Abstract
Human mitochondrial transcription requires the bacteriophage-related RNA polymerase, POLRMT, the mtDNA-binding protein, h-mtTFA/TFAM, and two transcription factors/rRNA methyltransferases, h-mtTFB1 and h-mtTFB2. Here, we determined the steady-state levels of these core transcription components and examined the consequences of purposeful elevation of h-mtTFB1 or h-mtTFB2 in HeLa cells. On a per molecule basis, we find an ∼6-fold excess of POLRMT to mtDNA and ∼3-fold more h-mtTFB2 than h-mtTFB1. We also estimate h-mtTFA at ∼50 molecules/mtDNA, a ratio predicted to support robust transcription, but not to coat mtDNA. Consistent with a role for h-mtTFB2 in transcription and transcription-primed replication, increased mitochondrial DNA and transcripts result from its over-expression. This is accompanied by increased translation rates of most, but not all mtDNA-encoded proteins. Over-expression of h-mtTFB1 did not significantly influence these parameters, but did result in increased mitochondrial biogenesis. Furthermore, h-mtTFB1 mRNA and protein are elevated in response to h-mtTFB2 over-expression, suggesting the existence of a retrograde signal to the nucleus to coordinately regulate expression of these related factors. Altogether, our results provide a framework for understanding the regulation of human mitochondrial transcription in vivo and define distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression that together likely fine-tune mitochondrial function.
INTRODUCTION
Mitochondria are multifunctional organelles in eukaryotic cells involved in numerous metabolic activities, the production of ATP, and regulation of apoptosis. Consistent with their bacterial ancestry and absolutely critical for their function, is the mitochondrial DNA (mtDNA) housed in the matrix of the organelle. In humans, the 16 569-bp circular mtDNA molecule contains 37 genes encoding thirteen essential integral membrane proteins of the ATP-producing oxidative phosphorylation (OXPHOS) system, two rRNA subunits of the mitochondrial ribosomes, and 22 tRNAs required for mitochondrial translation in the matrix. Therefore, the vast majority of the ∼1500 proteins that localize and function in mitochondria are not mtDNA-encoded, but rather are products of nuclear genes that are imported into the organelles. An important consequence of this arrangement is that there must be coordination of nuclear and mitochondrial gene expression in order to maintain organelle homeostasis and to properly regulate mitochondrial activities. This is uniquely pertinent to the assembly of the OXPHOS system and mitochondrial ribosomes, which are composed of subunits encoded by both the nuclear and mitochondrial genomes. That is, the coordination of synthesis of subunits in mitochondria (the 13 OXPHOS subunits and two rRNAs) with the import of those from the cytoplasm (the ∼70 OXPHOS subunits and ∼80 ribosomal proteins) is presumably critical to regulate the biogenesis of these large complexes. In this context, it is also important to consider that all of the machinery required for transcription and replication of mtDNA is a subset of the nucleus-encoded factors that are targeted to mitochondria (1). How the relative amounts and altered expression of these key regulatory factors influence expression of mtDNA-encoded genes and subsequent assembly of the mitochondrial OXPHOS complexes and ribosomes is largely unknown.
The core protein components required for human mitochondrial transcription have recently been defined as a mixed three-component system comprising a T-odd bacteriophage-related mitochondrial RNA polymerase (POLRMT), the high mobility-group-box mitochondrial transcription factor A (h-mtTFA/TFAM), and two dual-function transcription factors (h-mtTFB1/TFB1M and h-mtTFB2/TFB2M) that are orthologs of an ancestral bacterial rRNA methyltransferase (2–4). In terms of transcription, POLRMT/h-mtTFB1 and POLRMT/h-mtTFB2 complexes are each capable of initiation at the mitochondrial L-strand promoter (LSP) and H-strand promoter 1 (HSP1) in the presence of h-mtTFA, although POLRMT/h-mtTFB2 complexes are reported to be significantly more active than POLRMT/h-mtTFB1 complexes in vitro (2). However, at specific h-mtTFA: DNA ratios the activities of the two complexes are altered to differing degrees. This leaves open the likely possibility that the rates of transcription in vivo can be dictated by the relative concentrations of mtDNA and each of the transcription components in different cell types (1,5). Estimates of h-mtTFA abundance in cells of human and Xenopus have been reported by several groups, but with conflicting results (6–9), and the relative or absolute amounts of POLRMT, h-mtTFA, h-mtTFB1 and h-mtTFB2 have not been systematically examined. Thus a basic definition of the mitochondrial transcription system in vivo is still lacking, a situation that limits our understanding of how mitochondrial gene expression is regulated.
As already mentioned, the recently identified h-mtTFB1 and h-mtTFB2 transcription factors are homologous to rRNA methyltransferases. Specifically, they are related to a large family of site-specific methyltransferases that catalyzes the N6-dimethylation of two adjacent adenine residues in a conserved stem-loop found in small subunit rRNAs (10). In bacteria this modification, while not essential, influences both stability and fidelity of ribosomes and is thought to modulate translational output (11–13). In the original report describing h-mtTFB1, this transcription factor was postulated to have rRNA methyltransferase activity based on its ability to bind the methyl-group donating co-factor s-adenosylmethionine (3). Subsequent work showed that both h-mtTFB1 and h-mtTFB2 can functionally replace the homologous KsgA dimethyltransferase in Escherichia coli (5,14), demonstrating that both proteins have retained this enzymatic activity. Interestingly, in this assay, h-mtTFB2 has significantly lower activity than h-mtTFB1 (5). This, coupled to their differing relative activity with regard to transcription output and the concentration of h-mtTFA, again suggests a potentially interesting regulatory scenario in which differential effects on mitochondrial transcription and ribosome biogenesis/function (via differential rRNA methylation) are mediated by the relative abundance or activity of h-mtTFB1 and h-mtTFB2 (1,5). Interestingly, other functions for members of this class of rRNA methyltransferases have been assigned that are independent of the catalytic activity. For example, the yeast Dim1p cytoplasmic rRNA methyltransferase plays a role in rRNA processing (15,16). Likewise, we have shown that the transcription factor activity of h-mtTFB1 is unaffected by mutations that inactivate its rRNA methyltransferase activity (17). Thus, whether h-mtTFB1 and h-mtTFB2 have additional roles beyond transcription and rRNA methylation remains a formal possibility.
To date, the only studies regarding the function of metazoan mtTFB transcription factors in vivo were performed on the Drosophila proteins using cultured Schneider cells. Consistent with a role for dm-mtTFB2 in transcription and transcription-primed mtDNA replication, RNAi knock-down or over-expression results in decreased or increased steady-state levels of mitochondrial RNA and mtDNA, respectively (18). In contrast, no effect on mtDNA copy number or the steady-state levels of mitochondrial transcripts is observed when expression of dm-mtTFB1 is knocked down or over-expressed in this system (19). However, RNAi knock-down of dm-mtTFB1 does result in a reduction in the rate of mitochondrial protein synthesis, indicating a role for this protein in regulating mitochondrial translation (perhaps via its rRNA methyltransferase activity) (19). These results demonstrate that the functions of dm-mtTFB1 and dm-mtTFB2 are not fully redundant in vivo and suggest that the primary role of dm-mtTFB2 is to regulate transcription and/or mtDNA copy number and that of dm-mtTFB1 is to modulate translation. However, it is important to note that these studies do not discount the possibility of partially overlapping functions for each factor in transcription and rRNA methylation that one might predict based on the fact that both h-mtTFB1 and h-mtTFB2 have retained both of these activities over the course of evolution (5). Furthermore, given the significant differences between mammals and flies in general and with regard to mitochondrial genome organization and dynamics (20–22), it is important not to generalize any relationships described in this (or any other) model system without direct confirmation in human cells. Here, we describe our analysis of the human mitochondrial transcription system in HeLa cells that defines the relative steady-state levels of the entire human mitochondrial transcription machinery for the first time and elucidates novel properties and functions of h-mtTFB1 and h-mtTFB2 in vivo via their purposeful over-expression.
MATERIALS AND METHODS
Plasmids for bacterial and human cell expression of h-mtTFB1 and h-mtTFB2
Bacterial expression vectors for production of h-mtTFB1 and h-mtTFB2 in E. coli were created using pET21b (Promega). The h-mtTFB1 and h-mtTFB2 cDNAs were amplified using primers 5′-AACATATGGCTGCCTCCGG-3′ and 5′-AAGAGCTCGAGTCTGTAATTCTC-3′ or primers 5′-AACATATGTGGATCCCAGTGG-3′ and 5′-GCGGCCGCCCTATCTTCCAGGGTTC-3′, respectively, and the resulting PCR products were ligated into pGEMT-Easy (Promega). These plasmids were then digested with appropriate restriction enzymes for ligation in pET21b (NdeI and XhoI for h-mtTFB1 or NdeI and NotI for h-mtTFB2). To generate plasmids for over-expression in human cells, the h-mtTFB1 and h-mtTFB2 cDNAs were both cut from previously described vectors (5) using EcoRV and NotI and ligated into pcDNA3.1 zeo (+) (Invitrogen) cut with the same enzymes. The vector used to express POLRMT in bacteria was pProEX-Htb (Invitrogen). A portion of the human cDNA encoding amino acids 41–1250 and the stop codon was cloned into the BamH1 and XhoI of this vector via a BamH1–Sal1 restriction fragment. Amino acids 1–40 were deleted since they compose the mitochondrial localization sequence (MLS) that is predicted to be removed by proteases during import into mitochondria (23). However, in place of the MLS, there are 29 unnatural amino acids fused to POLRMT that include a initiator methionine, a His6 tag, a spacer of seven amino acids, a TEV protease cleavage site, and another spacer of six amino acids. The vector has an intact E. coli lacIq gene allowing POLRMT expression from the trc promoter to be regulated by addition of isopropylthiogalactoside (IPTG; Sigma).
Purification of recombinant proteins
BL21-Codonplus® E. coli (Stratagene) were transformed with pET21b vectors containing either h-mtTFB1 or h-mtTFB2. For purification of h-mtTFB1, an overnight culture of bacteria was diluted into 1 l of LB media and grown at 37°C until culture reached an OD600 of 0.8. IPTG was added to 1 mM for induction of expression and the culture was allowed to incubate at room temperature with shaking for 24 h. Cells were harvested by centrifugation, then pellets were resuspended in 80 ml of lysis buffer (50 mM sodium phosphate, pH 8.0; 0.3 M NaCl, 10% glycerol, 0.1% Tween-20 and 10 mM imidazole). Cells were lysed by sonication and the suspension was cleared by centrifugation. The clarified lysate was applied to a BD-Talon column (BD Biosciences). The column was washed with 10 column volumes of lysis buffer and eluted with three column volumes of lysis buffer plus 150 mM imidazole. Fractions were assayed for presence of mtTFB1 by SDS-PAGE followed by either coomassie staining or western blot. Peak fractions were combined, concentrated and dialyzed into storage buffer (20 mM Tris-HCl, pH 8.0; 0.5 mM EDTA, 0.25 M sucrose, 15% glycerol, 1 mM DTT and 1 mM PMSF). Protein concentration was determined with a protein assay kit from Bio-Rad and confirmed by SDS-PAGE and coomassie staining with comparison against a BSA standard curve. For purification of h-mtTFB2, the same procedure was followed for mtTFB1, except that the protein was found to be highly insoluble and present in the pellet after sonication. Pellets after sonication were resuspended in lysis buffer with the addition of 0.1% Triton, 5 mM BME, 1 mM PMSF and 6 M guanidine-HCl. The solution was stirred overnight at 4°C then clarified by centrifugation. The protein solution was added to a Talon column as before, then washed with 10 column volumes of the pellet resuspension buffer. The column was eluted with three column volumes of the same buffer plus 200 mM imidazole. Fractions were analyzed as above and peak fractions were collected. Concentrated samples were dialyzed extensively against storage buffer and protein concentration was determined. Purification of h-mtTFA was performed as described (24), except the process was stopped after the BioRex-70 column (Bio-Rad). The expression and purification of POLRMT was performed as described (25).
HeLa cell growth and transfection
For transfection of HeLa cells (Clonetech) with h-mtTFB1 or h-mtTFB2 over-expression plasmids, cell were seeded at 5 × 105 per 10 cm dish in DMEM (Sigma) plus 10% Bovine Growth Serum (Hyclone) and allowed to grow for 24 h. Cells were transfected with empty pcDNA3.1 vector, pcDNA3.1-mtTFB1 or pcDNA3.1-mtTFB2 using Effectene (Qiagen) according to manufacturer's suggestions. Cells were allowed to incubate with plasmid/reagent complexes for 24 h. Transfected cells were subcultured by diluting cells 1:50 or 1:100 in growth media with 400 μg/ml Zeocin (Invitrogen) and plated in 10 cm dishes. Cells were grown until individual colonies were visualized and 10–15 clones were individually selected with glass cylinders and transferred to 24-well plates. Cells were grown to confluence, subcultured into 6-cm dishes in growth media plus 100 μg/ml Zeocin and grown for 48 h. One dish for each clone was harvested and assayed for protein expression via western blot. Remaining plates were harvested and stored in growth media plus 10% DMSO at −80°C. Stably transfected HeLa lines were consistently plated at 5000 cells/cm2, grown for 72 h at 37°C and 5% CO2 before subculturing again. All experiments were performed on cells passaged at most 10 times.
Antibody production and western analysis of whole-cell and mitochondrial proteins
Four polyclonal peptide antibodies (two for h-mtTFB1 and two for h-mtTFB2) were generated for us by Multiple Peptide Systems. The peptides used as antigens were as follows: TFB1-1 H-CVPKPEVDVGVVHFTPLIQPKIE-NH2, TFB1-2 H-CREELKRRKSKNEEKEEDDAENYRL-NH2, TFB2-1 H-CWIPVVGLPRRLRLSALAGA-NH2, TFB2-2 H-CPQLWPEPDFRNPPRKASKASLD-NH2. Multiple Peptide Systems performed the synthesis of peptides, injection of rabbits and collection of serum, and also provided a small batch of peptide-affinity purified antibody. We also used antibodies that were purified from provided crude serum using protein-A sepharose (Amersham). Specificity of each antibody was determined by immunoblots of 200 ng of recombinant full-length h-mtTF1 and h-mtTFB2 proteins run alongside one another on the same gel. Antiserum used to detect human POLRMT was the same as that described previously (26) and polyclonal antibodies for detection of h-mtTFA were generously provided by Dr David Clayton.
For whole-cell extracts, 1 × 106 cells were suspended in 100 μl of cold lysis buffer (50 mM Tris-HCl pH 8.8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 2 mM EDTA, 10% glycerol, 5 mM DTT, 1 mM PMSF) and incubated at 4°C with rotation for 30 min. Protein concentration was determined using the BioRad Protein Assay Kit and indicated amounts of total protein were loaded on polyacrylamide gels for analysis. For mitochondrial extracts, mitochondria were harvested by differential centrifugation. Briefly, cells were resuspended in 10 pellet volumes of RSB buffer (10 mM NaCl, 1.5 mM MgCl2, 10 mM Tris-HCl pH 7.5), swelled on ice for 10 min, homogenized with a motorized Teflon pestle, then 2.5 × MS Buffer (525 mM mannitol, 175 mM sucrose, 125 mM Tris-HCl pH 7.5, 2.5 mM EDTA) was added to 1 ×. The homogenate was centrifuged at 980 g for 10 min twice to pellet nuclei and unbroken cells. The supernatant was transferred to a fresh tube and spun at 17 000 g for 30 min to pellet mitochondria. The mitochondrial pellet was washed three times with 1 × MS buffer then stored at −80°C until further use. Mitochondrial pellets were resuspended in lysis buffer equal to one volume of the original cell pellet. The protein extract was then treated and quantified as described above.
Protein extracts were separated on SDS–polyacrylamide gels and then transferred to PVDF membranes. Membranes were blocked with 5% milk/TBST (10 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.05% Tween-20) for 30 min at room temperature, then probed with primary antibody in 5% milk/TBST overnight at 4°C. The primary antibody was removed and the blot was washed three times for 10 min with TBST. The appropriate secondary antibody was applied in 5% milk/TBST and allowed to incubate at room temperature for 1 h. The blot was washed again, ECL reagent was added and blots were exposed to film. Several exposures were obtained, then films were photographed and analyzed with a BioRad VersaDoc and Quantity One software (v4.6.1). For serial western blots, membranes were stripped with stripping buffer (65 mM Tris-HCl pH 6.8, 2% SDS, 100 mM BME) at 50°C for 30 min, then reblocked with 5% milk/TBST.
Determination of the relative steady-state levels of the human mitochondrial transcription proteins
Mitochondria were harvested as described above but were purified additionally by centrifugation through a 1.0–1.5 M sucrose step gradient at 45 000 r.p.m. in a SW50.1 rotor for 2 h. Mitochondria were removed from the interface of the two sucrose solutions by pipetting and transferred to a fresh tube. Mitochondria were washed with 1 ml of 1 × MS Buffer and pelleted by centrifugation at 13 000 r.p.m. for 10 min. The final pellet was stored at −80°C until further use. Efficiency of mitochondrial harvest was measured by amount of HSP60 and h-mtTFA immunoblot signal obtained from fractions reserved throughout the process and compared to amounts from starting whole-cell lysates. Final mitochondrial pellets were lysed, extracts were quantified as described above, and samples ranging from 10 to 100 μg of total mitochondrial protein were separated alongside 50–150 µg of whole-cell lysate and a range of amounts of purified recombinant proteins (1–100 ng) on 6–20% SDS–polyacrylamide gradient gels. Proteins were transferred to PVDF and western blotting was performed as described above. A standard curve was generated from signal obtained from recombinant proteins and used to determine the amount of the respective protein in each of the sample lanes.
Nucleic acid extraction, northern blotting, reverse transcriptase real-time PCR and mtDNA copy number analysis
Cells were harvested after 72 h of growth in conditions described above and counted. For each cell line, 1 × 106 cells were used for RNA extraction using the RNEasy Kit (Qiagen) according to included instructions. During the process, DNA was digested with RNAse-free DNAse according to manufacturer's suggestions. RNA was eluted in the final step using dH2O, quantified and stored at −80°C until further use. Gel electrophoresis, blotting of RNA and probing of blots with 16S, 12S, ND2 and ND6 probes were performed as previously described (25).
For analysis of h-mtTFB1 transcript levels, first strand cDNA synthesis was performed by combining 2 μg of total RNA with 1 mM dNTPs and 8 μM oligodT15 in a 20 μl reaction volume. This mixture was heated to 70°C for 10 min then allowed to cool to 4°C over 10 min to anneal the oligodT primer to polyA tails of mRNAs. An equal volume of reverse transcriptase mix [200 units M-MuLV Reverse Transcriptase (NEB), 2X M-MuLV RT Buffer and 25 units of RNAseOUT (Invitrogen)] was added to each primer-RNA mix and incubated at 42°C for 1 h. The reactions were then heated to 70°C for 10 min to deactivate the reverse transcriptase. Finally, an equal volume of 3 mM Tris-HCl pH 8.5, 0.3 mM EDTA was added to each reaction and samples were stored at −20°C until further use. Samples of each cDNA were thawed and diluted from 16- to 32-fold in dH2O. A 10 μl of a diluted cDNA sample was added to 14 μl of SYBR Green reaction mix (27) to each well along with 0.5 μl of both appropriate 25 μM primers. β-Actin primers, β-actin RT F 5′ TGGCACCACACCTTCTACAATGAGC 3′ and β-actin RT R 5′ GCACAGCTTCTCCTTAATGTCACGC 3′, were used as controls for total cDNA in each reaction. To amplify the h-mtTFB1 transcript, primers TFB1 RT-F 5′ TCTGCAATGTTCGACACATC 3′ and TFB1 RT-R 5′ ACCTATATAAGAAGCTCCAC 3′ were used. Reaction conditions for the BioRad iCycler were as follows: 95°C, 10 min, 1 cycle; 95°C, 30 s, 56°C, 30 s, 72°C, 30 s, 40 cycles. Fluorescence was measured after each 56°C step. Melt-curve analysis to ensure single products was performed immediately after completion of steps above by using a temperature step gradient from 55 to 80°C in 0.5°C increments with fluorescence measured after a 10 s incubation at each temperature. Ct values for each reaction were obtained through the iCycler iQ software. h-mtTFB1 transcript levels were then normalized by β-actin transcript levels and compared to empty vector controls.
Total DNA was extracted from 1 × 106 cells by addition of 500 μl of extraction buffer (50 mM Tris-HCl pH 8.5, 0.25% SDS, 1 mM EDTA, 5 mM DTT) and boiling for 10 min. After cell lysis, tubes were allowed to cool to room temperature, 100 μg of RNAse A were added, and tubes were allowed to incubate at 37°C. Following a 3 h incubation, 100 μg of proteinase K were added and samples were placed at 55°C overnight. Samples were heated to 95°C for 5 min and allowed to cool to room temperature. Total DNA concentration was measured and samples were stored at -20°C until further use.
Relative and absolute mtDNA copy numbers were measured by SYBR Green fluorescence using a BioRad iCycler and accompanying software (v3.1). Total DNA samples were diluted to a range of DNA concentrations from 500 to 7.8 pg/µl and 10 μl were dispensed to appropriate wells of a 96-well PCR plate. 14 μl of SYBR Green reaction mix (27) were added to each well along with 0.5 μl of both appropriate 25 μM primers. For detecting relative levels of mtDNA, a region of the mitochondrial genome encompassing a portion of COX3 and a region of the multicopy nuclear 18S rDNA locus were used. Primers are as follows: RTQ COX3-F 5′ CACCCAAGAACAGGGTTTGT3′, RTQ COX3-R 5′ TGGCCATGGGTATGTTGTTAA 3′, RTQ 18S F 5′TAGAGGGACAAGTGGCGTTC3′ and RTG 18S R 5′CGCTGAGCCAGTCAGTGT3′. Reaction conditions for the iCycler were as follows: 95°C, 10 min, 1 cycle; 95°C, 15 s, 60°C, 1 min, 40 cycles. Fluorescence was measured after each 60°C step. Melt-curve analysis to ensure single products was performed immediately after completion of steps above by using a temperature step gradient from 55 to 80°C in 0.5°C increments with fluorescence measured after a 10 s incubation at each temperature. Standard curves to determine absolute copy number were constructed with known amounts (1 638 400 to 100 templates) of the plasmid pGEMT Easy (Promega) containing either one of the PCR products obtained using the mitochondrial and nuclear primers described above (COX3 or 18S).
Labeling of mitochondrial translation products in vivo
Labeling was performed as described (28) with modifications. HeLa cells were seeded and grown as described above in 10 cm dishes. After 72 h of growth, cells were washed three times with 5 ml of sulfur-free media (Gibco) without serum. Cells were allowed to incubate for 5 min at 37°C, 5% CO2 before removing media. After the last wash, 5 ml of sulfur-free media with 10% BGS plus 100 μg/ml emetine were added to each plate and allowed to incubate at 37°C, 5% CO2 for 5 min. After this incubation step, Expre35S35S Protein Labeling Mix (Perkin Elmer) was added to each plate to a concentration of 125 μCi/ml and labeling was allowed to proceed at 37°C and 5% CO2. After 1 h, the labeling media was removed and the plates were rinsed once with 5 ml of normal DMEM with 10% BGS. Plates were then washed twice with 10 ml of TD buffer (25 mM Tris pH 7.5, 137 mM NaCl, 10 mM KCl, 0.7 mM Na2HPO4), trypsinized and harvested by centrifugation at 1000 g. Cell pellets were washed once with 10 ml of TD buffer, then transferred to a 1.5 ml tube with 1 ml of TD buffer and harvested again by centrifugation. Mitochondria were harvested as above and protein was quantified. 50 μg of total protein were added to each lane of a 10–20% linear gradient SDS–polyacrylamide gel. Samples were separated and then the gel was dried and exposed to film at −80°C.
Kasugamycin sensitivity assays
HeLa cells were plated and grown as described above. After 48 h of growth, kasugamycin was added at indicated concentrations and cells were allowed to incubate for 72 h. Cells were washed, trypsinized and treated with 0.08% Trypan Blue in PBS. The total number of viable cells at each drug concentration was compared to that of zero-drug controls to yield a percentage of viable cells that is plotted as a function of the drug concentration.
FACS analysis
HeLa cultures were plated and grown as described above. After 72 h of growth, cells were stained in culture media with 70 nM Mitotracker Green and 90 nM Mitotracker Red CMXRos, both from Molecular Probes, for 30 min at 37°C, 5% CO2. After staining, dyes were removed and cells were washed three times with PBS. Cells were trypsinized and collected by centrifugation. Cell pellets were resuspended in 1 ml of PBS and analyzed on a BD FACS calibur instrument with accompanying software. Histograms and means for these data were obtained using FlowJo (v 8.0.1)
Mitochondrial localization sequence and cleavage prediction
Full-length sequences of h-mtTFB1, h-mtTFB1 and sc-mtTFB were obtained from NCBI. These sequences were submitted to cleavage site prediction by the SignalP program (v3.0, http://www.cbs.dtu.dk/services/SignalP/) (29). Hidden Markov modeling option was selected and the first 70 amino acids for each sequence were used for signal and cleavage site prediction (30).
RESULTS
Relative levels of the human mitochondrial transcription machinery in HeLa cells
To fully understand any transcription system, a critical parameter to know is the relative amounts of the basal transcription machinery in vivo. Our approach was to use immunoblotting to establish the levels of the mitochondrial transcription machinery on a total cell and mitochondrial basis. This method requires antibodies that are specific for their target, especially for homologous proteins that are very similar in size. Since h-mtTFB1 and h-mtTFB2 share a large degree of sequence similarity and are predicted to migrate similarly on polyacrylamide gels, it was unclear if antibodies we had generated would cross react with the paralogous protein and confound our analysis. Therefore, we first generated peptide antibodies against h-mtTFB1 and h-mtTFB2 (see Materials and Methods section) and determined their specificity by assessing reactivity toward purified full-length recombinant h-mtTFB1 and h-mtTFB2 by immunoblotting. We found that these antibodies were indeed highly specific for their targets showing no reactivity with the paralogous protein (Figure 1A). Having antibodies capable of specifically detecting each of the four human mitochondrial transcription proteins, as well as known amounts of corresponding recombinant proteins, allowed us to determine for the first time their relative abundance. We purified mitochondria from logarithmically growing HeLa cells and performed quantitative western blot analysis of POLRMT, h-mtTFA, h-mtTFB1 and h-mtTFB2 on known amounts of total mitochondrial protein. For each protein analyzed, multiple dilutions of total mitochondrial protein or total whole-cell lysate were probed in parallel with a dilution series of known amounts of recombinant protein to allow the amount of each transcription component relative to the total amount of mitochondrial lysate to be determined. Representative western blots and standard curves showing linearity of the assays employed are shown in Supplementary Figure S1.
Figure 1.
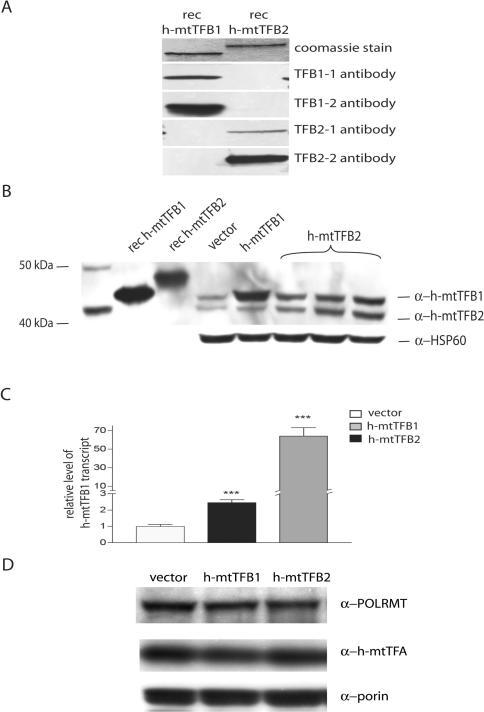
h-mtTFB2 is processed in vivo and its over-expression in HeLa cells results in a coordinated increase of h-mtTFB1, but not POLRMT or h-mtTFA. (A) Shown are western blots on 200 ng of recombinant h-mtTFB1 and h-mtTFB2 proteins probed with four peptide antibodies: TFB1-1, TFB1-2, TFB2-1 and TFB2-2. Antibodies were found to specifically recognize their full-length recombinant peptide and not cross react with the paralogous protein. Coomassie staining of full-length recombinant h-mtTFB1 and h-mtTFB2 (top panel) demonstrates loading and their difference in molecular weight. (B) Western blot of mitochondrial extracts (100 μg protein) from HeLa cell lines over-expressing h-mtTFB1 or h-mtTFB2 used in this study in parallel with recombinant h-mtTFB1 and h-mtTFB2 run as controls. The blot was probed as indicated using peptide antibodies that distinguish h-mtTFB1 and h-mtTFB2 (α-h-mtTFB1 and α-h-mtTFB2) and an antibody that recognizes HSP60 (α-HSP60) that was used as a mitochondrial loading control. The lanes are loaded as follows: lane 1, molecular weight markers; lane 2, recombinant h-mtTFB1; lanes 3, recombinant h-mtTFB2; lanes 4–8, mitochondrial extracts from an empty pcDNA 3.1 zeo (+) vector-control, h-mtTFB1 over-expression, and three different h-mtTFB2 stable over-expression HeLa cell lines, respectively. (C) Shown are the results of real-time RT-PCR measurement of h-mtTFB1 mRNA (see Supplementary Figure S3) in h-mtTFB1 and h-mtTFB2 over-expression cell lines relative to empty-vector control cells. Reverse transcriptase real-time PCR was used to measure Ct values for cDNA samples from listed cell lines. The Ct values for h-mtTFB1 mRNA were normalized to those of β-actin and the values shown were normalized to the ratio obtained in the empty-vector control, which was given a value of 1. Values shown are the mean ± SD for three separate measurements. (D) Western blot of mitochondrial lysates from the same cell lines described in B probed using antibodies that recognize h-mtTFA (α-h-mtTFA), POLRMT (α-POLRMT), and the outer mitochondrial membrane protein VDAC (α-VDAC/porin) as a mitochondrial loading control.
Based on this analysis, we calculated the following amounts of each transcription component/100 μg of total mitochondrial protein: 45.8 ± 9.7 ng of h-mtTFA, 4.03 ± .20 ng of h-mtTFB1, 14.9 ± .97 ng of h-mtTFB2 and 32.3 ± 2.1 ng of POLRMT. For whole-cell lysates, we were only able to accurately measure h-mtTFA levels and found 11.1 ± 2.4 fg of protein per cell (data not shown). Next, we determined the number of molecules/cell based on the amount of total mitochondrial protein isolated from 4.5 × 107 cells. We found there to be 25.2 ± 2.51 pg of mitochondrial protein per cell with recoveries of 75–80% of total mitochondria for three separate experiments based on HSP60 and h-mtTFA immunoblots (data not shown). These measurements result in 2.52 × 105, 1.35 × 104, 4.29 × 104 and 3.12 × 104 molecules per cell for h-mtTFA, h-mtTFB1, h-mtTFB2 and POLRMT, respectively (Table 1). This amount of h-mtTFA from mitochondrial samples agreed well with our measurement in whole-cell lysates at 2.78 × 105 molecules per cell (data not shown). Finally, we also determined the mtDNA copy number in the cells to be 5010 ± 386 using quantitative real-time PCR (Supplementary Figure S2). From these data, we calculated the relative levels of the four transcription proteins on a molecule/cell basis and as function of the number of mtDNA molecules/cell (Table 1). Interestingly, there is ∼3-fold more h-mtTFB2 than h-mtTFB1 in these cells, which results in a close to 1:1 relationship between h-mtTFB2 and POLRMT (1.37:1), but an excess of POLRMT to h-mtTFB1 (2.31:1). Furthermore, our measurements place POLRMT at an ∼6-fold molar excess over the number of mtDNA molecules and between 42 and 58 h-mtTFA molecules/mtDNA molecule in HeLa cells. Our h-mtTFA to mtDNA ratio is consistent with that observed by Wiesner and colleagues (8), but more than 30-fold lower than that proposed by Kang and colleagues (7).
Table 1.
Relative steady-state abundance of the mitochondrial transcription components in HeLa cells
| Molecules/cell | Molecules/ mtDNA | Molecules: h-mtTFA | Molecules: h-mtTFB1 | Molecules: h-mtTFB2 | Molecules:POLRMT | |
|---|---|---|---|---|---|---|
| h-mtTFA | 2.52 × 105 ± 4.86 × 104 | 50.3 ± 8.8 | – | 18.6 ± 3.59:1 | 5.87 ± 1.13:1 | 8.07 ± 1.56:1 |
| h-mtTFB1 | 1.35 × 104 ± 4.17 × 102 | 2.7 ± .08 | 1:18.6 ± 3.59 | – | 1:3.17 ± .21 | 1:2.31 ± .15 |
| h-mtTFB2 | 4.29 × 104 ± 2.80 × 103 | 8.57 ± .56 | 1:5.87 ± 1.13 | 3.17 ± 21.1 | – | 1:37 ± .09:1 |
| POLRMT | 3.12 × 104 ± 1.99 × 103 | 6.23 ± .39 | 1:8.07 ± 1.56 | 2.31 ± 15.1 | 1:1.37 ± .09 | – |
h-mtTFB2 is cleaved upon import and its over-expression results in an increase in h-mtTFB1 levels
With a determination of relative levels of the mitochondrial transcription machinery in hand, we next set out to examine the consequence of altering the relative amounts of these proteins on mitochondrial gene expression and to determine if their expression levels are coordinately regulated. To do this, we created stable HeLa cell lines that over-express either h-mtTFB1 or h-mtTFB2. From our initial analysis of these lines, we immediately made two novel observations. First, the mobility of h-mtTFB2 isolated from cells was significantly faster than that of the corresponding recombinant protein (Figure 1B). Since many matrix-localized mitochondrial proteins have an N-terminal localization sequence (MLS) that is often removed by proteases upon import, we hypothesized that this was the case for h-mtTFB2. Consistent with this, using SignalP (a MLS and cleavage site prediction program), a single high-probability mitochondrial peptidase cleavage site for h-mtTFB2 was found between amino acids 30 and 31 (data not shown). No such cleavage site was predicted for h-mtTFB1, consistent with no obvious change in mobility of this protein relative to its recombinant control (Figure 1B). Furthermore, mapping amino acids 5–22 onto a helical-wheel diagram reveals a pattern consistent with a putative amphipathic helix (data not shown), which is another common attribute of an MLS. Finally, using a peptide antibody (TFB2-1) that was made against amino acids 2–20 of h-mtTFB2 (which are predicted to be removed by SignalP), we were unable to successfully detect endogenous h-mtTFB2 by western blotting (data not shown). However using the peptide antibody (TFB2-2) generated against amino acids 49–70, which is used throughout this study, we readily detect endogenous h-mtTFB2 (Figure 1B). Taken together, these data suggest that amino acids 1–30 likely comprise a significant portion of the MLS for h-mtTFB2 and are removed upon import.
In addition to the increased mobility of h-mtTFB2 described above, we also found that in the cell lines that over-express h-mtTFB2, there was a corresponding increase in the steady-state level of h-mtTFB1 (Figure 1B, compare lanes 6, 7 and 8 to lane 4). However, the converse was not true. That is, there was no increase in h-mtTFB2 steady-state levels when h-mtTFB1 was over-expressed ∼10-fold (Figure 1B, compare lane 5 to lane 4). We also analyzed the steady-state levels of the remaining transcription machinery in the face of h-mtTFB1 or h-mtTFB2 over-expression. No obvious changes in the amounts of POLRMT or h-mtTFA per mitochondrion were observed in either case (Figure 1D).
To begin to determine the mechanism of increased h-mtTFB1 levels due to h-mtTFB2 over-expression we employed reverse transcriptase real-time PCR to measure the steady-state amounts of h-mtTFB1 mRNA. In the h-mtTFB2 over-expression cell lines, h-mtTFB1 mRNA was increased ∼2.5-fold (Figure 1C and Supplementary Figure S3), in good correspondence with the observed increase in h-mtTFB1 protein levels (Figure 1B). As expected there was a large (∼64-fold) increase in h-mtTFB1 mRNA in the h-mtTFB1 over-expression lines (Figure 1C). However, this did not correspond with the only ∼10-fold over-expression of the protein (Figure 1B), suggesting that post-transcriptional mechanisms may limit h-mtTFB1 expression or accumulation.
Over-expression of h-mtTFB2, but not h-mtTFB1, increases the steady-state levels of mtDNA-encoded transcripts, OXPHOS subunits and mtDNA and enhances the rate of mitochondrial translation
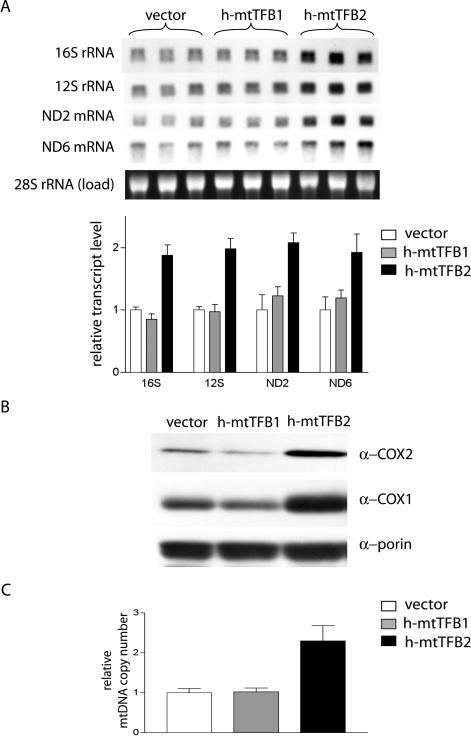
To address how altered levels of h-mtTFB1 and h-mtTFB2 affect mitochondrial gene expression, we next examined the h-mtTFB1 and h-mtTFB2 over-expression HeLa cell lines for changes in the steady-state levels of mtDNA-encoded transcripts and proteins, as well as for alterations in mtDNA copy number. Northern analysis of the mitochondrial 16S and 12S rRNAs and of ND2 and ND6 transcripts (representing mRNAs transcribed from each strand of mtDNA) revealed a ∼2-fold increase in their steady-state levels in the h-mtTFB2 over-expression cell line, but no change in the h-mtTFB1 over-expression line (Figure 2A). Similar results were obtained when immunoblots of the mtDNA-encoded COX1 and COX2 proteins was performed and mtDNA copy number was measured. That is, over-expression of h-mtTFB2, but not h-mtTFB1, led to a significant increase in the steady-state levels of COX1 and COX2 proteins (Figure 2B) and a doubling of the mtDNA copy number (Figure 2C). Altogether, these results are consistent with a role for h-mtTFB2 in transcription and in transcription-primed mtDNA replication. However, the fact that h-mtTFB1 is up-regulated in the h-mtTFB2 over-expression lines makes it difficult to assign these functions to h-mtTFB2 acting independently of h-mtTFB1.
Figure 2.
Over-expression of h-mtTFB2 increases the steady-state levels of mtDNA-encoded transcripts and proteins, and doubles mtDNA copy number. (A) Northern analysis of the mtDNA-encoded 12S, 16S, ND2 and ND6 from the same cell lines described in Figure 1B. Total RNA (2 μg) from the indicated cell line was loaded in each lane and the analysis was performed in triplicate on samples from three independent cultures. Ethidium bromide staining of the cytoplasmic 28S rRNA is shown as a loading control. Results of a quantification of the blots are graphed below. The relative transcript level (ratio of the mitochondrial signal to that of the 28S control) is plotted with the ratios obtained in the h-mtTFB1 and h-mtTFB2 cell lines normalized to that obtained from the empty-vector control cells, which was given a value of 1. The values are the mean ± SD. (B) Western blot of mitochondrial extracts from the indicated cell lines as in Figure 1B, probed using antibodies that recognize the mtDNA-encoded COX1 and COX2 protein or porin (VDAC) as a loading control. This demonstrates that there is an increase (on a per mitochondria basis) of these components unlike h-mtTFA and POLRMT. (C) Plotted is the relative mtDNA copy number (mtDNA relative to the nuclear 18S rDNA) normalized to that of the empty-vector control cells, whose ratio was given a value if 1. The analysis was done in triplicate and values shown are the mean ± SD.
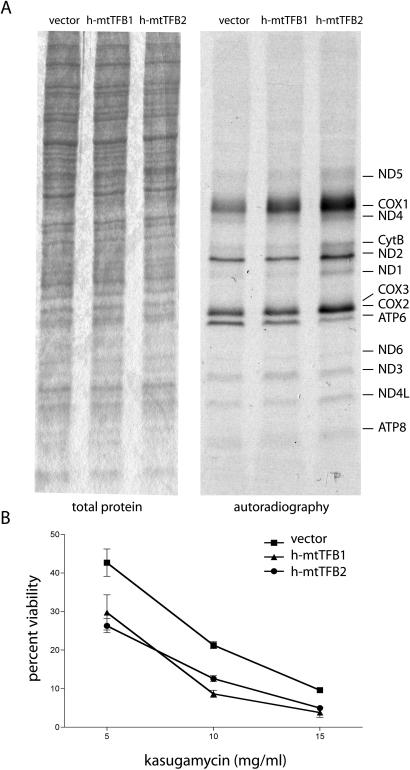
Both h-mtTFB1 and h-mtTFB2 have retained rRNA methyltransferase activity that is postulated to modulate mitochondrial ribosome biogenesis and/or translation (5). This, coupled to the fact that a function for Drosophila mtTFB1 in mitochondrial translation has been documented (19), we next measured mitochondrial translation rates in the h-mtTFB1 and h-mtTFB2 over-expression HeLa cell lines using an in vivo-radiolabeling approach. Similar to the analysis of the other mitochondrial gene expression parameters (Figure 2), we again saw major differences only in the h-mtTFB2 over-expression cell line, where a significant increase in the overall rate of translation was observed (Figure 3A). Interestingly, however, the labeling of specific products was not uniform (e.g. COX1, COX2 and ATP6), suggesting that increasing the amount of h-mtTFB2 and/or mitochondrial mRNAs is leading to increased translation of some mRNAs to the exclusion of others.
Figure 3.
Analysis of mitochondrial translation rates and kasugamycin sensitivity in h-mtTFB1 and h-mtTFB2 over-expression HeLa cell lines. (A) Mitochondrial translation products were labeled in vivo with 35S-methionine in the presence of the cytoplasmic translation inhibitor emetine. Mitochondria were purified after labeling and 100 μg of mitochondrial protein were loaded on a linear gradient polyacrylamide gel. Shown on the left is a coomassie stain of the resulting gel demonstrating equal protein loading and on the right is an autoradiogram of the radiolabeled mitochondrial proteins (specific proteins are indicated on the right). Changes in the profile of labeled products only occur during over-expression of h-mtTFB2. (B) Plotted is the percent viable cells of the indicated cell lines after growth for 72 h in the indicated amount of the aminoglycoside kasugamycin. The analysis was done in triplicate and values shown are the mean ± SD.
Over-expression h-mtTFB1 increases sensitivity to the aminoglycoside antibiotic kasugamycin
In all of the analyses described thus far, no obvious consequences of over-expressing h-mtTFB1 alone were observed. We were somewhat surprised that over-expression of h-mtTFB1 did not influence mitochondrial translation rates given that it is more active as a rRNA methyltransferase (5) and that its Drosophila ortholog has been implicated in translation efficiency (19). It remained a formal possibility that h-mtTFB1 is affecting mitochondrial ribosome function or biogenesis in manner that is not read out as an increase in the overall rate of translation. In bacteria, methylation of the small subunit rRNA by the h-mtTFB1 ortholog KsgA results in sensitivity to the aminoglycoside antibiotic kasugamycin and we have shown that h-mtTFB1 and h-mtTFB2 can functionally replace KsgA in E. coli (5). Furthermore, the human mitochondrial 12S rRNA and the bacterial 16S rRNA are highly conserved at the site that is methylated (a stem-loop at the 3′ end). We therefore determined whether over-expression of h-mtTFB1 alters the sensitivity of human mitochondrial ribosomes to kasugamycin. We found that over-expression of h-mtTFB1 resulted in a significant decrease in viability of HeLa cells grown in the presence of high concentrations of this drug (Figure 3B). Similar results were obtained in the h-mTFB2 over-expression line (Figure 3B). However, given that h-mtTFB1 is also up-regulated when h-mtTFB2 is over-expressed (Figure 1B), whether this effect is due to h-mtTFB1 or h-mtTFB2 or perhaps both cannot be distinguished.
Over-expression of h-mtTFB1 or h-mtTFB2 increases mitochondrial biogenesis, but their coordinate up-regulation also results in increased membrane potential
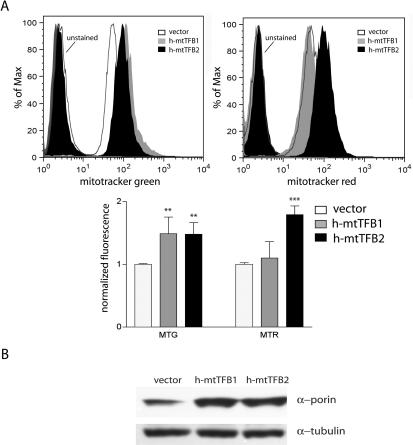
Given that over-expression of h-mtTFB2 (and as a result also h-mtTFB1, Figure 1B) results in an increase in multiple mitochondrial gene expression parameters (Figures 2 and 3), we next determined whether this signaled cells to increase overall mitochondrial biogenesis. In parallel, we also analyzed the h-mtTFB1 over-expression line. Surprisingly, we found that both cell lines exhibited a ∼50% increase in mitochondrial mass as measured by Mitotracker Green staining and FACS analysis (Figure 4A). Thus, despite the fact that over-expression of h-mtTFB1 does not increase mitochondrial gene expression in any manner examined thus far (Figures 2 and 3), remarkably, it does invoke a mitochondrial biogenesis response. The results of the Mitotracker Green staining were largely confirmed by immunoblots, where mitochondrial biogenesis was assayed as the ratio of the amount of the mitochondrial outer membrane protein VDAC to that of tubulin (Figure 4B). We also analyzed the h-mtTFB1 and h-mtTFB2 over-expression cell lines for mitochondrial membrane potential using Mitotracker Red staining and FACS analysis. Here we observed an ∼80% increase in membrane potential in the h-mtTFB2 over-expression line, but no statistically significant change as the result of h-mtTFB1 over-expression alone (Figure 4A). These results suggest the interesting possibility that the simultaneous up-regulation of both h-mtTFB1 and h-mtTFB2 (as is the case in the h-mtTFB2 over-expression lines) is required to fine-tune mitochondrial activity in accordance with changes in mitochondrial biogenesis and gene expression.
Figure 4.
Analysis of mitochondrial biogenesis and membrane potential in h-mtTFB1 and h-mtTFB2 over-expression HeLa cell lines. (A) Shown are representative results from FACS analysis of the indicated cell lines (see key in figure) stained with Mitotracker Green FM, as a measure of mitochondrial mass, or Mitotracker Red, as a measure of mitochondrial membrane potential. Plotted below is a quantification of a triplicate analysis of these parameters, with the mean fluorescence of the vector control cell line given a value of 1. Error bars represent SD of three experiments and asterisks indicate statistically significant differences in mean fluorescence as determined by t-tests, **P < 0.005 and ***P < 0.0005. (B) Western blot of whole-cell lysate (100 μg protein) from empty-vector control, h-mtTFB1 over-expression, and h-mtTFB2 over-expression cell lines with antibodies that recognize tubulin (α-tubulin) and α-porin (VDAC). The ratio of the VDAC signal to the tubulin signal is interpreted as a measure of the amount of mitochondria per cell.
DISCUSSION
In this study, we have performed a detailed analysis of the human mitochondrial transcription machinery in HeLa cells. Specifically, we have determined for the first time the relative in vivo steady-state levels of the four core components of this system: the human mitochondrial RNA polymerase, POLRMT; the mtDNA binding transcription factor, h-mtTFA; and the two dual-function transcription factors/rRNA methyltransferases, h-mtTFB1 and mtTFB2. In addition, we have clearly demonstrated that the two h-mtTFB paralogs have unique attributes and distinct functions with regard to mitochondrial gene expression and biogenesis. We will discuss the primary findings and the main conclusion we reach based on the results of this study below.
The first goal of this study was to define the relative amounts of the human mitochondrial transcription machinery in vivo. Previously, groups have estimated the amounts of h-mtTFA (6–8), but a simultaneous assessment of the entire core transcription system had not been described. We now have antibodies that recognize the four core human mitochondrial transcription components and, importantly, we generated peptide antibodies that are capable of distinguishing the two related h-mtTFB paralogs, h-mtTFB1 and h-mtTFB2 (Figure 1A). Using these antibodies, we were able to detect each component in mitochondrial extracts containing known amounts of total mitochondrial protein by western blotting and compare these to the signals obtained from known amounts of cognate recombinant protein analyzed in parallel. Since we also quantified the total number of cells from which the extracts were derived and the copy number of mtDNA, we are able to express the results for each individual component as the number of molecules/cell or the number of molecules/molecule of mtDNA (Table 1).
Our results revealed a number of novel and salient points about the relative abundance of the mitochondrial transcription system in HeLa cells (see Table 1). First, relative to the mtDNA copy number, which we found to be 5010 ± 386 (Supplementary Figure S2), POLRMT is in ∼6-fold excess of the mtDNA on a per-molecule basis. In principle, this is sufficient to allow all templates to be engaged in transcription simultaneously. The second main conclusion that we reach is that there is roughly three times more h-mtTFB2 than h-mtTFB1 molecules/cell. This difference is perhaps most relevant when compared to the amount of POLRMT, from which it becomes clear that there is an excess of h-mtTFB2 to POLRMT (1.3:1), but a limiting amount of h-mtTFB1 relative to POLRMT (0.43:1). While the relevance of these differences is difficult to predict, it is noteworthy that for both h-mtTFB1 and h-mtTFB2 the levels relative to POLRMT are not too far removed from 1:1, which is consistent with the predicted optimal stoichiometry in the core transcription complexes for transcription in vitro (2). Finally, we arrive at a value of ∼25 000 molecules of h-mtTFA per cell, which places it in ∼5–18 fold excess of the other core transcription components (Table 1).
Given that h-mtTFA has been postulated to have an mtDNA-packaging role in addition to its transcription factor function (7,31–34), its abundance relative to mtDNA is important to discuss. Based on our measurement of mtDNA copy number, the ratio of h-mtTFA:mtDNA molecule we observe is 50 ± 8:1. This value is in good agreement with that of 35:1 reported by Wiesner and colleagues (8), but substantially lower than that proposed by Kang and colleagues, who suggest a ratio of ∼1700:1 (7). Kang and colleagues cite the ratio of mtTFA to mtDNA in Xenopus oocytes at 2000:1 (9) to support of their findings (7). However, ratios of X. laevis mtTFA (xl-mtTFA):mtDNA are greatly up-regulated during Xenopus oocyte maturation ranging from a resting immature oocyte level of ∼200:1 (35) to the noted ratio of ∼2000:1 (9), which occurs only in mature oocytes. Given that xl-mtTFA binds mtDNA as a tetramer (36), there is effectively ∼50 xl-mtTFA complexes per genome in immature oocytes, which we argue is a cell type that is more relevant for comparison to mammalian cell types than a mature oocyte, which has dramatically up-regulated mitochondria and mtDNA in preparation for fertilization and development. Finally, if h-mtTFA levels were indeed high enough to completely coat the mtDNA genome as suggested by Kang and colleagues (7,31,33,34), this would seem incompatible with any significant transcriptional output based on in vitro transcription studies (2,37). Furthermore, it becomes difficult to explain why over-expression of h-mtTFA increases mitochondrial transcription and mtDNA copy number (32,38) if, in fact, mtDNA is already fully saturated with h-mtTFA (31). It is noteworthy that we observe less than one-sixth the amount of h-mtTFA and five times more mtDNA per cell than Takamatsu et al. measured in HeLa cells (7). These differences likely begin to account for the apparent overestimation of the mtTFA:mtDNA ratio by Kang and colleagues compared to that reported herein and by Wiesner and colleagues, which largely corroborate each other.
With the new knowledge of the relative levels of the human mitochondrial transcription system in HeLa cells, we went on to analyze the consequences of over-expressing each of the h-mtTFB paralogs on mitochondrial gene expression and biogenesis in vivo. We established stable HeLa cell lines that over-express h-mtTFB1 or h-mtTFB2 by ∼10-fold or up to ∼3-fold, respectively (Figure 1B). Characterization of these lines by western blotting immediately revealed a salient difference between these two factors; that h-mtTFB2 is processed in vivo (Figure 1B). Several additional lines of evidence strongly indicate that the processing of h-mtTFB2 is via cleavage by mitochondrial proteases upon import of the protein into mitochondria, including a strong predicted mitochondrial protease cleavage site between amino acids 30 and 31, the presence of a predicted amphipathic alpha helix (stereotypical of mitochondrial localization sequences) spanning amino acids 5–22, and an inability to detect h-mtTFB2 with a peptide antibody that was directed against amino acids 2–20, which would be removed by the predicted cleavage event (data not shown). No obvious mobility differences were observed between recombinant h-mtTFB1 and that isolated from cells. Furthermore, SignalP does not predict a mitochondrial cleavage site for h-mtTFB1 (data not shown). Thus, h-mtTFB2 and h-mtTFB1 are handled quite differently upon import, which is consistent with their distinct evolutionary history subsequent to the putative gene duplication event that created the two protein families early in eukaryotic lineage (4,5). It is also tempting to speculate that the use of different modes of import for these two transcription factors could provide a mechanism to control their relative levels in the organelle in response to different conditions.
A second important observation that came from the initial analysis of the h-mtTFB1 and h-mtTFB2 over-expression lines is that there is an increase in h-mtTFB1 when h-mtTFB2 is over-expressed (Figure 1B). However, the converse was not true. That is, in the h-mtTFB1 over-expression line, there is no change in the steady-state amounts of h-mtTFB2 per mitochondrion (Figure 1B). Furthermore, there are no obvious changes in the levels of POLRMT or h-mtTFA per mitochondrion in either of the h-mtTFB factor over-expression lines (Figure 1D). We conclude that there is some form of one-way communication between h-mtTFB2 and h-mtTFB1. The fact that this upregulation occurs, at least in part, via increased levels of the h-mtTFB1 mRNA (Figure 1C), suggests that this involved a retrograde signal transduction mechanism from the mitochondria to the nucleus (39). It is tempting to speculate that this response is initiated via signals generated by alterations in mitochondrial transcriptional output or the amount of 12S rRNA methylation.
We next examined multiple mitochondrial parameters in the h-mtTFB1 and h-mtTFB2 over-expression cell lines. In the h-mtTFB2 over-expression cell lines (where it is important to keep in mind that there is also a compensatory increase in mtTFB1 as discussed above; Figure 1B), there is a ∼2-fold increase in overall mitochondrial transcript levels as evidenced by northern analysis of the two rRNAs (12S and 16S) and two mRNAs (ND2 and ND6) encoded by mtDNA and representing transcripts derived from both strands (Figure 2A). This was accompanied by a corresponding doubling of the mtDNA copy number (Figure 2C). Given the documented roles for the h-mtTFB factors in directing transcription initiation efficiency (2), these changes most likely represent an increase in the rate of mitochondrial transcription that is also driving a increase in transcription-primed mtDNA replication (1). However, it remains a formal possibility that the increase in steady-state levels of mitochondrial transcripts is due to enhanced RNA stability and/or similar rates of transcription from the increased number of mtDNA templates. Also evident in the h-mtTFB2 over-expression lines was an increased rate of mitochondrial translation of most, but not all mitochondrial gene products (Figure 3A) that, at least in the case of COX1 and COX2, results in significantly increased steady-state amounts of protein (Figure 2B). However, the rates of mitochondrial translation were not uniform for all of the subunits. For example, there is apparently a reduced rate of translation of ATP6 and no apparent change in the rate of synthesis of ND3 or ND4L (Figure 3A). Thus, artificially raising the levels of h-mtTFB2 (and in response, also h-mtTFB1) does increase mitochondrial gene expression and mtDNA replication, but may come at the cost of imbalanced relative rates of mitochondrial protein synthesis. We suspect that most of the described effects on mitochondrial gene expression just listed are driven primarily by the increased levels of h-mtTFB2 since no major changes in mtDNA copy number, transcript levels, translation rates or steady-state levels of COX1 and COX 2 were observed in the cell line in which h-mtTFB1 alone was over-expressed (Figures 2 and 3). However, it is also just as likely that up-regulation of both factors is necessary to coordinately mount all of the responses we observe.
The lack of a significant ‘mitochondrial gene expression’ response in the h-mtTFB1 over-expression lines is somewhat surprising given the documented ability of this protein to activate transcription in vitro (2,3). One potential reason for this may be explained via the relative levels of the transcription system we established in these cells. For example, the ratio of h-mtTFA to h-mtTFB1 required for optimal transcription in vitro is ∼1:2 (2). Thus, the corresponding ratio we determined in vivo of 18:1 would likely not allow much of a contribution of h-mtTFB1 to the total transcriptional output. Furthermore, to reach the optimal ratio of 1:2 would require nearly 40-fold over-expression of h-mtTFB1. We were only able to increase h-mtTFB1 by ∼10-fold, which likely explains why no alterations in mitochondrial transcripts were observed in the h-mtTFB1 over-expression cell lines (Figure 2A). In contrast, the ratio of h-mtTFA to h-mtTFB2 in vivo of 5:1 (Table 1) is well within the range of 2:1–40:1 that is optimal for mitochondrial transcription in vitro (2). Increasing this by ∼3-fold in the h-mtTFB2 over-expression cell line would keep this ratio within this optimal range, entirely consistent with our results. Whether the relative amounts of the transcription components vary in different cell or tissue types such that h-mtTFB1 also contributes to mitochondrial transcription remains an open question.
While over-expression of h-mtTFB1 does not show any major effects on the mitochondrial transcription and translation parameters we measured, it did result in increased sensitivity of HeLa cells to the aminoglycoside antibiotic kasugamycin (Figure 3B). Sensitivity to this antibiotic in E. coli is modulated by dimethylation of two adenine residues in the 3′ terminal stem loop of the small subunit ribosomal RNA by KsgA (40,41), the bacterial homolog of h-mtTFB1 and h-mtTFB2. We have shown previously that h-mtTFB1 and h-mtTFB2 are able to methylate the homologous stem-loop in bacteria and restore sensitivity to kasugamycin (5,14). Thus, we interpret the ability of increased levels of h-mtTFB1 to sensitize HeLa cells to kasugamycin to indicate that h-mtTFB1 is increasing the number of methylated ribosomes that are targets for inhibition by the drug. This provides the first in vivo confirmation that h-mtTFB1 is likely the 12S rRNA methyltransferase in human mitochondria as we predicted from our earlier studies (5,14). Interestingly, we observed a similar sensitivity to kasugamycin in the h-mtTFB2 over-expression lines (Figure 3B). While this is most likely due to the fact that h-mtTFB1 is also up-regulated in this cell line, we cannot exclude the possibility that h-mtTFB2 is also contributing to the methylation of the 12S rRNA since it too possesses rRNA methyltransferase activity, albeit at a much lower level (5). In this regard it is interesting to note that others have shown that different methylation combinations on the E. coli 16S rRNA (adenines in the stem loop being monomethylated, dimethylated or mixed) result in different levels of aminoglycoside resistance (10). Thus, the idea that h-mtTFB1 and h-mtTFB2 might orchestrate different states of 12S rRNA methylation with different functional outcomes is an intriguing possibility.
Finally, we have elucidated a new role for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis. Over-expression of h-mtTFB1 alone results in a ∼50% increase in mitochondrial membrane as judged by Mitotracker Green staining and confirmed by western analysis of a mitochondrial marker protein, porin (a.k.a. VDAC, Figure 4). Thus, even in the absence of a direct effect on mitochondrial transcription or translation, increased h-mtTFB1 somehow signals a change in mitochondrial biogenesis. It will be of interest to determine the nature of this signal and whether it involves, for example, a sensing of the amount of ongoing mitochondrial ribosome assembly through the rRNA methylation status. A similar increase in mitochondrial biogenesis is observed in the h-mtTFB2 over-expression cell line (Figure 4), in which both h-mtTFB1 and h-mtTFB2 are elevated. However, here, unlike h-mtTFB1 over-expression alone, it was accompanied by a significant (∼80%) increase in mitochondrial membrane potential (Figure 4A). Taken together, these results suggest that, while h-mtTFB1 can alone induce a mitochondrial biogenesis response, an increase in both h-mtTFB1 and h-mtTFB2 is needed to coordinate an increase in mitochondrial gene expression with the increase in mitochondrial mass. We propose that the lack of an increase in membrane potential in the h-mtTFB1 over-expression cell line (Figure 4A) is a result of a defect in this response that leads to more mitochondria with fewer OXPHOS complexes per mass of organelle.
This study provides the first analysis of the relative levels of the human mitochondrial transcription system and functional roles of the h-mtTFB1 and h-mtTFB2 paralogs in vivo. Our results revealed distinct, but possibly coordinated functions of each of these factors in mitochondrial gene expression, biogenesis and activity that provide a new framework for future studies aimed at understanding the regulation of human mitochondrial transcription system in vivo and its potential as a therapeutic target for human diseases and aging.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health grant HL-059655 from the National Heart, Lung, and Blood Institute awarded to G.S.S. The authors wish to thank Dr Sharen Mckay for critical assistance on key aspects of the project. Funding to pay the Open Access publication charges for this article was provided by HL-059655.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 3.McCulloch V, Seidel-Rogol BL, Shadel GS. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol. Cell. Biol. 2002;22:1116–1125. doi: 10.1128/MCB.22.4.1116-1125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shutt TE, Gray MW. Homologs of mitochondrial transcription factor B, sparsely distributed within the eukaryotic radiation, are likely derived from the dimethlyadenosine methyltransferase of the mitochondrial endosymbiont. Mol. Biol. Evol. 2006;23:1169–1179. doi: 10.1093/molbev/msk001. [DOI] [PubMed] [Google Scholar]
- 5.Cotney J, Shadel GS. Evidence for an early gene duplication event in the evolution of the mitochondrial transcription factor B family and maintenance of rRNA methyltransferase activity in human mtTFB1 and mtTFB2. J. Mol. Evol. 2006;63:707–717. doi: 10.1007/s00239-006-0075-1. [DOI] [PubMed] [Google Scholar]
- 6.Fisher RP, Lisowsky T, Breen GA, Clayton DA. A rapid, efficient method for purifying DNA-binding proteins. Denaturation-renaturation chromatography of human and yeast mitochondrial extracts. J. Biol. Chem. 1991;266:9153–9160. [PubMed] [Google Scholar]
- 7.Takamatsu C, Umeda S, Ohsato T, Ohno T, Abe Y, Fukuoh A, Shinagawa H, Hamasaki N, Kang D. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002;3:451–456. doi: 10.1093/embo-reports/kvf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiesner RJ, Zsurka G, Kunz WS. Mitochondrial DNA damage and the aging process: facts and imaginations. Free Radic. Res. 2006;40:1284–1294. doi: 10.1080/10715760600913168. [DOI] [PubMed] [Google Scholar]
- 9.Shen EL, Bogenhagen DF. Developmentally-regulated packaging of mitochondrial DNA by the HMG-box protein mtTFA during Xenopus oogenesis. Nucleic Acids Res. 2001;29:2822–2828. doi: 10.1093/nar/29.13.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Farrell HC, Pulicherla N, Desai PM, Rife JP. Recognition of a complex substrate by the KsgA/Dim1 family of enzymes has been conserved throughout evolution. RNA (New York, N.Y) 2006;12:725–733. doi: 10.1261/rna.2310406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Igarashi K, Kishida K, Hirose S. Stimulation by polyamines of enzymatic methylation of two adjacent adenines near the 3′ and end of 16S ribosomal RNA of Escherichia coli. Biochem. Biophys. Res. Commun. 1980;96:678–684. doi: 10.1016/0006-291x(80)91408-4. [DOI] [PubMed] [Google Scholar]
- 12.Igarashi K, Kishida K, Kashiwagi K, Tatokoro I, Kakegawa T, Hirose S. Relationship between methylation of adenine near the 3′ end of 16-S ribosomal RNA and the activity of 30-S ribosomal subunits. Eur. J. Biochem. 1981;113:587–593. doi: 10.1111/j.1432-1033.1981.tb05103.x. [DOI] [PubMed] [Google Scholar]
- 13.Poldermans B, Bakker H, Van Knippenberg PH. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3′ end of 16S ribosomal RNA of Escherichia coli. IV. The effect of the methylgroups on ribosomal subunit interaction. Nucleic Acids Res. 1980;8:143–151. doi: 10.1093/nar/8.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seidel-Rogol BL, McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat. Genet. 2003;33:23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]
- 15.Carnahan RH, Feoktistova A, Ren L, Niessen S, Yates JR, III, Gould KL. Dim1p is required for efficient splicing and export of mRNA encoding lid1p, a component of the fission yeast anaphase-promoting complex. Eukaryot. Cell. 2005;4:577–587. doi: 10.1128/EC.4.3.577-587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lafontaine D, Vandenhaute J, Tollervey D. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev. 1995;9:2470–2481. doi: 10.1101/gad.9.20.2470. [DOI] [PubMed] [Google Scholar]
- 17.McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol. Cell. Biol. 2003;23:5816–5824. doi: 10.1128/MCB.23.16.5816-5824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsushima Y, Garesse R, Kaguni LS. Drosophila mitochondrial transcription factor B2 regulates mitochondrial DNA copy number and transcription in schneider cells. J. Biol. Chem. 2004;279:26900–26905. doi: 10.1074/jbc.M401643200. [DOI] [PubMed] [Google Scholar]
- 19.Matsushima Y, Adan C, Garesse R, Kaguni LS. Drosophila mitochondrial transcription factor B1 modulates mitochondrial translation but not transcription or DNA copy number in Schneider cells. J. Biol. Chem. 2005;280:16815–16820. doi: 10.1074/jbc.M500569200. [DOI] [PubMed] [Google Scholar]
- 20.Clary DO, Goddard JM, Martin SC, Fauron CM, Wolstenholme DR. Drosophila mitochondrial DNA: a novel gene order. Nucleic Acids Res. 1982;10:6619–6637. doi: 10.1093/nar/10.21.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goddard JM, Wolstenholme DR. Origin and direction of replication in mitochondrial DNA molecules from Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 1978;75:3886–3890. doi: 10.1073/pnas.75.8.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubenstein JL, Brutlag D, Clayton DA. The mitochondrial DNA of Drosophila melanogaster exists in two distinct and stable superhelical forms. Cell. 1977;12:471–482. doi: 10.1016/0092-8674(77)90123-4. [DOI] [PubMed] [Google Scholar]
- 23.Tiranti V, Savoia A, Forti F, D'Apolito MF, Centra M, Rocchi M, Zeviani M. Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the Expressed Sequence Tags database. Hum. Mol. Genet. 1997;6:615–625. doi: 10.1093/hmg/6.4.615. [DOI] [PubMed] [Google Scholar]
- 24.Dairaghi DJ, Shadel GS, Clayton DA. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Cotney J, Shadel GS. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J. Biol. Chem. 2007;282:12610–12618. doi: 10.1074/jbc.M700461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seidel-Rogol BL, Shadel GS. Modulation of mitochondrial transcription in response to mtDNA depletion and repletion in HeLa cells. Nucleic Acids Res. 2002;30:1929–1934. doi: 10.1093/nar/30.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor SD, Zhang H, Eaton JS, Rodeheffer MS, Lebedeva MA, O’Rourke TW, Siede W, Shadel GS. The conserved Mec1/Rad53 nuclear checkpoint pathway regulates mitochondrial DNA copy number in Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:3010–3018. doi: 10.1091/mbc.E05-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chomyn A. In: Methods in Enzymology. Attardi GM, Chomyn A, editors. San Diego, CA, USA: Vol. 264: Mitochondrial Biogenesis and Genetics Part B. Academic Press; 1996. [Google Scholar]
- 29.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen H, Krogh A. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology (ISMB 6); Manlo Park, CA, USA: AAAI Press; 1998. pp. 122–130. [Google Scholar]
- 31.Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31:1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 33.Kanki T, Nakayama H, Sasaki N, Takio K, Alam TI, Hamasaki N, Kang D. Mitochondrial nucleoid and transcription factor A. Ann. N. Y. Acad. Sci. 2004;1011:61–68. doi: 10.1007/978-3-662-41088-2_7. [DOI] [PubMed] [Google Scholar]
- 34.Kanki T, Ohgaki K, Gaspari M, Gustafsson CM, Fukuoh A, Sasaki N, Hamasaki N, Kang D. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol. Cell. Biol. 2004;24:9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antoshechkin I, Bogenhagen DF. Distinct roles for two purified factors in transcription of Xenopus mitochondrial DNA. Mol. Cell. Biol. 1995;15:7032–7042. doi: 10.1128/mcb.15.12.7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antoshechkin I, Bogenhagen DF, Mastrangelo IA. The HMG-box mitochondrial transcription factor xl-mtTFA binds DNA as a tetramer to activate bidirectional transcription. EMBO J. 1997;16:3198–3206. doi: 10.1093/emboj/16.11.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dairaghi DJ, Shadel GS, Clayton DA. Human mitochondrial transcription factor A and promoter spacing integrity are required for transcription initiation. Biochim. Biophys. Acta. 1995;1271:127–134. doi: 10.1016/0925-4439(95)00019-z. [DOI] [PubMed] [Google Scholar]
- 38.Garstka HL, Schmitt WE, Schultz J, Sogl B, Silakowski B, Perez-Martos A, Montoya J, Wiesner RJ. Import of mitochondrial transcription factor A (TFAM) into rat liver mitochondria stimulates transcription of mitochondrial DNA. Nucleic Acids Res. 2003;31:5039–5047. doi: 10.1093/nar/gkg717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol. Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 40.Helser TL, Davies JE, Dahlberg JE. Mechanism of kasugamycin resistance in Escherichia coli. Nat. New Biol. 1972;235:6–9. doi: 10.1038/newbio235006a0. [DOI] [PubMed] [Google Scholar]
- 41.Van Buul CP, Damm JB, Van Knippenberg PH. Kasugamycin resistant mutants of Bacillus stearothermophilus lacking the enzyme for the methylation of two adjacent adenosines in 16S ribosomal RNA. Mol. Gen. Genet. 1983;189:475–478. doi: 10.1007/BF00325912. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.