of the loops. G-rich sequences with the potential to form quadruplex structures are common in genomic DNA and these have been identified in several biologically important regions (7–9). The most widely studied is telomeric DNA, which in higher eukaryotes is composed of repeats of the sequence GGGTTA (10,11) and for which about 50–100 bases at the 3′-end are single stranded. A number of other non-telomeric G-rich DNA sequences may also form quadruplexes and these have been identified in the promoters of c-myc (12–15), Ki-ras (16), bcl2 (17–19), c-kit (20), VEGF gene (21) and HIF 1α (22), as well as in fragile X-syndrome (23) and other trinucleotide repeat sequences (24), the retinoblastoma susceptibility gene (25), the chicken β-globin gene (26) and the insulin gene (27). G-rich sequences are especially abundant in gene promoter regions (8) and there is an overabundance of G-rich sequences in the regulatory regions of muscle-specific genes (28).
For intramolecular quadruplexes, the four G-tracts are separated by loops. These are of various lengths and can be as short as a single nucleotide (29–31). Genomic searches (7,9) have revealed many G-rich sequences which may be able to adopt these structures, the most common of which are successive G-tracts that are separated by single T or A residues. The loops can be arranged in several different ways; double-chain reversal (propeller) loops link two adjacent parallel strands (32), while edgewise or diagonal loops link two antiparallel strands (33). Some structures contain both edge-wise and propeller loops (34–36). In the all-parallel (propeller) structures, the nucleotides are in the anti conformation, while the other structures have different combinations of anti or syn glycosidic bonds (3,4). It is known that loop length and sequence affect quadruplex stability and structure (29,37–40). Sequences with single nucleotide loops between the G3 tracts only adopt a parallel structure, while longer loops can also adopt an antiparallel arrangement of the strands. Quadruplex stability is also affected by the sequence of the loops (39–41), and the bases that flank the quadruplex (42–44).
There is considerable variation in quadruplex structure, depending on the DNA sequence and the ionic conditions. The biological function of quadruplexes may well depend on the folded conformation that is adopted, especially if this involves interaction with specific proteins. Such an effect has been suggested for the NHE element of the c-myc promoter, which can in principle adopt multiple conformations. Since the loops can have a considerable effect on quadruplex folding and stability, we have examined how changes in loop length affect quadruplex properties. One very stable intramolecular quadruplex contains four G3 tracts that are linked by single T residues (30,41,45) and this is known to be an inhibitor of HIV integrase. We have used variations on this sequence to examine the importance of loop length on quadruplex folding and stability. In this study, we have systematically replaced each of the single T loops with T4 and have used CD, fluorescence melting, 1D-NMR, gel electrophoresis and kinetic studies to examine the effect of loop length and position on quadruplex folding and stability.
MATERIAL AND METHODS
Oligonucleotides
All oligonucleotides were synthesized on an Applied Biosystems ABI 394 automated DNA/RNA synthesiser on the 0.2 µmole scale using the standard cycles of acid-catalysed detritylation, coupling, capping and iodine oxidation procedures. Phosphoramidite monomers and other reagents were purchased from Applied Biosystems, Proligo and Link Technologies. The sequences of the oligonucleotides used in this work are shown in Table 1. Fluorescently labelled oligonucleotiodes were used in all the experiments. These were labelled at the 5′-end with 6-amidohexylfluorescein (FAM), and at the 3′-end with dabcyl using C7 dabcyl cpg (Link Technologies). Oligonucleotides were purified by gel filtration using Nap10 columns (GE Healthcare) and analysed by gel electrophoresis. The bases adjacent to the fluorophore and quencher were the same (T) for all the oligonucleotides to avoid any differences in their effects on quadruplex formation and stability.
Table 1.
Sequences of the quadruplex-forming oligonucleotides used in this work
| Name | Sequence | ||||||
|---|---|---|---|---|---|---|---|
| Loop 1 | Loop 2 | Loop 3 | |||||
| G3T | d-F-TGGG | T | GGG | T | GGG | T | GGGT-Q |
| G3T-T4-T | d-F-TGGG | T | GGG | TTTT | GGG | T | GGGT-Q |
| G3T4-T-T | d-F-TGGG | TTTT | GGG | T | GGG | T | GGGT-Q |
| G3T4-T-T4 | d-F-TGGG | TTTT | GGG | T | GGG | TTTT | GGGT-Q |
| G3T4-T4-T | d-F-TGGG | TTTT | GGG | TTTT | GGG | T | GGGT-Q |
| G3T4 | d-F-TGGG | TTTT | GGG | TTTT | GGG | TTTT | GGGT-Q |
F = FAM; Q = dabcyl.
Fluorescence melting studies
The thermal melting temperatures of the quadruplexes were determined using the fluorescence melting technique that we have developed (46) and have used previously for assessing the stability of related quadruplexes (39,41,44,47). When the sequence adopts a folded structure the quencher and fluorophore are in close proximity and the fluorescence is quenched. When the structure melts, these groups become separated and there is a large increase in fluorescence. Since the fluorophore and quencher are anchored on relatively long aliphatic tethers the quenching does not depend on the quadruplex topology and the fluorescence is quenched for both parallel and antiparallel complexes. Fluorescence melting experiments were conducted in a Roche LightCycler as previously described (39,41,44,46,47) in a total reaction volume of 20 µl. Oligonucleotides (final concentration 0.25 µM) were prepared in 10 mM lithium phosphate pH 7.4, which was supplemented with various concentrations of potassium chloride or sodium chloride. The LightCycler has one excitation source (488 nm) and the changes in fluorescence were measured at 520 nm. For several of the oligonucleotides initial experiments revealed that there was considerable hysteresis between the heating and annealing profiles when the temperature was changed at 0.2°C.s−1, indicating that the process was not at thermodynamic equilibrium. Melting experiments were therefore performed at a much slower rate of heating and cooling (0.2°C.min−1) by changing the temperature in 1°C steps, leaving the samples to equilibrate for 5 min at each temperature before recording the fluorescence. Under these conditions, no hysteresis was observed (except for some experiments with G3T4). In a typical experiment, the oligonucleotides were first denatured by heating to 95°C for 5 min. They were then annealed by cooling to 30°C at 0.2°C.min−1 and melted by heating to 95°C at the same rate. The fluorescence was recorded during both the annealing and melting steps. In some instances, the formation of intramolecular or intermolecular complexes was examined by determining the melting curves using a range of oligonucleotide concentrations (0.1–10 µM). Melting temperatures (Tm values) were determined from the first derivatives of the melting profiles using the Roche LightCycler software.
Thermodynamic and kinetic analysis
Tm values were obtained from the maxima of the first derivatives of the melting profiles using the LightCycler software or, together with ΔH, from van't Hoff analysis of the melting profiles using FigP for Windows. The fraction folded was calculated as previously described (48) from the difference between the measured fluorescence and the upper and lower baselines. All reactions were performed at least twice and the calculated Tm values usually differed by <0.5°C with a 5% variation in ΔH. Since ΔG = 0 at the Tm, ΔS was estimated as ΔH/Tm. Values for ΔG at 310 K were then estimated from ΔG = ΔH – TΔS. The van't Hoff analysis assumes that ΔH is independent of temperature (i.e. ΔCp = 0), that the reaction is only a two-step process (i.e. that there are no significant reaction intermediates) and that there is only one folded form of the quadruplex. The number of specifically bound monovalent cations (Δn), was calculated from the slopes of plots of ΔG against log[M+] as previously described (30,49).
Hysteresis between the melting and annealing profiles occurs when the reaction is not at thermodynamic equilibrium as a result of the slow folding and/or unfolding kinetics. Individual folding (k1) and unfolding (k−1) rate constants can be derived from this hysteresis as previously described (47,48,50,51).
Temperature jump kinetics
The kinetics of quadruplex unfolding were also determined by measuring the rate of change of fluorescence after rapidly increasing the temperature (47,52). The quadruplexes were equilibrated at a temperature around the Tm, which was then rapidly increased by 5°C at the fastest rate on the LightCycler (20°C.s−1). This temperature change causes the quadruplex to partially unfold, moving along the melting curve. Although the theoretical dead-time under these conditions is only 0.25 s, all fluorescence changes that occurred in the first 2 s were ignored, during equilibration to the new temperature. Successive temperature-jumps were then recorded on the same sample by further increasing the temperature by 5°C. Each experiment was repeated at least twice. The time-dependent changes in fluorescence were fitted by an exponential function Ft = Ff × (1 − e−kt) + F0, using SigmaPlot 10, where Ft is fluorescence at time t, F0 is the initial fluorescence and Ff is total change in fluorescence (the final fluorescence is Ff + F0). The relaxation rate constant (k) obtained from this analysis is equal to the sum of the folding (k1) and unfolding (k−1) rate constants. Arrhenius plots of ln(k) against 1/T were constructed from these data and used to estimate the activation energy Ea and pre-exponential factor A [k = A × exp(−Ea/RT)].
Gel electrophoresis
Non-denaturing gel electrophoresis was performed using 14% polyacrylamide gels, which were run in TBE buffer that had been supplemented with 20 mM KCl. Bands in the gels were visualised under UV light. The oligonucleotide concentration was 20 µM.
Circular dichroism
CD spectra were measured on a Jasco J-720 spectropolarimeter as previously described (39). Oligonucleotide solutions (5 µM) were prepared in 10 mM lithium phosphate pH 7.4, containing either 200 mM potassium chloride or 200 mM sodium chloride. The samples were heated to 95°C and annealed by slowly cooling to 15°C over a period of 12 h. Spectra were recorded between 220 and 320 nm in 5 mm path length cuvettes. Spectra were averaged over 10 scans, which were recorded at 100 nm.min−1 with a response time of 1 s and a bandwidth of 1 nm. A buffer baseline was subtracted from each spectrum and the spectra were normalized to have zero ellipticity at 320 nm.
Proton NMR
One-dimensional 1H NMR experiments were performed on a Varian Inova 600 MHz spectrometer. Oligonucleotides were prepared in 200 mM potassium phosphate pH 7.4 and were annealed by heating to 95°C before slowly cooling to 15°C. 300 µl of the oligonucleotide sample was mixed with 20 µl D2O and placed in a Shigemi NMR tube. The final strand concentration was 100 µM. 1D proton NMR spectra were recorded at 25°C with a sweep width of 25 p.p.m., WATERGATE water suppression, an acquisition time of 0.5 s and 32 k scans. Data were processed using VNMR software (Varian Inc.) with zero filling and resolution enhancement.
RESULTS
A variety of physical techniques were used to examine the folding, stability and kinetics of the intramolecular quadruplexes that are formed by sequences containing four G3 tracts separated by either single T or T4 loops, in different combinations. The sequences of these oligonucleotides are shown in Table 1.
Circular dichroism
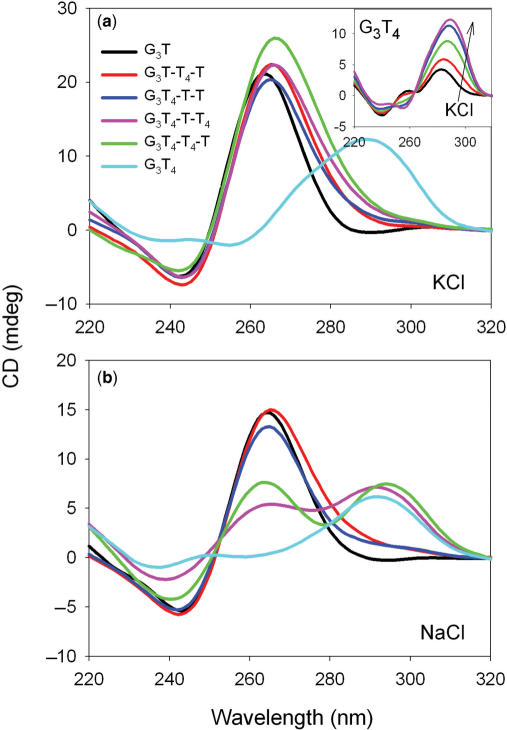
Intramolecular quadruplexes can adopt a variety of different topologies in which the strands run in different orientations with lateral, edgewise or diagonal loops. Circular dichroism has frequently been used to indicate whether these fold in a parallel or antiparallel configuration (43,53,54). Parallel quadruplexes, in which the glycosidic bonds are all anti, display a positive CD signal around 265 nm, with a negative peak at 240 nm. In contrast, antiparallel topologies, with both syn and anti bonds, exhibit a positive signal at around 295 nm, with a negative signal or shoulder around 260 nm. CD spectra for these oligonucleotides, in the presence of sodium or potassium ions, are shown in Figure 1 [other studies with related sequences have shown that the fluorophores do not affect the CD spectra (55)].
Figure 1.
CD spectra of the fluorescently-labelled quadruplex-forming oligonucleotides in the presence of 10 mM lithium phosphate pH 7.4 containing 200 mM KCl (a) or 200 mM NaCl (b). G3T, black; G3T-T4-T, red; G3T4-T-T, blue; G3T4-T-T4, pink; G3T4-T4-T, green; G3T4, cyan. The inset to the upper panel shows the CD spectrum of G3T4 in the presence of different concentrations of KCl: black 1 mM; red, 5 mM; green 20 mM; blue 50 mM; pink, 200 mM.
In the presence of potassium (Figure 1) all the sequences, except G3T4 show CD spectra with a positive peak around 265 nm and a minimum around 240 nm, which is typical of the parallel configuration. In contrast, G3T4 displays a positive peak at 295 nm, indicative of an antiparallel topology. Quadruplexes with single nucleotide loops are thought to be only able to form ‘propeller-type’ fold-back loops generating parallel-stranded complexes, while longer T4 loops can form lateral, edgewise or diagonal loops. These CD spectra suggest that the presence of only one single T loop is sufficient to induce the formation of a parallel-stranded structure and that the complexes only adopt an antiparallel arrangement when all the loops are longer. In general, these CD spectra were independent of the potassium concentration in the range 20–200 mM K+, though pronounced changes were observed for G3T4 (inset to Figure 1a). For this sequence a secondary peak is visible around 260 nm at low potassium ion concentrations, which disappears as the potassium ion concentration is increased; this is accompanied by an increase in the peak at 295 nm. The presence of isoelliptic points in these spectra suggests that this sequence may adopt two distinct structural forms in the presence of low or high potassium ion concentrations.
In the presence of sodium ions, the CD spectra for G3T, G3T-T4-T and G3T4-T-T are again typical of a parallel topology, with peaks around 265 nm (Figure 1b). However, the addition of a second T4 loop results in spectra with equal-sized peaks at 265 nm and 295 nm. It has been suggested that sodium ions promote the formation of antiparallel topologies and it is possible that the two longer T4 loops are laterally arranged, while the single T loop is in a fold-back arrangement. This mixed spectrum could indicate the co-existence of parallel and antiparallel topologies in solution, but a hybrid structure containing both syn and anti bonds seems more likely; this will be considered further in the Discussion. The spectrum of G3T4 is similar in the presence of sodium and potassium ions, with a peak at 295 nm, suggesting an antiparallel topology.
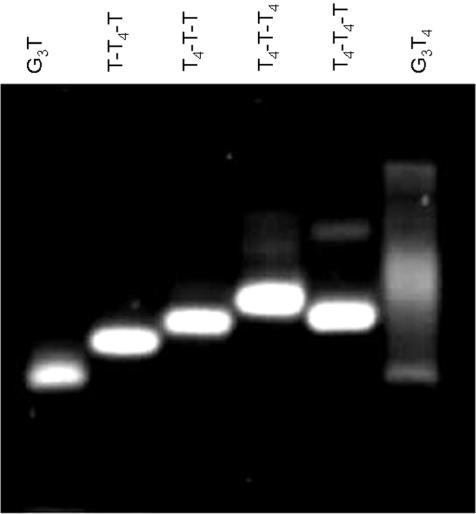
Gel mobility
We further compared the global structures of these sequences by examining their mobilities in polyacrylamide gels that had been supplemented with 20 mM KCl (Figure 2). Each of the sequences ran as a single band, with the exception of G3T4 which is smeared, possibly because this sequence is less stable under these conditions (Tm ∼ 40°C, see below). G3T has the greatest mobility, as expected as it has the lowest molecular weight. Surprisingly we find that G3T4-T-T has a lower mobility than G3T-T4-T and similarly G3T4-T-T4 is slower than G3T4-T4-T. It appears that the presence of a single T in the central loop reduces the mobility. This will be considered in the Discussion.
Figure 2.
Mobility of the quadruplex-forming oligonucleotides on a 14% polyacrylamide gel supplemented with 20 mM KCl.
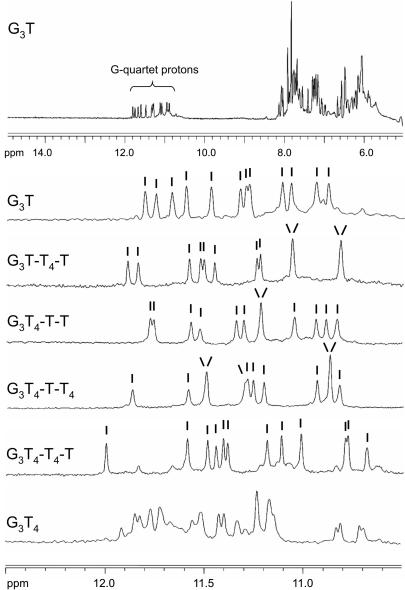
Imino proton NMR spectra
One of the defining features of structures that contain G-quartets is the appearance of imino proton resonances between 10.5 and 12.0 p.p.m. in NMR spectra (56). Examination of this region of NMR spectra has often been used to assess whether the sequence adopts a unique structure (20,34,36,56) and the presence of multiple or ill-defined peaks is evidence for the existence of multiple structures. The imino proton spectra for each of these sequences are shown in Figure 3. It can be seen that the spectra of G3T, G3T-T4-T, G3T4-T-T and G3T4-T-T4 display between 10 and 12 well-resolved and sharp peaks, indicative of well-defined structures. In the cases where 10 or 11 peaks are resolved, the intensities indicate that one or two imino protons have degenerate chemical shifts. Hence, the number of hydrogen-bonded imino protons is 12, as expected for three stacked G-quartets. G3T4-T4-T also shows 12 major peaks, though the spectrum contains some minor peaks, which might indicate the presence of a small amount of a second structure. In contrast, the imino proton spectrum of G3T4 shows multiple peaks confirming that it adopts more than one stable conformation.
Figure 3.
1D imino proton NMR spectra of the quadruplex-forming oligonucleotides. The samples (100 µM) were prepared in 200 mM potassium phosphate pH 7.4. The top panel shows the 1D-NMR spectrum for G3T between 5 and 15 p.p.m., while the other panels show the imino proton region for each oliogonucleotide. The individual peaks are indicated.
Fluorescence melting curves
Representative fluorescent melting curves for these sequences are shown in Figure 4 in the presence of potassium and sodium ions. The Tm values at different ionic strengths, along with the calculated values for ΔH, are shown in Table 2. The samples were melted and annealed at 0.2°C.min−1; no hysteresis was observed at this rate of temperature change (except for G3T4 at low ionic strengths). The melting temperatures were all independent of concentration (between 0.1 and 10 µM; Supplementary material Figure 1) confirming that these oligonucleotide sequences form intramolecular (not intermolecular) complexes. As expected, all the complexes are more stable in potassium than sodium ions. G3T is the most stable and, in the presence of potassium, substituting a T4 loop instead of a loop with a single T decreases the Tm by about 20°C, irrespective of whether the replacement is in a central (G3T-T4-T) or peripheral loop (G3T4-T-T), though G3T-T4-T is about 2–3°C more stable than G3T4-T-T. Replacing a second T loop with T4 causes a further 20°C decrease in Tm and G3T4-T4-T is about 2–4°C more stable than G3T4-T-T4. In each case, the sequence with a single T in the central loop is slightly less stable than the equivalent sequence with T4 in the same position. Replacing all three single T loops with T4 decreases the Tm by a further 10°C, though the melting and annealing curves with this sequence show hysteresis at low ionic strengths and the melting (but not the annealing) profiles are biphasic.
Figure 4.
Fluorescence melting profiles for the quadruplex-forming oligonucleotides. The reactions were performed in 10 mM lithium phosphate pH 7.4 containing either 20 mM KCl (left hand panel) or 200 mM NaCl (right hand panel). The temperature was changed at 0.2°C.min−1. The curves show the fraction folded (α) as a function of temperature, calculated as described in the Methods section. G3T, black; G3T-T4-T, red; G3T4-T-T, blue; G3T4-T-T4, pink; G3T4-T4-T, green; G3T4, cyan.
Table 2.
Tm and ΔH values for the fluorescently labelled quadruplex-forming oligonucleotides, determined in the presence of 10 mM lithium phosphate pH 7.4 containing different concentrations of KCl. The samples were heated and cooled at a rate of 0.2°C.min−1
| [KCl] mM | G3T | G3T-T4-T | G3T4-T-T | G3T4-T-T4 | G3T4-T4-T | G3T4 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tm°C | ΔH kJ.mol−1 | Tm°C | ΔH kJ.mol−1 | Tm°C | ΔH kJ.mol−1 | Tm°C | ΔH kJ.mol−1 | Tm°C | ΔH kJ.mol−1 | Tm°C | ΔH kJ.mol−1 | |
| 0 | 46.6 | |||||||||||
| 0.1 | 57.2 | −242 ± 4 | 37.7 | |||||||||
| 1 | 73.5 | −271 ± 5 | 53.8 | −227 ± 7 | 51.3 | −208 ± 7 | ||||||
| 5 | 85.7 | −275 ± 5 | 65.3 | −259 ± 11 | 63.4 | −250 ± 15 | ||||||
| 10 | 70.7 | −266 ± 9 | 67.3 | −258 ± 9 | 46.3 | −184 ± 13 | 48.1 | −205 ± 5 | ||||
| 20 | 75.1 | −267 ± 7 | 73.1 | −262 ± 10 | 52.5 | −207 ± 3 | 54.1 | −227 ± 11 | 44.5/37.6* | |||
| 50 | 81.4 | −284 ± 7 | 79.8 | −275 ± 8 | 59.5 | −225 ± 13 | 60.5 | −246 ± 8 | 47.0/51.0* | −234 ± 11 | ||
| 100 | 87.5 | 84.6 | 65.3 | −247 ± 8 | 67.1 | −269 ± 13 | 56.3 | −266 ± 10 | ||||
| 200 | 72.8 | −256 ± 9 | 74.2 | −276 ± 10 | 63.3 | −293 ± 9 | ||||||
| Δn | 2.13 ± 0.10 | 2.29 ± 0.11 | 2.31 ± 0.16 | 2.75 ± 0.11 | 2.89 ± 0.12 | 4.01 ± 0.16 | ||||||
*Indicates a biphasic melting profile. All reactions were performed at least twice and the calculated Tm values usually differed by <0.5°C. ΔH values were typically calculated for melting profiles for which the Tm was between 40°C and 80°C. Missing values at low concentrations of KCl correspond to complexes for which the Tms were too low to measure (<30°C), while those at high ionic strengths (especially G3T) were too stable (Tm > 85°C).
The relative order of stability is the same in the presence of sodium ions (Supplementary material Table 1). Replacing one T loop with T4 decreases the Tm by 15–20°C, and a second substitution causes a further 10°C decrease. Replacing all three loops with T4 does not affect the stability any further and G3T4 has a similar Tm to G3T4-T4-T and G3T4-T-T4. As seen with potassium, sequence isomers with a central T4 loop are slightly more stable than those with a central T loop (i.e. G3T4-T4-T > G3T4-T-T4 and G3T-T4-T > G3T4-T-T).
ΔH values for the quadruplex single-strand transition in the presence of potassium were derived from these melting profiles by van't Hoff analysis, assuming that the reaction is a two-state equilibrium, and the values are shown in Table 2. These are typical of those for similar quadruplexes and show a general decrease in ΔH as the overall loop length increases. As previously reported for other quadruplexes, ΔH increases with ionic strength, consistent with the presence of specific cation binding sites within the quadruplex (30). The slopes of plots of ΔG against log[M+] can be used to determine the stoichiometry of cation binding (30) yielding values of Δn (the difference between the number of ions bound in the folded and unfolded states) and the values of Δn in the presence of potassium are listed in Table 2. For an antiparallel structure containing three stacked G-quartets, Δn would be expected to be either two (the number of potassium ions located between the stacked quartets) or four (including two more ions that may be coordinated between the loops and the terminal quartets). A value of two seems more likely for a parallel topology with single nucleotide loops, in which the loops do not interact with the terminal quartets. The values of Δn are between two and three for all the complexes that contain at least one loop with a single T residue, though there is a steady increase in this value with increased numbers of T4 loops, which will be considered further in the Discussion. Δn is larger for G3T4, consistent with the suggestion that it adopts a different topology, though this value may not be accurate as there is some hysteresis in its melting profiles at low potassium concentrations and the NMR data suggest that it adopts more than one conformation.
Kinetics of quadruplex formation
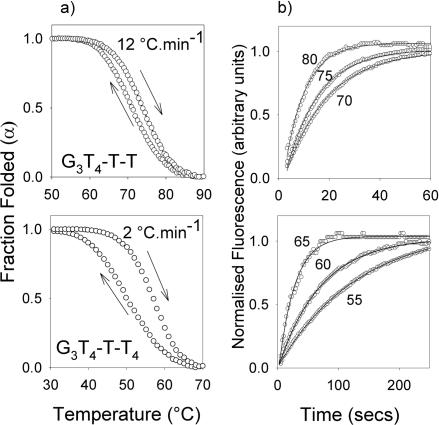
Hysteresis
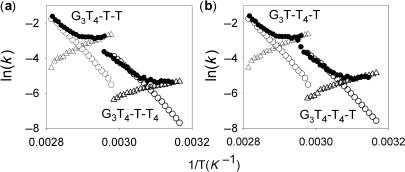
The fluorescence melting experiments shown in Figure 4 were performed at a rate of temperature change of 0.2°C.min−1 and only G3T4 showed hysteresis between the melting and annealing profiles. On increasing the rate to 2°C.min−1 there was a 7–10°C hysteresis for the sequences with two T4 loops in the presence of potassium, though the melting and annealing profiles were identical for the sequences with single T4 loops. The sequences with single T4 loops only displayed hysteresis when the rate of heating was increased to 12°C.min−1, while the melting and annealing curves for G3T were always superimposable. Representative heating and annealing curves at different rates of heating and cooling in the presence of 20 mM potassium are shown in Supplementary material Figure 2 and the different Tm values are summarized in Supplementary Table 1. No hysteresis was observed for any of these sequences in the presence of sodium at even the fastest rate of heating and cooling. Differences between the melting and annealing curves arise because the reaction is not at thermodynamic equilibrium and indicate that either the folding or the unfolding process is slow. The folding (k1) and unfolding (k−1) rate constants for the unimolecular folding reaction can be obtained by analysis of these data as previously described (47,48). Figure 5a shows the melting and annealing profiles for G3T4-T-T and G3T4-T-T4, determined at 12°C.min−1 and 2°C.min−1, respectively, while similar plots for G3T-T4-T and G3T4-T4-T are included in Supplementary material, Figure 3. Figure 6 shows Arrhenius plots for the folding and unfolding rates constructed from these data for G3T-T4-T and G3T4-T4-T (Figure 6a) and G3T4-T-T and G3T4-T-T4 (Figure 6b). The kinetic parameters derived from these Arrhenius plots are presented in Table 3. Several factors are apparent from these kinetic data. Firstly, the association reaction shows unusual temperature dependence, with an apparent negative activation energy, i.e. the reaction is faster a lower temperatures. This has been noted by others and is consistent with the reaction occurring by a nucleation-zipper mechanism (47,48). Secondly, the data for G3T4-T4-T are very similar to G3T4-T-T4 and G3T4-T-T is similar to G3T-T4-T, suggesting that the distribution of the different loops is less important than their length. Thirdly, the unfolding parameters are very similar for all four oligonucleotides, while the folding parameters vary according to the loop lengths. For the association reaction both ln(A) and Ea are less negative for the complexes with longer loops. This kinetic analysis was not performed for G3T as it showed no hysteresis and for G3T4 as the melting and annealing curves were biphasic.
Figure 5.
(a) Hystersis between the melting and annealing profiles for G3T4-T-T (upper panel, with a temperature change of 12°C.min−1) and G3T4-T-T4 (lower panel, with a temperature change of 2°C.min−1 in the presence of 10 mM lithium phosphate pH 7.4 containing 20 mM KCl. (b) temperature-jump relaxation profiles for G3T4-T-T (upper panel) and G3T4-T-T4 (lower panel. The traces show the rate of approach to a new equilibrium following a rapid 5°C increase in temperature to the value shown. The profiles have been normalized to show the fractional change in fluorescence with time.
Figure 6.
Arrhenius plots showing the temperature dependence of the kinetic parameters for G3T4-T-T and G3T4-T-T4 (a) and G3T-T4-T and G3T4-T4-T (b). Open symbols were derived from the hysteresis between the melting and annealing profiles; k−1, open circles; k1, open triangles. Filled circles show the time constants obtained from the temperature-jump experiments (k1 + k−1).
Table 3.
Kinetic parameters for folding (k1) and unfolding (k−1) of the quadruplex-forming oligonucleotides determined from analysis of the hysteresis between melting and annealing curves
| Sequence | k1 | k−1 | ||
|---|---|---|---|---|
| Ea (kJ mo1−1) | ln (A) s−1 | Ea (kJ mo1−1) | ln (A) s−1 | |
| G3T | - | - | - | - |
| G3T-T4-T | −93 ± 2 | −36 ± 1 | 182 ± 3 | 60 ± 1 |
| G3T4-T-T | −88 ± 3 | −34 ± 1 | 178 ± 2 | 58 ± 1 |
| G3T4-T-T4 | −48 ± 4 | −24 ± 1 | 173 ± 2 | 58 ± 1 |
| G3T4-T4-T | −59 ± 2 | −27 ± 1 | 174 ± 2 | 59 ± 1 |
| G3T4 | - | - | - | - |
The experiments were performed in 10 mM lithium phosphate pH 7.4 containing 20 mM potassium chloride. Ea is the activation energy (kJ.mol−1) and A is the pre-exponential factor from the equation k = Ae(–Ea/RT). No values are presented for G3T as it does not show any hysteresis, or G3T4 as more than one folded configuration exists in solution.
Temperature-jump kinetics
In order to confirm the kinetic data obtained from the hysteresis experiments we performed temperature-jump relaxation kinetics on these complexes. In this technique, the temperature of the complex (maintained around the Tm) is rapidly increased (by 5°C) and the time-dependent changes in fluorescence are recorded as the reaction relaxes to a new equilibrium. Representative temperature-jump relaxation profiles for these complexes are shown in Figure 5b and reveal a slow time-dependent relaxation to the new equilibrium, which is clearly faster for the complexes with two short loops than the ones with two long loops. The kinetic curves at different temperatures were fitted with single exponential functions and the rate constants obtained are presented as Arrhenius plots in Figure 6. For this unimolecular reaction the apparent rate constant for the relaxation is equal to the sum of the folding and unfolding rate constants (k−1 + k1). Although it is not possible to resolve these individual components, at low temperatures the sum is dominated by k1, while the sum approximates to k−1 at high temperatures. It can be seen that there is excellent agreement between the rate constants determined by the two independent methods for each of the sequences, confirming that sequences with longer loops display slower rates of folding with little effect on the rate of unfolding.
DISCUSSION
Topology
Circular dichroism is often used to indicate the folding topology of DNA quadruplexes (20,38,53,54). Antiparallel quadruplexes typically have a positive CD signal at around 295 nm, while parallel quadruplexes display a positive signal around 260 nm. These differences reflect both the arrangements of the strands and the syn/anti orientations around the glycosidic bonds. Parallel topologies have all-anti glycosidic angles, while antiparallel ones have both syn and anti in varying ratios. However, it is clear that these spectral signatures are not necessarily an indicator of quadruplex folding as some exceptions have been noted (57,58). Nonetheless, CD spectra are useful indicators of changes in global quadruplex configuration for series of related oligonucleotides. In the presence of potassium, all the oligonucleotides that contain at least one single T loop exhibit a similar CD spectrum that is indicative of a parallel topology. It therefore appears that in potassium the presence of only one single T loop, in any position, is sufficient to promote all the other loops to form a fold-back propeller-like structure. In principle, these oligonucleotides could adopt several different folded configurations, yet the NMR and gel electrophoresis experiments suggest that only one predominates. When all three loops contain T4, there is a dramatic change in the CD spectrum to a form that is consistent with antiparallel folding, though the details are dependent on the ionic strength suggesting that G3T4 can adopt multiple configurations. This again is consistent with the NMR and electrophoresis experiments, which suggest the presence of multiple folded forms. Previous studies have suggested that the quadruplex formed by d(G3T4G3)2 adopts an antiparallel hairpin dimer in the presence of both sodium and potassium (59,60).
A similar effect is seen for G3T, G3T4-T-T and G3T-T4-T in the presence of sodium ions and these display CD spectra that are consistent with parallel topologies. However, the greater propensity to form antiparallel structures in the presence of sodium is seen with T4-T4-T and T4-T-T4 loops, which have CD spectra with peaks at both 260 nm and 295 nm. This may indicate the presence of multiple structural forms, but it is more likely due to the formation of a structure that contains both edgewise (T4) and fold-back (T) loops, as observed with other sequences (34–36).
Stability
The fluorescence melting experiments show that the number of short loops, rather than their position, has the greatest effect on quadruplex stability. In the presence of potassium ions, G3T is the most stable and in concentrations above 5 mM it does not display a melting transition. Substituting a T4 into either the first or second loop decreases the Tm by about 20°C, with a further 20°C decrease on introducing a second T4 substitution. The same effect is seen in the presence of sodium ions though there is only a small decrease in stability on changing the third loop to T4, consistent with the CD spectra which show that G3T4-T-T4, G3T4-T4-T and G3T4 display some antiparallel characteristics in contrast to all the other oligonucleotides.
It is noticeable that sequences with a single T loop in the central position are less stable and have lower gel mobilities than their sequence isomers with T4 in this position (i.e. compare T4-T-T4 loops with T4-T4-T and T4-T-T with T-T4-T). It appears that folded structures with a central T4 loop are more compact and have higher thermal stability. There is then a further decrease in stability when all three loops are composed of T4, which as noted above adopts a different configuration.
Potassium ion binding
The variation of ΔG with ionic strength allows us to estimate the difference in the number of potassium ions specifically bound to the folded and unfolded structures. This value is close to two for G3T as expected, since two potassium ions can bind between the three stacked quartets. Although the precise values of Δn should be interpreted with caution, it is noticeable that there is a steady increase in this value as the number of longer loops is increased. The value of Δn is similar for G3T-T4-T and G3T4-T-T and is lower than for both G3T4-T4-T and G3T4-T-T4. These results suggest that the longer loops are involved in cation binding. The larger value of Δn seen with G3T4 may not be significant, as this sequence adopts multiple configurations.
Kinetic analysis
Comparing the kinetic parameters for the sequences with one or two T4 loops (Table 3) reveals that the unfolding parameters are very similar, while there are clear differences in the folding reaction. Complexes with longer loops have higher (less negative) activation energies for the association reaction and larger values for the pre-exponential factor (which is related to the entropy of the transition state). In comparison, no hysteresis is observed with G3T and temperature-jump experiments showed a very fast re-equilibration, while G3T4 has slower folding and unfolding parameters, though not as slow as G4T4 (47). It is clear that the folding of intramolecular structures with only single T loops is fast, and we imagine that when one loop is composed of a single nucleotide the G-tracts on either side rapidly associate, forming a platform to which the other G-tracts can bind. The results with the oligonucleotides containing one or two T4 loops suggest that the position of the single-base loop has little effect on the kinetics and that the most important factor is the number of longer loops.
It is interesting to note that none of these sequences show any hysteresis in the presence of sodium ions, even though G3T and those with two single T loops appear to adopt a similar global structure. The higher stability and slower kinetics in the presence of potassium may therefore reflect conformational changes subsequent to the initial folding events (45).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
PAR is supported by a research studentship from BBSRC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis JT. G-quartets 40 years later: From 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 3.Phan AT, Kuryavyi V, Patel DJ. DNA architecture: from G to Z. Curr. Opin. Struct. Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simonsson T. G-quadruplex DNA structures - Variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 5.Sen D, Gilbert W. A sodium-potassium switch in the formation of 4-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 6.Williamson JR, Raghuraman MK, Cech TR. Mono-valent cation induced atructure of telomeric DNA - the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 7.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sen D, Gilbert W. Formation of parallel 4-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 11.Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 12.Ambrus A, Chen D, Dai JX, Jones RA, Yang DZ. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 13.Rangan A, Fedoroff OY, Hurley LH. Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J. Biol. Chem. 2001;276:4640–4646. doi: 10.1074/jbc.M005962200. [DOI] [PubMed] [Google Scholar]
- 14.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai JX, Chen D, Jones RA, Hurley LH, Yang DZ. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai JX, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang DZ. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun DY, Guo KX, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Armond R, Wood S, Sun DY, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1 alpha promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 23.Fry M, Loeb LA. The fragile-X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsugami A, Okuizumi T, Uesugi S, Katahira M. Intramolecular higher order packing of parallel quadruplexes comprising a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad of GGA triplet repeat DNA. J. Biol. Chem. 2003;278:28147–28153. doi: 10.1074/jbc.M303694200. [DOI] [PubMed] [Google Scholar]
- 25.Murchie AIH, Lilley DMJ. Retinoblastoma susceptibility genes contain 5′ sequences with a high propensity to form guanine-tetrad structures. Nucleic Acids Res. 1992;20:49–53. doi: 10.1093/nar/20.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howell RM, Woodford KJ, Weitzmann MN, Usdin K. The chicken beta-globin gene promoter forms a novel “cinched” tetrahelical structure. J. Biol. Chem. 1996;271:5208–5214. doi: 10.1074/jbc.271.9.5208. [DOI] [PubMed] [Google Scholar]
- 27.Lew A, Rutter WJ, Kennedy GC. Unusual DNA structure of the diabetes susceptibility locus IDDM2 and its effect on transcription by the insulin promoter factor Pur-1/MAZ. Proc. Natl Acad. Sci. USA. 2000;97:12508–12512. doi: 10.1073/pnas.97.23.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yafe A, Etzioni S, Weisman-Shomer P, Fry M. Formation and properties of hairpin and tetraplex structures of guanine-rich regulatory sequences of muscle-specific genes. Nucleic Acids Res. 2005;33:2887–2900. doi: 10.1093/nar/gki606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hazel P, Parkinson GN, Neidle S. Topology variation and loop structural homology in crystal and simulated structures of a bimolecular DNA quadruplex. J. Am. Chem. Soc. 2006;128:5480–5487. doi: 10.1021/ja058577+. [DOI] [PubMed] [Google Scholar]
- 30.Jing NJ, Rando RF, Pommier Y, Hogan ME. Ion selective folding of loop domains in a potent anti-HIV oligonucleotide. Biochemistry. 1997;36:12498–12505. doi: 10.1021/bi962798y. [DOI] [PubMed] [Google Scholar]
- 31.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 34.Ambrus A, Chen D, Dai JX, Bialis T, Jones RA, Yang DZ. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: An intramolecular (3+1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cevec M, Plavec J. Role of loop residues and cations on the formation and stability of dimeric DNA G-quadruplexes. Biochemistry. 2005;44:15238–15246. doi: 10.1021/bi0514414. [DOI] [PubMed] [Google Scholar]
- 38.Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 39.Risitano A, Fox KR. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smirnov I, Shafer RH. Effect of loop sequence and size on DNA aptamer stability. Biochemistry. 2000;39:1462–1468. doi: 10.1021/bi9919044. [DOI] [PubMed] [Google Scholar]
- 41.Risitano A, Fox KR. Stability of intramolecular DNA quadruplexes: Comparison with DNA duplexes. Biochemistry. 2003;42:6507–6513. doi: 10.1021/bi026997v. [DOI] [PubMed] [Google Scholar]
- 42.Guo Q, Lu M, Kallenbach NR. Effect of thymine tract length on the structure and stability of model telomeric sequences. Biochemistry. 1993;32:3596–3603. doi: 10.1021/bi00065a010. [DOI] [PubMed] [Google Scholar]
- 43.Lu M, Guo Q, Kallenbach NR. Structure and stability of sodium and potassium complexes of dT4G4 and dT4G4T. Biochemistry. 1992;31:2455–2459. doi: 10.1021/bi00124a003. [DOI] [PubMed] [Google Scholar]
- 44.Merkina EE, Fox KR. Kinetic stability of intermolecular DNA quadruplexes. Biophys. J. 2005;89:365–373. doi: 10.1529/biophysj.105.061259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jing NJ, Gao XL, Rando RF, Hogan ME. Potassium-induced loop conformational transition of a potent anti-HIV oligonucleotide. J Biomol. Struct. Dyn. 1997;15:573–585. doi: 10.1080/07391102.1997.10508967. [DOI] [PubMed] [Google Scholar]
- 46.Darby RAJ, Sollogoub M, McKeen C, Brown L, Risitano A, Brown N, Barton C, Brown T, Fox KR. High throughput measurement of duplex, triplex and quadruplex melting curves using molecular beacons and a LightCycler. Nucleic Acids Res. 2002;30:e39. doi: 10.1093/nar/30.9.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown NM, Rachwal PA, Brown T, Fox KR. Exceptionally slow kinetics of the intramolecular quadruplex formed by the Oxytricha telomeric repeat. Org. Biomol. Chem. 2005;3:4153–4157. doi: 10.1039/b511706b. [DOI] [PubMed] [Google Scholar]
- 48.Mergny JL, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 49.Cantor CR, Schimmel PR. Biophysical Chemistry. New York: W. H. Freedman and Company; 1980. [Google Scholar]
- 50.Bernal-Mendez E, Leumann CJ. Stability and kinetics of nucleic acid triplexes with chimaeric DNA/RNA third strands. Biochemistry. 2002;41:12343–12349. doi: 10.1021/bi025937m. [DOI] [PubMed] [Google Scholar]
- 51.Rougee M, Faucon B, Mergny JL, Barcelo F, Giovannangeli C, Garestier T, Helene C. Kinetics and thermodynamics of triple-helix formation - effects of ionic-strength and mismatches. Biochemistry. 1992;31:9269–9278. doi: 10.1021/bi00153a021. [DOI] [PubMed] [Google Scholar]
- 52.James PL, Brown T, Fox KR. Thermodynamic and kinetic stability of intermolecular triple helices containing different proportions of C+.GC and T.AT triplets. Nucleic Acids Res. 2003;31:5598–5606. doi: 10.1093/nar/gkg782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balagurumoorthy P, Brahmachari SK, Mohanty D, Bansal M, Sasisekharan V. Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 1992;20:4061–4067. doi: 10.1093/nar/20.15.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 55.Rachwal PA, Brown T, Fox KR. Effect of G-tract length on the structure and stability of intramolecular DNA quadruplexes. Biochemistry. 2007;46:3036–3044. doi: 10.1021/bi062118j. [DOI] [PubMed] [Google Scholar]
- 56.Feigon J, Koshlap KM, Smith FW. H-1 NMR spectroscopy of DNA triplexes and quadruplexes. Methods Enzymol. 1995;261:225–255. doi: 10.1016/s0076-6879(95)61012-x. [DOI] [PubMed] [Google Scholar]
- 57.Dapic V, Abdomerovic V, Marrington R, Peberdy J, Rodger A, Trent JO, Bates PJ. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Esposito V, Randazzo A, Piccialli G, Petraccone L, Giancola C, Mayol L. Effects of an 8-bromodeoxyguanosine incorporation on the parallel quadruplex structure [d(TGGGT)]4. Org. Biomol. Chem. 2004;2:313–318. doi: 10.1039/b314672c. [DOI] [PubMed] [Google Scholar]
- 59.Keniry MA, Strahan GD, Owen EA, Shafer RH. Solution structure of the Na+ form of the dimeric guanine quadruplex [d(G3T4G3)]2. Eur. J. Biochem. 1995;233:631–643. doi: 10.1111/j.1432-1033.1995.631_2.x. [DOI] [PubMed] [Google Scholar]
- 60.Strahan GD, Keniry MA, Shafer RH. NMR structure refinement and dynamics of the K+-[d(G3T4G3)]2 quadruplex via particle mesh Ewald molecular dynamics simulations. Biophys. J. 1998;75:968–981. doi: 10.1016/S0006-3495(98)77585-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.