Abstract
In the last years, RNA interference (RNAi)-mediated gene knockdown has developed into a routine method to assess gene function in cultured mammalian cells in a fast and easy manner. For the use of this technique in developing or adult mice, short hairpin (sh)RNA vectors expressed stably from the genome are a faster alternative to conventional knockout approaches. Here we describe an advanced strategy for conditional gene knockdown in mice, where we used the Cre/loxP system to activate RNAi in a time and tissue dependent manner in the adult mouse brain. By placing conditional RNAi constructs into the defined genomic Rosa26 locus and by using recombinase mediated cassette exchange (RMCE) instead of laborious homologous recombination, we developed a fast, easy and reproducible approach to assess gene function in adult mice. We applied this technique to three genes of the MAPK signaling pathway—Braf, Mek1 and Mek2—and demonstrate here the potential of this new tool in mouse mutagenesis.
INTRODUCTION
RNA interference (RNAi) has become an extremely useful genetic tool to study gene function in mammalian cells. The discovery that short double-stranded RNAs, known as short interfering (si)RNAs, avoid an interferon response and the global shutdown of translation has enabled the wide use of transient gene silencing in cultured cells and specific tissues of mice upon local administration (1). To elicit permanent gene silencing, short hairpin (sh)RNA expression vectors can be used. These vectors consist of an RNA polymerase III promoter producing short RNA fragments, which form hairpin structures. These shRNAs are processed by the RNAi machinery in the same way as linear double-stranded RNAs such that sequence-specific gene silencing occurs (2). Mice transgenic for shRNA vectors produce an all-over knockdown phenotype, similar to conventional knockout mice (3). To overcome the embryonic lethality of many mutants and to investigate gene functions in specific tissues or in a time dependent manner, conditional vectors have to be used. ShRNA expression can either be regulated by an inducing compound-like doxycycline acting on artificial regulatory sequences in the polymerase III promoter (4) or shRNA production is blocked by a transcriptional stop element that can be deleted through Cre mediated recombination. The latter Cre/loxP approach is similar to conditional knockout or knock in strategies where it is widely used. Various vector designs for Cre/loxP regulated RNAi have been described (5,6). Here, we show a fast and highly reproducible system to generate mice expressing shRNAs under the control of Cre recombinase. This tool can be applied to a great variety of biological questions since a large collection of mouse strains that express Cre recombinase in specific cell types is available and can be used to activate conditional shRNA vectors at different developmental stages and in selected cell types of mice (7).
Mitogen activated protein kinases (MAPKs), also called extracellular signal-regulated kinases (ERKs), are a group of serine/threonine terminal protein kinases evolutionarily conserved from yeast to human. They form an intracellular signaling cascade regulating fundamental cellular functions including proliferation, cell survival and differentiation (8). The MAPK pathway plays also an important role in neurons as it is involved in synaptic plasticity, neuronal survival and regeneration (9). A regulatory function of MAPK signaling for anxiety and depression-like behavior of adult mice has been proposed (10) but not tested with genetic models in vivo. Here, we show that a conditional knockdown of Braf and Mek1 and Mek2 in the adult murine brain is possible with our RNAi approach, which gives the possibility to test the role of these genes in anxiety and depression-like behavior in vivo.
MATERIALS AND METHODS
LacZ and Luciferase shRNA vectors
H1 promoter vectors
The human H1 promoter was recovered as 119 bp SalI/KpnI fragment from the plasmid H1-RasGap (3) and ligated into the SalI/KpnI sites of pNEB193 (New England Biolabs). Plasmid H1-shLacZ coding for a lacZ specific shRNA was obtained by ligation of an oligonucleotide pair (5′-cccctggcgttacccaacttaatcgaagcttggattaagttgggtaacgccagggtttttggaaag-3′, 5′-aattctttccaaaaaccctggcgttacccaacttaatccaagcttcgattaagttgggtaacgccagggggtac-3′; targeting nt 57–79 of E.coli lacZ gene) into the KpnI/EcoRI sites of pNEB-H1. pNEB-H1-shLacZ was opened with PsiI/KpnI and ligated with an oligonucleotide pair (5′-taacttcgtatagcatacattatacgaagttatggtac-3′, 5′-cataacttcgtataatgtatgctatacgaagtta-3′) to create H1-shLacZ-loxP1 or opened with KpnI and ligated with another oligonucleotide pair (5′-ataacttcgtatagcatacattatacgaagttatatatactagtcgacgtac-3′, 5′-gtcgactagtatatataacttcgtataatgtatgctatacgaagttatgtac-3′) to create H1-shLacZ-loxP3. To create H1-shLacZ-lox2, that contains a lox2272 sequence (11), pNEB-H1-shLacZ was opened with SalI/KpnI and ligated with a 120 bp DNA fragment from an oligonucleotide pair (5′-acgcgtcgacggatcgggagatccaatatttgcatgtcgctatgtgttctgggaaatcaccataaacgtgaaatata-3′, 5′-ggggtaccgagtggtctcaataacttcgtataaggtatcctatacgaagttatatttcacgtttatggtgattt-3′) that was annealed, extended with Klenow polymerase, and digested with SalI/KpnI. Upon removal of a 5′ HindIII site from H1-shLacZ by SapI/PmeI digestion, end filling and religation, H1-shLacZ was opened within the shRNA loop region by HindIII and ligated with an oligonucleotide pair (5′-agctataacttcgtatagcatacattatacgaagttatggatcc-3′, 5′-agctggatccataacttcgtataatgtatgctatacgaagttat-3′) to create H1-shLacZ-loxP4 or with an other oligonucleotide pair (5′-agctataacttcgtatagcatacattatacgaagttat-3′, 5′-agctataacttcgtataatgtatgctatacgaagttat-3′) to obtain H1-shLacZ-loxP5.
U6 promoter vectors
The pSHAG plasmid containing the human U6 promoter (12) was opened with BseRI/BamHI and ligated with a lacZ specific shRNA oligonucleotide pair (5′-gcgttacccaacttaatcgccttggaagcttgcaaggcgattaagttgggtaacgccttttttggaaa-3′, 5′-gatctttccaaaaaaggcgttacccaacttaatcgccttgcaagcttccaaggcgattaagttgggtaacgccg-3′; targeting nt 61–85 of E. coli lacZ gene) to generate U6-shLacZ or ligated with a humanized F-Luciferase specific shRNA oligonucleotide pair (5′-tgcgctgctggtgccaacgaagcttggttggcaccagcagcgcacttttttggaaa-3′, 5′-gatctttccaaaaaagtgcgctgctggtgccaaccaagcttcgttggcaccagcagcgcacg-3′) to generate U6-shLuc. To create U6-shLacZ-loxP3, the U6-shLacZ segment was recloned into a modified pBluescript plasmid, opened with SgrAI at the transcriptional start site and ligated with an oligonucleotide pair (5′-ccggataacttcgtatagcatacattatacgaagttatatatactagtcgac-3′, 5′-ccgggtcgactagtatatataacttcgtataatgtatgctatacgaagttat-3′). For U6-shLacZ-loxP4, U6-shLacZ was opened in the shRNA loop region with HindIII and ligated with a pair of oligonucleotides (5′-agctataacttcgtatagcatacattatacgaagttatggatcc-3′, 5′-agctggatccataacttcgtataatgtatgctatacgaagttat-3′) that introduce a loxP sequence and a 5′ BamHI site. For U6-shLacZ-loxP5, U6-shLacZ was opened with HindIII and ligated with a pair of oligonucleotides (5′-agctataacttcgtatagcatacattatacgaagttat-3′, 5′-agctataacttcgtataatgtatgctatacgaagttat-3′) that inserts a loxP sequence into the shRNA loop in a symmetric fashion.
To generate the conditional lacZ and Luciferase specific shRNA vectors U6-lox-lox-shluc and U6-lox-lox-shLacZ, plasmids U6-shLuc and U6-shLacZ were opened in the loop region with HindIII, the ends were filled with Klenow fragment and ligated with a 338 bp MlyI fragment serving as loxP flanked stop cassette. This MlyI fragment was cut from a pNEB193 based vector that contained a 19 bp R-Luciferase antisense and two polythymidine sequences, serving as termination signals, and that has been flanked with loxP sequences from oligonucleotide pairs that were ligated into the HindIII site (5′-agctgagtcgactgataacttcgtatagcatacattatacgaagttatggatcc-3′, 5′-agctggatccataacttcgtataatgtatgctatacgaagttatcagtcgactc-3′) and NdeI site (5′-tattttttggatccataacttcgtatagcatacattatacgaagttatgactggactc-3′, 5′-tagagtccagtcataacttcgtataatgtatgctatacgaagttatggatccaaaaaa-3′). The sequence of the 338 bp stop cassette fragment is (loxP-sites underlined): 5′-ataacttcgtatagcatacattatacgaagttatggatccagcttggtagcgcggtgtattatactttttggaaagaattcactggccgtcgttttacaacgtcgtga ctgggaaaaccctggcgttacccaacttaatcgccttgcagcacatccccctttcgccagctggcgtaatagcgaagaggcccgcaccgatcgcccttcc caacagttgcgcagcctgaatggcgaatggcgcctgatgcggtattttctccttacgcatctgtgcggtatttcacaccgcatattttttggatccataacttcgtatagcatacattatacgaagttat-3′.
To delete the loxP flanked stop cassette from U6-lox-lox-shluc and U6-lox-lox-shLacZ, the plasmids were transformed into Cre expressing bacteria (294cre; 13) and recombined subclones (U6-loxP5-shLuc, U6-loxP5-shLacZ) were retransformed into E. coli strain DH5α. All plasmids were grown in DH5α, isolated with Qiagen plasmid Maxiprep columns and the integrity of the promoter and shRNA regions was confirmed by DNA sequencing.
Braf and Mek1/2 shRNA vectors
The pSHAG plasmid containing the human U6 promoter (12) was opened with BseRI/BamHI and ligated with a Braf specific shRNA oligonucleotide pair (5′-gagaggagttacatgttgaagaagcttgttcaacatgtaactcctctccttttttggaaa-3′, 5′-gatctttccaaaaaaggagaggagttacatgttgaacaagcttcttcaacatgtaactcctctccg-3′, targeting 5′-ggagaggagttacatgttgaag-3′ in Braf Exon 5) to generate pU6-shBraf or ligated with a Mek1/2 specific shRNA oligonucleotide pair (5′-acggcgagatcagcatctgcatgaagcttgatgcagatgctgatctcgccgtcttttttggaaa-3′, 5′-gatctttccaaaaaagacggcgagatcagcatctgcatcaagcttcatgcagatgctgatctcgccgtcg-3′, targeting 5′-gacggcgagatcagcatctgcatg-3′ in Mek1 Exon 2 and Mek2 Exon 2) to generate pU6-shMek1/2. To create pU6-shBraf-flox and pU6-shMek1/2-flox, the vector pU6-shBraf or pU6-shMek1/2, respectively, was opened with HindIII in the loop region of the hairpin sequence, ends were filled with Klenow fragment, followed by ligation with the above described MlyI fragment containing the loxP-flanked stop cassette.
Tissue culture
Mouse F1 ES cells (IDG3.2) were used for transient and stable transfections. Cells were grown in DMEM medium (Gibco) containing 15% fetal calf serum, 20 mM HEPES, 1× non-essential aminoacids, 0.1 mM β-mercaptoethanol and 1.5 × 103 U/ml leukemia inhibiting factor on gelatin coated culture dishes for transient transfections or on mitomycin c treated embryonic fibroblasts for stable transfections and blastocyst injections.
Transient transfections
Transient transfections with the lacZ and Luciferase specific shRNA vectors were performed with Fugene6 transfection reagent following the manufacturer's protocol (Roche Diagnostics). One day before transfection 60 000 ES cells were plated into each well of a 24 well culture plate. 150 ng of each supercoiled plasmid, a total amount of 450 ng, were complexed with 2 µl Fugene and transfected in duplicate into each well in 500 µl growth medium. For β-Galactosidase and Luciferase measurements 150 ng of the β-Galactosidase expression vector CMVβ (Promega) were transfected together with 150 ng shRNA vector and 150 ng of the F-Luciferase expression vector pCMV-hLuc. pCMV-hLuc was generated by ligation of the CMV promoter (770 bp XhoI/EcoRI fragment) from pUHD17-1 (14) into the XhoI/HindIII sites of pGL3 basic (Promega) that contains a codon optimized F-Luciferase gene. The medium was changed after 24 h and the cells were lysed and chemiluminescence was detected 48 h after transfection.
Chemiluminescence reporter gene assays
The preparation of lysates and measurement of β-Galactosidase activity were performed with the β-Galactosidase analysis kit (Roche Diagnostics) following the manufacturer's protocol. Fifty microliter of freshly prepared lysates were measured for 5 s in an Orion I plate luminometer (Berthold). For the detection of F-Luciferase activity, 20 µl of each lysate were mixed with 100 μl Luciferase assay buffer (25 mM glycylglycine, KH2PO4, 4 mM EGTA, 2 mM ATP, 1 mM DTT, 100 µM Coenzyme A, 75 µM Luciferin, pH8) and measured for 5 s in the Orion I plate luminometer.
For lacZ shRNA vectors, the relative light units (RLU) from β-Galactosidase measurements were normalized to the Luciferase RLU of each sample, and for Luciferase shRNA vectors, RLU from β-Galactosidase were used for normalization. The measurements of β-Galactosidase RLU in Rosa26.5/26.9 ES cells were normalized to the total amount of protein in the lysates. Protein determinations were performed with the BCA protein assay kit (Pierce) following the manufacturer's instruction.
Homologous recombination into Rosa26
The Rosa26 gene targeting vector pRosa26.5 was assembled from Rosa26-1 (15) by insertion of a splice acceptor element (16), the β-Galactosidase (lacZ) coding region and polyA signal from CMVβ (Promega), and the hygromycin resistance gene cassette from plasmid pgk-hygro-pA (a gift from P. Krimpenfort) into the XbaI site. The Rosa26 gene targeting vector pRosa26.9 was assembled from Rosa26-1 by insertion of a splice acceptor element, a SV40 late polyA signal, a neomycin resistance cassette from pgk-neo-pA (a gift from P. Soriano) and the conditional shRNA vector U6-lox-lox-shLacZ into the XbaI site; further details on cloning are available on request. The linearized vectors were electroporated into mouse F1-ES cells followed by selection with G418 (140 µg/ml) or hygromycin (125 U/ml) for 7 days. Resistant colonies were analyzed by Southern blotting of EcoRV digested genomic DNA using the Rosa26 5′-probe. One of the Rosa26.5 ES cell clones was further transfected with pRosa26.9 to introduce the conditional shRNA vector into the second Rosa26 locus.
The Rosa26 5′-probe for Southern blotting is a 448 bp fragment of the Rosa26 locus 5′ to the first exon, amplified with the following primer pair: 5′-aaggatactggggcatacg-3′ and 5′-cttctcagctacctttacacacc-3′. With this probe, recombined Rosa26.5 ES cell clones were recognized by the presence of a 3.6 kb fragment in addition to the 11.5 kb Rosa26 wild type band after EcoRV digestion. Recombined Rosa26.9 ES cell clones were recognized by the presence of a 3.1 kb fragment in addition to the 3.6 kb Rosa26.5 band; a remaining 11.5 kb wild type band indicated a trisomy of chromosome 6 in these ES cells (data not shown).
To activate the shRNA vector through Cre mediated recombination, double-targeted Rosa26.5/26.9 ES cells were transiently transfected with the Cre expression plasmid pCAG-cre-bpA. It was generated from pgk-cre-bpA (a gift from W. Müller) by EcoRI/PstI digestion to remove the pgk promoter and by insertion of the CAGGS promoter from CAGGS-FLPe (17) as a 1673 bp SalI/AlwNI fragment. Transfected subclones were expanded and analyzed by Southern blotting of BamHI digested genomic DNA for Cre mediated deletion of the stop cassette. The Rosa26 5′-probe detects a 6.5 kb band from the Rosa26.5 allele and a 5.8 kb band from the non-recombined Rosa26.9 allele, the same size is produced by the wild type locus. Cre mediated deletion can be recognized by the shift of the 5.8 kb Rosa26.9 band to a size of 9 kb since the deleted stop cassette contains a BamHI site (data not shown).
Three non-recombined and two recombined subclones were further expanded without feeder cells on gelatin coated plates and analyzed for β-Galactosidase activity by detection of chemiluminescence or by histochemical staining of fixed cells.
β-Galactosidase staining
Cells were washed with phosphate buffered saline (PBS) and fixed for 5 min at room temperature in 4% paraformaldehyde in PBS. Next, the cells were washed twice with PBS and incubated in X-Gal staining solution (5 mM K3(Fe(CN)6), 5 mM K4(Fe(CN)6), 2 mM MgCl2, 1 mg/ml X-Gal (BioMol) in PBS) for 24 h at 37°C. Images were taken using an AxioCam HRc camera and the AxioVision program (Carl Zeiss).
Cassette exchange vectors
The cassette exchange acceptor vector pRosa26.10 was assembled from Rosa26-1 (15) by insertion of a splice acceptor sequence and a modified hygromycin resistance gene derived from pgk-hygro-pA (a gift from P. Krimpenfort) into the XbaI site. One 50 bp attP recognition site (5′-gtagtgccccaactggggtaacctttgagttctctcagttgggggcgtag-3′) for φC31 integrase (18) was placed between the pgk-promoter and the hygro-coding region. A second attP site was cloned in the same orientation downstream of the hygro-polyA region. Homologous recombination of pRosa26.10 in ES cells was achieved as reported above and hygromycin resistant colonies were analyzed for homologous recombination by Southern blotting of EcoRV digested genomic DNA using the Rosa26 5 ‘-probe (see above). Homologous recombination of the pRosa26.10 vector is indicated by a 4.5 kb band in addition to the 11.5 kb band from the wild type Rosa26 allele. The ES cell clone IDG26.10-3 was selected for further work, and the ability of these cells to colonize the germline in chimeric mice was confirmed (data not shown).
The cassette exchange donor vector pRMCE was assembled within the backbone of pBluescript containing a modified polylinker region. One 53 bp attB recognition site (5′-ccgcggtgcgggtgccagggcgtgcccttgggctccccgggcgcgtactccac-3′) of C31Int was cloned upstream of the neomycin coding and polyA region derived from pgk-neo-bpA, and a second attB site was cloned downstream in the same orientation. The multiple cloning site from pBluescript was inserted as a SacI/KpnI fragment between the polyA signal and the second attB site for subcloning of shRNA constructs.
The expression vector pCAG-C31Int was generated by PCR amplification of the 1900 bp coding region of φC31 integrase from phage DNA (DSM-49156 received from DSMZ—Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany) with primers C31N (5′-taagtctagaccgatatgacacaaggggttgtgaccggggtg-3′) and C31C (5′-cgactctagactaaaccttcctcttcttcttaggcgccgctacgtcttccgtgccgtcctgg-3′), digestion with XbaI and cloning into the XbaI site of pNEB193. Primer C31C introduces a SV40 derived nuclear localization signal to the C-terminus of φC31 integrase as described (19). Upon sequence confirmation, the C31Int coding region was cloned as PacI-PstI fragment into pNEB-CAG downstream of the CAGGS promoter resulting in pCAG-C31Int(NLS). pNEB-CAG was generated by cloning the CAGGS promoter as 1673 bp blunt ended SalI/AlwNI fragment from pCAGGS-FLPe (17) into the filled BamHI site of pNEB193. A bovine polyA region from pgk-neo-bpA was added into the PmeI site of pCAG-C31Int(NLS) as 300 bp XbaI/NotI fragment resulting in pCAG-C31Int(NLS)-bpA. Further details on cloning strategies are available on request. The sequences and maps of pRosa26.10 and pCAG-C31(NLS)-bpA and a protocol for cassette exchange in ES cells are available at http://www.rnai.ngfn.de/index_4.htm.
Recombinase mediated cassette exchange
Braf and Mek1/2 shRNA constructs were cloned as NotI/EcoRV fragment from pU6-shBraf-flox or pU6-shMek-flox, respectively, into pRMCE to generate the shRNA donor vector for RMCE. The donor vector was coelectroporated with a C31Int expression vector into mouse acceptor ES-cells (clone IDG26.10-3) followed by selection with G418 for 7 days. Resistant colonies were isolated and analyzed for cassette exchange by Southern blotting of ScaI digested genomic DNA using the Rosa26 5′-probe (see above). A 9.2 kb band in addition to the 6.1 kb band from the wild type Rosa26 locus indicated correct cassette exchange, whereas the original RMCE locus gives rise to a 7.6 kb band.
Generation of mutant mice, breeding and genotyping
Positive ES cells containing the shBraf-flox or the shMek-flox construct, respectively, were injected into C57BL/6 blastocysts. Resulting chimeras were bred to C57BL/6 mice and offspring were tested for germline transmission. Heterozygous mice for the modified Rosa26 allele were crossed to CamKII-cre and Nestin-cre mice (on C57BL/6 background), respectively (20,21), to obtain shBraf+/flox/CamKII-cre mice and shMek1/2+/flox/Nestin-cre mice. Experiments on animals were carried out in accordance with national and institutional guidelines.
Cre transgenes were genotyped by PCR using the primer pair 5′-ATGCCCAAGAAGAAGAGGAAGGT-3′ and 5′-GAAATCAGTGCGTTCGAACGCTAGA-3′ to amplify a 447 bp fragment from the Cre gene. The shBraf and shMek1/2 constructs in the Rosa26 locus were genotyped by Southern blotting of EcoRV digested genomic DNA using the Rosa26 5′-probe (see above). ShRNA-alleles are indicated by a 5.2 kb band in addition to the 11.5 kb band of the wildtype Rosa26 allele.
Southern blotting
DNA was extracted from brain tissue with the Wizard genomic DNA extraction kit (Promega). Ten micrograms of genomic DNA were digested with BamHI, run on a 0.8% agarose gel and blotted on a nylon membrane (Hybond N+, Amersham). Hybridization was performed overnight at 65°C with the Rosa26 5′-probe, labeled with 32P (5′-[α-32P]dCTP, Amersham). After washing, the membrane was exposed to a Kodak BioMax MS film.
Northern blotting
Whole brains or half brains (with forebrain regions only, for Braf) were used for RNA extraction with TriReagent (Ambion). Northern blots for mRNA were performed with the NorthernMax-Gly system (Ambion). Membranes were hybridized overnight at 65°C with a 32P-labeled DNA probe (5′-[α-32P]dCTP, Amersham) against the 3′UTR of Mek1 or β-Actin (Ambion). After washing, the membrane was exposed to a Kodak BioMax film. Quantification of band intensities was performed with an Imaging Plate and the FLA-8000 instrument (Fujifilm Life Science). For analysis of siRNAs, forebrain regions of shBraf+/flox/CamKII-cre and half brains of shMek1/2+/flox/Nestin-cre were used for extraction of small RNAs with the mirVana miRNA isolation kit (Ambion). Two or three micrograms of small RNA were separated on a Novex® TBE-urea gel (15%, Invitrogen). As positive control 0.05 pmol of an oligonucleotide corresponding to the antisense region of the hairpin was used (5′-ATGCAGATGCTGATCTCGCCGTC-3′ for shMek1/2 and 5′-TTCAACATGTAACTCCTCTCC-3′ for shBraf). RNA was blotted onto a nylon membrane (Hybond N+, Amersham), which was hybridized at 40°C with an oligonucleotide probe (5′-GACGGCGAGATCAGCATCTGCAT-3′ for shMek1/2 and 5′-GGAGAGGAGTTACATGTTGAA-3′ for shBraf), labeled with 32P (5′-[γ-32P]dATP, Amersham). After washing, the membrane was exposed to a Kodak BioMax film.
In situ hybridization
Mice were asphyxiated with CO2 and perfused intracardially, after a brief rinse with ice-cold PBS, with ice-cold 4% paraformaldehyde in 0.1 M phosphate buffer (PB, pH 7.5). Brains were dissected and post-fixed for 2 h in 4% paraformaldehyde-PB. For embedding, the brains were dehydrated through an ascending ethanol scale (30%, 50%, 70%, 95%, 2× 100%, 1 h each passage), clarified in Rotihistol (Roth) 2× 45 min, equilibrated in 50% Rotihistol/50% paraffin for 1 h, and then transferred into paraffin at +65°C, 2× 1 h; they were equilibrated to room temperature in the last paraffin passage and kept at +4°C until cutting. Eight-micrometer sections were cut coronally and mounted on SuperFrost® Plus slides (Menzel-Glaeser); slides were dried overnight at 37°C and put at +4°C until proceeding with in situ hybridization (ISH).
For preparation of the Mek1 riboprobe, a DNA template was prepared by RT-PCR on total RNA extracted from adult brains of C57BL/6J mice. Total RNA was extracted using Trizol (Invitrogen) following manufacturer's instructions. Reverse transcription was performed using random hexamers and SuperScriptII (Invitrogen Kit) following manufacturer's instructions. PCR amplification was performed with the following oligonucleotides: 5′-GTCTGAGAGGGAGCCTTGTG-3′ and 5′-GCCAGCATCTGAGCCTTTAG-3′ (from NCBI acc #BC051137, nt 1303–2151). After linearization with the appropriate enzyme and purification (PCR purification Kit, QIAgen), two micrograms of DNA template were used for the synthesis of radiolabeled transcript by in vitro transcription with 35S-UTP (Amersham). After 20 min of DNase I (Roche) treatment, the probe was purified by the RNeasy Clean up protocol (QIAgen) and measured in a scintillation counter. For in situ hybridization, sections were dewaxed, pretreated and prehybridized as described previously (22). Subsequently, they were hybridized overnight with a probe concentration of 7 × 107 c.p.m./ml at 57°C and washed at 65°C in 0.1× SSC and 0.1 mM DTT. The hybridized slides were dipped in autoradiographic emulsion (Kodak, NTB2), developed after 3 weeks, counterstained with cresyl violet, dehydrated in an ascending ethanol scale and Xylol, and lidded with DPX. Images were taken using an AxioCam MRc camera and the AxioVision program (Carl Zeiss).
Detailed protocols for the preparation of radiolabeled probes and ISH procedures are given at http://www.empress.har.mrc.ac.uk/EMPReSS/servlet/EMPReSS.Frameset.
Western blotting
Total protein was extracted from brain tissue. Tissue was homogenized in RIPA buffer (50 mM Tris-HCl pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, protease inhibitor), sonificated and centrifuged. Fifty micrograms protein of each sample were run on a 10% Tris-HCl gel (Biorad) and blotted on a PVDF membrane (Pall). After blocking with 4% skim milk the membrane was incubated with the first antibody (1–3 h), washed with TBST, incubated with the second horseradish-peroxidase-conjugated antibody (1 h) and washed with TBST. The detection reaction was initiated with ECL detection reagents (Amersham) and the membrane was exposed to Hyperfilm (Amersham). For quantification of band intensities ECL plus was used instead of ECL detection reagent and chemifluorescent bands were detected with the FLA-8000 fluorescent image analyzer (Fujifilm Life Science). The antibodies used were anti-β-ACTIN (AC-15, #ab6276, Abcam, 1 : 100 000, 1 h incubation), anti-MEK1 (sc-219, Santa Cruz Biotechnology, 1 : 1000, 1 h incubation), anti-MEK2 (# 610235, BD Transduction Laboratories, 1:2500, 1 h incubation) and anti-BRAF (sc-166, Santa Cruz Biotechnology, 1 : 600, 3 h incubation).
RESULTS
Development of conditional shRNA vectors
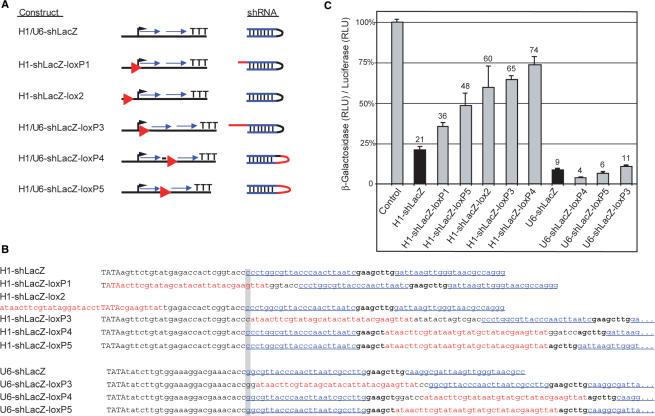
To control the activity of shRNA vectors through Cre/loxP mediated recombination, their transcription must be initially aborted by a loxP flanked stop element. The shRNA vector should be activated upon Cre mediated deletion of the stop segment. Since a single 34 bp loxP site remains in the activated vector after Cre recombination, we first tested whether this additional sequence disturbs RNAi efficiency. Therefore, we used vector configurations with different positions for the remaining loxP site, where the shRNA is driven by the human U6 or H1 promoter (Figure 1A and B). We found that the loxP site in all configurations of H1 promoter driven constructs strongly diminished the efficiency of gene silencing whereas the loxP site in all constructs driven by the U6 promoter did not significantly affect RNAi-mediated gene silencing (Figure 1C). Thus, the U6 but not the H1 promoter is of use for Cre mediated control of shRNA expression and we chose the loxP5 configuration (Figure 1A) as one effective configuration that includes the loxP sequence within the loop region.
Figure 1.
RNAi activity of H1 or U6 promoter based shRNA expression vectors modified by loxP sites. (A) Overview of the tested shRNA vectors. The position of the loxP sites (red triangle) in relation to the start of transcription (black arrow) and the shRNA region (blue arrows) are indicated. In the resulting shRNAs loxP derived sequences are shown in red color. All constructs contain identical sense and antisense shRNA sequences with specificity to β-Galactosidase (lacZ). (B) Sequences of the H1 and U6 promoter constructs. The different positions of the loxP and lox2272 sites (red), respectively, within or outside of the promoter are shown. The nucleotide of the transcriptional start is highlighted in gray. In capital letters: TATA box of the promoter; bold: original loop region and in blue and underlined: shRNA sense and antisense regions. (C) Silencing activity of β-Galactosidase specific shRNA vectors upon transient cotransfection with β-Galactosidase and F-Luciferase expression plasmids into ES cells. Values are expressed as ratio of β-Galactosidase to Luciferase activity in RLU in comparison to the positive control. The results obtained with the parental shRNA vectors H1-shLacZ and U6-shLacZ are shown as black columns. Numbers above the columns indicate the efficiency of silencing of β-Galactosidase activity in percent in comparison to the positive control. All results from duplicate samples are expressed as mean values with standard deviation.
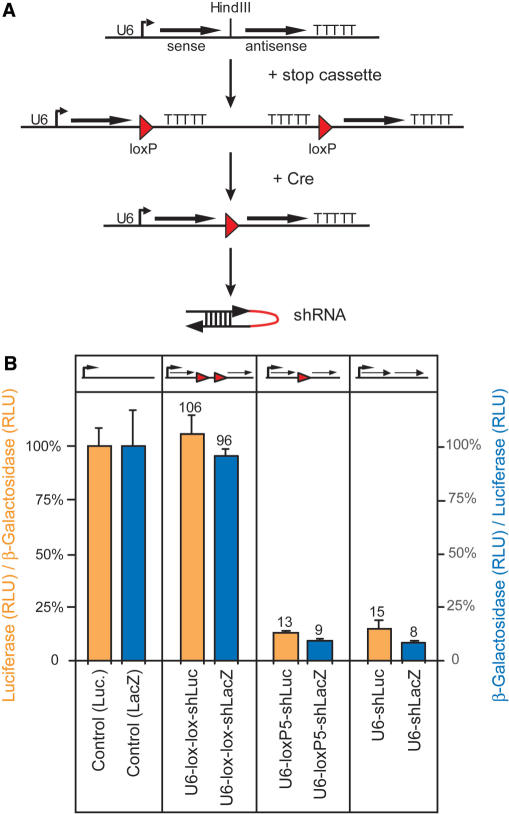
According to this design we inserted a loxP-flanked 270 bp DNA segment, which includes termination sequences and serves as transcriptional stop element, in our parental U6 driven shRNA vector. The presence of the stop element in the ‘off ’ configuration of the conditional shRNA vector allows only the transcription of the sense region that does not induce RNA interference, provided that the shRNA construct follows the order 5′-sense-loop-antisense-3′ (Figure 2A). Upon Cre mediated deletion of the stop cassette, the shRNA vector acquires the configuration of loxP5 with the remaining loxP site in the loop sequence. To assess the functionality of the stop element, it was first inserted into the loop region of U6-shRNA vectors against the β-Galactosidase or F-Luciferase reporter genes (Figure 2A). Upon transient cotransfection with reporter gene expression plasmids into mouse ES cells, the efficiency of the stop cassette in the ‘off ’ state and RNAi efficiency in the ‘on’ state were analyzed and compared to the non-modified parental vectors U6-shLuc and U6-shLacZ (Figure 2B). The measurement of F-Luciferase and β-Galactosidase activity from transfected cells showed that the ‘off ’ versions did not induce gene silencing whereas the ‘on’ versions induced the same level of RNAi as the parental, non-conditional vectors (Figure 2B). These results show that the stop element and our loxP5 configuration provide a way to control shRNA expression through Cre mediated recombination.
Figure 2.
Cre/loxP controlled conditional shRNA vectors. (A) Scheme for the modification of an shRNA vector into a Cre/loxP controlled version by insertion of a loxP flanked stop cassette into the shRNA loop region. The conditional shRNA vector can be activated by excision of the stop cassette through Cre mediated deletion. A single loxP site remains within the shRNA loop. (B) Transient transfection of β-Galactosidase (lacZ, blue columns) or F-Luciferase (Luc, orange columns) specific conditional shRNA vectors before (U6-lox-lox-shLuc; U6-lox-lox-shLacZ) and after excision of the loxP flanked stop cassette (U6-loxP5-shLuc; U6-loxP5-shLacZ) into ES cells. For F-Luciferase shRNA vectors values are expressed as ratio of Luciferase to β-Galactosidase activity (left Y axis) and for β-Galactosidase shRNA vectors as ratio of β-Galactosidase to Luciferase activity (right Y axis), in comparison to the appropriate positive control. Results are expressed as mean values from duplicate samples with SD. U6-shLuc, U6-shLacZ: parental shRNA vectors; RLU: relative light units.
Single copy shRNA expression from the Rosa26 locus
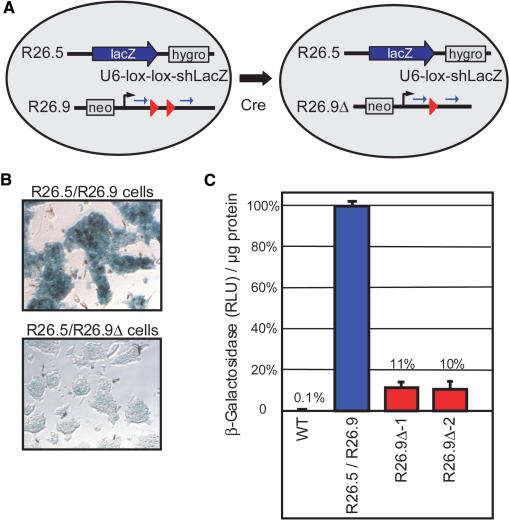
For the genomic integration of conditional shRNA vectors, a single-copy approach is preferable since multi copy integrations could undergo unpredictable and non-functional rearrangements upon Cre mediated recombination. We tested the efficiency of a single shRNA vector copy in the Rosa26 locus (15) of murine ES cells with β-Galactosidase as reporter gene. We first inserted a splice acceptor sequence and the β-Galactosidase coding region by gene targeting downstream of the first exon of one Rosa26 allele of murine ES cells. These modified R26.5 ES cells express β-Galactosidase from the endogenous, ubiquitously active Rosa26 promoter. Next, the second Rosa26 allele of R26.5 ES cells was targeted with a single copy of the conditional shRNA vector U6-lox-lox-shLacZ giving rise to R26.5/R26.9 ES cells. In the double-targeted R26.5/R26.9 cells, the shRNA vector stop cassette was deleted by transient transfection with a Cre expression plasmid (Figure 3A). Three ES cell clones harboring the non-activated and two clones carrying the recombined shRNA construct (R26.5/R26.9Δ cells) were isolated and analyzed for β-Galactosidase activity by histochemical staining (X-Gal) and by a chemiluminescence assay using cell lysates. X-Gal staining revealed a strong reduction of β-Galactosidase activity in R26.5/R26.9Δ ES cells as compared to non-recombined R26.5/R26.9 cells (Figure 3B). The quantitative analysis revealed that the β-Galactosidase activity in R26.5/R26.9Δ cells is reduced by 90% (Figure 3C). Thus, a single genomic shRNA vector copy within the Rosa26 locus is sufficient to induce RNAi in ES cells and the extent of gene silencing can reach a similar level as obtained by transient transfections (compare to Figure 2B).
Figure 3.
Cre mediated activation of a single copy conditional shRNA vector within the Rosa26 locus of murine ES cells. (A) One Rosa26 allele of ES cells was modified by a gene-targeting vector (R26.5) that introduced a splice acceptor-lacZ cassette and a hygromycin resistance gene such that β-Galactosidase (lacZ) is expressed from the endogenous Rosa26 promoter. R26.5 ES cells were further modified with a gene targeting vector (R26.9) that introduced a neomycin resistance gene and the conditional shRNA vector U6-lox-lox-shLacZ. The R26.5/R26.9 ES cells were transiently transfected with a Cre expression vector and subclones that recombined the R26.9 allele (R26.9Δ) were isolated. (B) X-Gal staining of fixed R26.5/R26.9 ES cells in comparison to a R26.5/R26.9Δ clone shows highly reduced β-Galactosidase activity in the latter cells (magnification 20×). (C) Comparison of β-Galactosidase activity in lysates of R26.5/R26.9 ES cells (mean of three non-deleted subclones, blue column) in comparison to two deleted subclones (R26.9Δ-1, R26.9Δ-2, red columns) and wild type ES cells (WT). Values are shown as β-Galactosidase activity in RLU per micrograms protein of the lysates in comparison to the non-deleted clones and are expressed as mean values with SD.
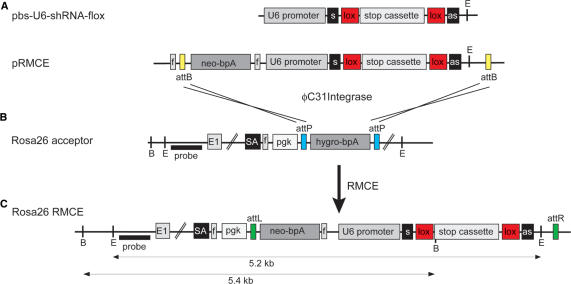
To facilitate the insertion of shRNA vectors into the Rosa26 locus of ES cells for the fast generation of multiple knockdown mouse lines, we developed an approach for recombinase mediated cassette exchange (RMCE) using the integrase of phage φC31 (C31Int). For this purpose, ES cells with a Rosa26 acceptor allele were generated. These cells harbor a pgk promoter driven hygromycin resistance gene, of which the coding and polyA region is flanked by attP recognition sites, in the Rosa26 locus (Figure 4B). The acceptor allele is designed such that the attP-flanked segment can be replaced by an attB-flanked construct from a donor plasmid (pRMCE) upon transfection of ES cells together with a C31Int expression plasmid (Figure 4A). The donor vector contains a promoterless neomycin resistance coding and polyA region and the shRNA expression cassette between the attB recognition sites. The shift from hygromycin to neomycin resistance in ES cells selects for RMCE events (Figure 4C), such that correct cassette exchange occurs at a frequency of 40–60% among the neomycin resistant ES cell colonies (data not shown).
Figure 4.
Vector construction and RMCE for the generation of shBraf-flox and shMek-flox mice. (A) The conditional shRNA expression cassette from the vector pbs-U6-shRNA-flox contains the U6 promoter in front of the sense (s) sequence of the shRNA, the loxP (lox) flanked stop cassette in the loop region and the antisense (as) shRNA sequence. The shRNA expression cassette is cloned into the RMCE donor vector pRMCE behind the promoterless neomycin selection marker (neo-bpA) so that the two attB sites from pRMCE flank the selection marker as well as the shRNA expression cassette. (B) Acceptor ES cells for RMCE harbor a Rosa26 allele where in intron 1 a splice acceptor (SA) is inserted followed by a pgk promoter (pgk) driving a hygromycin selection marker (hygro-bpA) which is flanked by attP sites. (C) Upon RMCE with C31Int, the attP flanked cassette in the acceptor ES cells from B is replaced by the attB flanked cassette from the donor vector in A. FRT (f) sites allow to excise the pgk promoter and the neomycin marker in recombined ES cells or mice. E: EcoRV; B: BamHI; probe: 5′-Rosa26 probe and E1: exon 1.
Conditional Braf and Mek1/2 shRNA mice
To study the effects of RNA interference against components of the MAPK cascade in vivo, we generated two conditional knockdown mouse lines. For the first mouse line, shBraf-flox, expressing an shRNA against Braf, we generated six shRNA vectors and tested their knockdown efficiency by transient transfection in cell culture (data not shown). In the vector containing the most efficient shRNA sequence, we inserted the stop cassette to block shRNA transcription before Cre recombination (pbs-U6-shBraf-flox). The shRNA expression cassette of this vector was in turn inserted into the donor vector pRMCE to generate recombinant ES cells via RMCE (Figure 4A).
The second mouse line, shMek-flox, was generated in the same way as shBraf-flox. The shRNA sequence was chosen in a way that it targets Mek1 as well as Mek2, in a region where both mRNAs show 100% homology (in Exon 2 of both genes). Due to the restricted targeting region, here only two different shRNA vectors were generated and tested for efficiency.
Recombinant ES cells were injected into blastocysts to generate shBraf-flox and shMek-flox mice. Upon germline transmission, heterozygous shBraf+/flox mice were crossed to CamKII-cre mice to activate RNAi in forebrain neurons of adult mice (20). Mice carrying one modified Rosa26 allele with the shMek-flox construct were crossed to Nestin-cre mice, which express Cre recombinase in neuronal and glia cell precursors, to activate RNAi in the nervous system (21). All animals, containing the conditional shRNA construct or the activated shRNA together with the Cre transgene, were viable, fertile and showed no obvious phenotype.
Analysis of knockdown in shBraf-flox mice
Cre recombinase under the CamKII promoter activates the shRNA only in neurons of the adult forebrain (20). To show the deletion of the stop cassette in the U6-shBraf-flox element, a Southern blot analysis with genomic DNA from different brain regions was performed. A shift of the 5.4 kb band with the stop cassette to an 8.6 kb band after Cre recombination indicates activation of RNAi (Figure 5A). This larger band is visible only in forebrain regions of mutant mice, like the olfactory bulb, the hippocampus, striatum, cortex and very weak in the thalamus, but not in more posterior parts of the brain of these mice or in control mice. As expected, the 5.4 kb band from the inactive shRNA allele is still visible in all the forebrain regions of mutant mice, resulting from non-neuronal cells in the forebrain expressing no Cre recombinase.
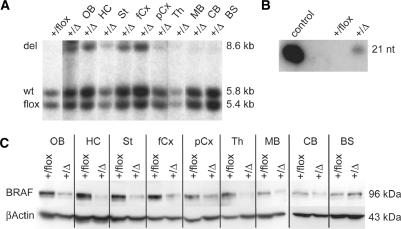
Figure 5.
Analysis of RNA interference in shBraf+/flox/CamKII-cre mice. (A) Tissue-specific activation of shRNA in the brain with CamKII-cre. Southern blot analysis of BamHI digested genomic DNA from different brain regions of adult shBraf+/flox/CamKII-cre mice. Lane 1: DNA from whole brains of shBraf+/flox control mice; lane 2–10: DNA from indicated regions of shBraf+/flox/CamKII-cre mice. The wild type Rosa26 allele (wt) gives a 5.8 kb band and the band from the shRNA allele is shifted from 5.4 kb with the stop cassette (flox) to 8.6 kb after Cre recombination (del). (B) Expression of shRNA against Braf in adult forebrain. On a Northern blot with small RNAs against the sequence of shBraf the 21 nt band of the processed siRNA against Braf is only detectable with the control oligonucleotide and in mutant mice but not in control mice. (C) BRAF protein reduction in forebrain regions of shBraf+/flox/CamKII-cre mice. On a Western blot with protein from the indicated brain regions from adult mouse brain, knockdown of BRAF protein in forebrain regions of mutant mice is shown in comparison to the protein level from control mice. β-ACTIN was used as a loading control. OB: olfactory bulb, HC: hippocampus, St: Striatum, fCx: frontal cortex, pCx: posterior cortex, Th: Thalamus, MB: midbrain, CB: cerebellum, BS: brainstem, +/flox: shBraf+/flox and +/Δ: shBraf+/flox/CamKII-cre.
Consistent with the situation on the genomic level, we could show expression of the Braf shRNA in mutant mice, but not in control mice without Cre transgene, on a Northern blot with short RNAs (Figure 5B). At the protein level, reduction of BRAF protein is visible in brain regions of mutant mice where Cre recombination occurred (Figure 5C). Slight protein knockdown of BRAF in midbrain and cerebellum, where no Cre recombination on the DNA level is visible, may be due to recombination in a small fraction of cells that remains undetected by Southern blot analysis (Figure 5A). Since the BRAF expression in these regions is low and restricted (23), the limited Cre activity could still result in visible protein knockdown if a substantial fraction of Braf expressing neurons also coexpress Cre recombinase. No protein knockdown is visible in the brainstem. Estimation of levels of protein reduction in forebrain regions revealed a knockdown of ∼70% compared to protein expression in control mice.
Analysis of knockdown in shMek-flox mice
In contrast to the CamKII promoter, the Nestin promoter drives the expression of Cre recombinase in all nervous tissue (21). To show the deletion of the stop cassette from the U6-shMek-flox vector in the whole brain of adult mice, a Southern blot was performed in the same way as for the shBraf-flox mice. Also here the band from the shRNA allele is shifted from 5.4 kb with the stop cassette to 8.6 kb after Cre recombination (Figure 6A). The larger band is visible in all brain regions of mutant mice, but not in control mice. The 5.4 kb band from the inactive shRNA allele is absent in all brain regions from mutant mice, indicating complete Cre mediated recombination and therefore shRNA activation in the whole adult brain. Hence, we could show high expression of the shMek transcript in mutant mice but not in control mice on a Northern blot with short RNAs from whole brains (Figure 6B).
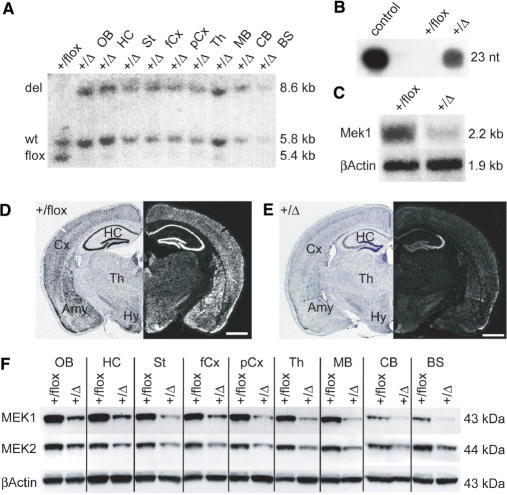
Figure 6.
Analysis of RNA interference in shMek+/flox/Nestin-cre mice. (A) Tissue specific activation of shRNA in the brain with Nestin-cre. Southern blot analysis of BamHI digested genomic DNA from different brain regions of adult shMek+/flox/Nestin-cre mice. Lane 1: DNA from whole brains of shMek+/flox control mice; lane 2–10: DNA from indicated regions of shMek+/flox/Nestin-cre mice. The wild type Rosa26 allele (wt) gives a 5.8 kb band and the band from the shRNA allele is shifted from 5.4 kb with the stop cassette (flox) to 8.6 kb after Cre recombination (del). (B) Expression of shRNA against Mek1 and Mek2 in whole adult brain. On a Northern blot with small RNAs against the sequence of shMek the 23 nt band of the processed siRNA against Mek1 and Mek2 is highly detectable with the control oligonucleotide and in mutant mice but not in control mice. (C–E) Knockdown of Mek1 mRNA. On a Northern blot of total RNA from whole adult brain (C) Mek1 mRNA from mutant mice is decreased ∼65% compared to control mice. β-Actin was used as a loading control. In situ hybridization against Mek1 mRNA on coronal brain sections shows the knockdown effect throughout the brain of mutant mice (E) in contrast to the high wildtype expression of Mek1 mRNA in control mice (D). (F) MEK1 and MEK2 protein reduction in the brain of shMek+/flox/Nestin-cre mice. On a Western blot with protein from the indicated regions from adult brain, knockdown of MEK1 and MEK2 in mutant mice is shown in comparison to control mice. β-ACTIN was used as a loading control. OB: olfactory bulb, HC: hippocampus, St: Striatum, fCx: frontal cortex, pCx: posterior cortex, Th: Thalamus, MB: midbrain, CB: cerebellum, BS: brainstem, Cx: cortex, Amy: amygdala, Hy: hypothalamus, +/flox: shMek+/flox, +/Δ: shMek+/flox/Nestin-cre; scale bar in D and E: 1 mm.
Since the shRNA of these mice is specific for both Mek1 and Mek2 mRNA, knockdown should occur at the mRNA and protein level of both genes. But due to the very low expression level of Mek2 mRNA in the adult brain (23), detection of mRNA is possible only for Mek1 (Figure 6C). Quantification of the Northern signal revealed a 65% reduction of mRNA in mutant brains as compared to wild type controls. In situ the reduction of Mek1 mRNA can be observed in the whole brain of mutant mice compared to control mice (Figure 6D and E). On the protein level, reduction of MEK1 and MEK2 protein was observed in the whole brain of mutant mice (Figure 6F). Estimation of levels of protein reduction revealed a knockdown of about ∼65% for MEK1—consistent with the mRNA knockdown—and ∼50% for MEK2 compared to control expression each.
DISCUSSION
Here, we describe a novel technique to inactivate one or two related genes in the adult murine brain with RNA interference in a tissue-specific manner. We developed conditional shRNA expression vectors that can be activated upon Cre mediated recombination. After testing various configurations for the positional effect of a transcriptional stop cassette within H1 or U6 promoter driven shRNA vectors, we selected one construct with high knockdown efficiency after Cre recombination. Due to the position of the stop cassette, the loop region of the shRNA transcribed from this expression construct is elongated by 34 nt. In contrast to former studies asserting a 9 nt loop sequence being the most efficient configuration (24), our elongated loop sequence does not interfere with shRNA efficiency. Essential for this technique, we could show that the insertion of a loxP flanked stop segment into the loop region of shRNA vectors disrupts RNAi induction and that such vectors can be activated through Cre mediated recombination.
An obvious application for conditional RNAi are shRNA vector transgenic mice since a large collection of mouse strains that express Cre recombinase in specific cell types is available and can be used to activate conditional shRNA in different developmental stages and cell types (7). For the genomic integration of such expression vectors, different approaches are available (25). Transgenic RNAi mice have been generated by pronuclear injection (26–28), by lentiviral infection (29) or electroporation of ES cells (3). All these approaches result in random, multicopy integrants of the shRNA vector and therefore require a laborious analysis of multiple lines due to the influence of the genomic environment and the vector copy number on transgene expression (3,27). Apart from the time and resources needed for this initial screening of mouse lines, it is not applicable to conditional shRNA vectors since multicopy integrations could undergo unpredictable and non-functional rearrangements through Cre mediated recombination. Hence, a single-copy approach using a defined and well-characterized genomic locus is preferable.
The HPRT and the Rosa26 locus are frequently used for the genomic integration of expression vectors and have also been used for shRNA vectors (30–32). Since the HPRT locus is affected by X-inactivation in female mice, we chose the Rosa26 locus for integration of our shRNA vectors. Using a reporter gene, we showed in homologous recombinant ES cells that our conditional shRNA expressed from one vector copy in the Rosa26 locus gives rise to highly efficient gene knockdown after Cre mediated recombination, comparable to transient transfections of multiple vector copies. Thus, the Rosa26 locus allows effective and reproducible U6 promoter driven transcription of shRNA and the amount of shRNA transcribed from one single vector integrant is sufficient for efficient gene knockdown. The functionality of the U6 promoter within a defined chromosomal locus, like Rosa26, lays the basis for conditional RNAi in mice using a single vector copy that is recombined by Cre recombinase into a single, predictable product.
To circumvent the laborious and inefficient homologous recombination step for mouse generation, we further used RMCE to produce ES cells harboring one copy of the shRNA expression vector in the Rosa26 locus. With this approach, the frequency of properly recombined ES cell clones among selection positive clones rises from ∼1% with homologous recombination at Rosa26 to 40–60% with RMCE.
Our RNAi mice generated with these techniques harbor a conditional shRNA vector either against Braf or Mek1 and Mek2 at once. These mice are viable, fertile, show no overt phenotype, and do not express the specific shRNA before Cre mediated recombination. We used two different Cre expressing mouse lines—both expressing Cre recombinase in a brain-specific manner—to activate shRNA transcription. We showed that in shBraf+/flox/CamKII-cre mice Cre mediated recombination occurs only in neurons of forebrain regions of the adult brain and in shMek+/flox/Nestin-cre mice in all cells of the adult brain, as described previously (20,21). Consequently, shRNA expression and reduction of mRNA and protein is detectable in these tissues from mice expressing Cre recombinase. The extent of specific mRNA and protein knockdown reaches ∼70% in case of Braf and Mek1 and ∼50% for Mek2. Important to mention here is, that shMek+/flox/Nestin-cre mice produce one shRNA targeting Mek1 and Mek2 at once. Although the efficiency of RNAi is not identical for both genes, we show that it is feasible to knockdown several related genes simultaneously.
So, this technique will facilitate functional studies of gene families where the loss of one gene may be compensated by other family members (33,34). Addressing such questions with conventional knockout strategies implies an enormous effort of breeding and genotyping to obtain double or even triple knockout animals. With RNAi, generating multiple knockdown mice is not more effort as compared to single knockdown mice. Furthermore, the production of shRNA expressing mice is much faster and easier than that of knockout mice, especially with the use of RMCE. Thus, in 3–4 months even conditional knockdown mice can be generated whereas at least 12 months are required for the production of a classical unconditional knockout strain. Moreover, the effort of breeding shRNA mice is significantly decreased since only one shRNA allele is required to exert the knockdown of the targeted gene and breeding for homozygosity (as for knockout mice) is not required.
In the conditional shRNA mice against MAPKs reported here, we reached knockdown levels of up to 70% after Cre mediated recombination. In the reporter gene experiments, we showed that in ES cells a higher knockdown level of up to 90% is possible with the single-copy approach in the Rosa26 locus. Using shRNAs against the CRHR1 (corticotropin releasing hormone receptor 1) and LRRK2 (leucine-rich repeat kinase 2) genes we indeed obtained knockdown efficiencies of 80 and 90%, respectively, in the adult brain (R. Kühn, unpublished data). Therefore, the level of knockdown that can be reached in vivo with the Rosa26-shRNA approach is not limited to 70% but rather depends on the intrinsic efficacy of the specific target sequence of an individual shRNA, able to elicit either higher or lower levels of gene silencing. Using public available siRNA prediction programs as paradigm for the design of five shRNA vectors for each gene, we usually find one vector that induces in vitro a knockdown of 90%. This average efficiency should improve in future with the further development of siRNA and shRNA prediction algorithms. To obtain higher knockdown levels for Braf and Mek1/2 the selection of new, more efficient shRNA sequences would be necessary. Another possibility to enhance the efficiency of less potent shRNA sequences may be to increase the number of active siRNAs in the cell. To test whether increased shRNA levels result in improved knockdown, we compared the residual target gene expression in single copy heterozygous and double copy homozygous shRNA mice. In the three tested mouse lines we found in homozygous shRNA mice a moderate (∼5–10%) but not dramatic increase of knockdown efficiency (C. Hitz, P. Steuber-Buchberger, unpublished data). But even a less potent protein knockdown can result in a phenotype giving interesting insights to the mode of function of the gene. For example Shalin et al. (35) showed impaired fear conditioning in mice with a 40% reduction in ERK1/2 activation. Furthermore, constitutive knockdown with our shRNA against Mek1/2, indeed, does not lead to a placental defect as described for the constitutive knockout of Mek1 (36), but it leads to dwarfism and death at the age of 6 weeks (data not shown). All in all, the doubling of the shRNA copy number and the use of different shRNA target sequences offer the option to exploit differences in efficiency of shRNA sequences to produce allelic series as refined models of genetic diseases beyond the ‘all or nothing’ principle of knockout mice (37).
We demonstrate that RNAi is a powerful tool for the generation of conditional mouse mutants to study the function of single or related genes in vivo. Recently, Seibler et al. (38) described a new approach for conditional RNAi using the tetO/tetR system. In contrast to our cell type-specific Cre/loxP regulated system the tetR regulated system is reversible and can be switched on and off optionally. However, the tetO/tetR system is not cell type specific and acts simultaneously in all organs and cell types. Since more than 150 strains of tissue specific Cre transgenic mice are available, we expect that our conditional shRNA approach can be applied to a wide range of biological questions in a variety of tissues. Here, we used this approach to generate brain-specific knockdown mice for members of the MAPK pathway to gain insight into the function of this signaling cascade that has been proposed to play a role in mood disorders (10). In a first behavioral characterization of Mek1/2 knockdown mice, we observed a contribution of these kinases to the expression of exploratory and anxiety related behavior (data not shown). Further, more detailed studies using conditional knockdown mice are in progress to establish the relation of this phenotype to human anxiety disorders.
ACKNOWLEDGEMENTS
We thank S. Michailidou, S. Kareth, C. Birke, R. Kneuttinger and A. Tasdemir for excellent technical help and G. Hannon for the pSHAG vector. This work has been funded by the Volkswagen Foundation and the Federal Ministry of Education and Research (BMBF) in the framework of the National Genome Research Network (FKZ:01GR0404). The authors are responsible for the contents of this publication. Funding to pay the Open Access publication charges for this article was provided by the GSF National Research Center.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lieberman J, Song E, Lee SK, Shankar P. Interfering with disease: opportunities and roadblocks to harnessing RNA interference. Trends Mol. Med. 2003;9:397–403. doi: 10.1016/S1471-4914(03)00143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 3.Kunath T, Gish G, Lickert H, Jones N, Pawson T, Rossant J. Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype. Nat. Biotechnol. 2003;21:559–561. doi: 10.1038/nbt813. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Stamatoyannopoulos G, Song CZ. Down-regulation of CXCR4 by inducible small interfering RNA inhibits breast cancer cell invasion in vitro. Cancer Res. 2003;63:4801–4804. [PubMed] [Google Scholar]
- 5.Kasim V, Miyagishi M, Taira K. Control of siRNA expression using the Cre-loxP recombination system. Nucleic Acids Res. 2004;32:e66. doi: 10.1093/nar/gnh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tiscornia G, Tergaonkar V, Galimi F, Verma IM. CRE recombinase-inducible RNA interference mediated by lentiviral vectors. Proc. Natl Acad. Sci. USA. 2004;101:7347–7351. doi: 10.1073/pnas.0402107101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagy A, Mar L. Creation and use of a Cre recombinase transgenic database. Methods Mol. Biol. 2001;158:95–106. doi: 10.1385/1-59259-220-1:95. [DOI] [PubMed] [Google Scholar]
- 8.Rubinfeld H, Seger R. The ERK cascade: a prototype of MAPK signaling. Mol. Biotechnol. 2005;31:151–174. doi: 10.1385/MB:31:2:151. [DOI] [PubMed] [Google Scholar]
- 9.English JD, Sweatt JD. A requirement for the mitogen-activated protein kinase cascade in hippocampal long term potentiation. J. Biol. Chem. 1997;272:19103–19106. doi: 10.1074/jbc.272.31.19103. [DOI] [PubMed] [Google Scholar]
- 10.Coyle JT, Duman RS. Finding the intracellular signaling pathways affected by mood disorder treatments. Neuron. 2003;38:157–160. doi: 10.1016/s0896-6273(03)00195-8. [DOI] [PubMed] [Google Scholar]
- 11.Siegel RW, Jain R, Bradbury A. Using an in vivo phagemid system to identify non-compatible loxP sequences. FEBS Lett. 2001;499:147–153. doi: 10.1016/s0014-5793(01)02541-8. [DOI] [PubMed] [Google Scholar]
- 12.Paddison PJ, Cleary M, Silva JM, Chang K, Sheth N, Sachidanandam R, Hannon GJ. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nat. Methods. 2004;1:163–167. doi: 10.1038/nmeth1104-163. [DOI] [PubMed] [Google Scholar]
- 13.Buchholz F, Angrand PO, Stewart AF. A simple assay to determine the functionality of Cre or FLP recombination targets in genomic manipulation constructs. Nucleic Acids Res. 1996;24:3118–3119. doi: 10.1093/nar/24.15.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 15.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 16.Choi T, Huang M, Gorman C, Jaenisch R. A generic intron increases gene expression in transgenic mice. Mol. Cell Biol. 1991;11:3070–3074. doi: 10.1128/mcb.11.6.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaft J, Ashery-Padan R, van der Hoeven F, Gruss P, Stewart AF. Efficient FLP recombination in mouse ES cells and oocytes. Genesis. 2001;31:6–10. doi: 10.1002/gene.1076. [DOI] [PubMed] [Google Scholar]
- 18.Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl Acad. Sci. USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andreas S, Schwenk F, Kuter-Luks B, Faust N, Kuhn R. Enhanced efficiency through nuclear localization signal fusion on phage PhiC31-integrase: activity comparison with Cre and FLPe recombinase in mammalian cells. Nucleic Acids Res. 2002;30:2299–2306. doi: 10.1093/nar/30.11.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minichiello L, Korte M, Wolfer D, Kuhn R, Unsicker K, Cestari V, Rossi-Arnaud C, Lipp HP, Bonhoeffer T, et al. Essential role for TrkB receptors in hippocampus-mediated learning. Neuron. 1999;24:401–414. doi: 10.1016/s0896-6273(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 21.Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 22.Dagerlind A, Friberg K, Bean AJ, Hokfelt T. Sensitive mRNA detection using unfixed tissue: combined radioactive and non-radioactive in situ hybridization histochemistry. Histochemistry. 1992;98:39–49. doi: 10.1007/BF00716936. [DOI] [PubMed] [Google Scholar]
- 23.Di Benedetto B, Hitz C, Holter SM, Kuhn R, Vogt Weisenhorn DM, Wurst W. Differential mRNA distribution of components of the ERK/MAPK signalling cascade in the adult mouse brain. J. Comp. Neurol. 2007;500:542–556. doi: 10.1002/cne.21186. [DOI] [PubMed] [Google Scholar]
- 24.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 25.Gordon JW. Production of transgenic mice. Methods Enzymol. 1993;225:747–771. doi: 10.1016/0076-6879(93)25048-7. [DOI] [PubMed] [Google Scholar]
- 26.Carmell MA, Zhang L, Conklin DS, Hannon GJ, Rosenquist TA. Germline transmission of RNAi in mice. Nat. Struct. Biol. 2003;10:91–92. doi: 10.1038/nsb896. [DOI] [PubMed] [Google Scholar]
- 27.Coumoul X, Shukla V, Li C, Wang RH, Deng CX. Conditional knockdown of Fgfr2 in mice using Cre-LoxP induced RNA interference. Nucleic Acids Res. 2005;33:e102. doi: 10.1093/nar/gni100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasuwa H, Kaseda K, Einarsdottir T, Okabe M. Small interfering RNA and gene silencing in transgenic mice and rats. FEBS Lett. 2002;532:227–230. doi: 10.1016/s0014-5793(02)03680-3. [DOI] [PubMed] [Google Scholar]
- 29.Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl Acad. Sci. USA. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oberdoerffer P, Kanellopoulou C, Heissmeyer V, Paeper C, Borowski C, Aifantis I, Rao A, Rajewsky K. Efficiency of RNA interference in the mouse hematopoietic system varies between cell types and developmental stages. Mol. Cell Biol. 2005;25:3896–3905. doi: 10.1128/MCB.25.10.3896-3905.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seibler J, Kuter-Luks B, Kern H, Streu S, Plum L, Mauer J, Kuhn R, Bruning JC, Schwenk F. Single copy shRNA configuration for ubiquitous gene knockdown in mice. Nucleic Acids Res. 2005;33:e67. doi: 10.1093/nar/gni065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J, McMahon AP. Reproducible and inducible knockdown of gene expression in mice. Genesis. 2006;44:252–261. doi: 10.1002/dvg.20213. [DOI] [PubMed] [Google Scholar]
- 33.Cammalleri M, Cervia D, Dal Monte M, Martini D, Langenegger D, Fehlmann D, Feuerbach D, Pavan B, Hoyer D, et al. Compensatory changes in the hippocampus of somatostatin knockout mice: upregulation of somatostatin receptor 2 and its function in the control of bursting activity and synaptic transmission. Eur. J. Neurosci. 2006;23:2404–2422. doi: 10.1111/j.1460-9568.2006.04770.x. [DOI] [PubMed] [Google Scholar]
- 34.Grady RM, Teng H, Nichol MC, Cunningham JC, Wilkinson RS, Sanes JR. Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: a model for Duchenne muscular dystrophy. Cell. 1997;90:729–738. doi: 10.1016/s0092-8674(00)80533-4. [DOI] [PubMed] [Google Scholar]
- 35.Shalin SC, Zirrgiebel U, Honsa KJ, Julien JP, Miller FD, Kaplan DR, Sweatt JD. Neuronal MEK is important for normal fear conditioning in mice. J. Neurosci. Res. 2004;75:760–770. doi: 10.1002/jnr.20052. [DOI] [PubMed] [Google Scholar]
- 36.Giroux S, Tremblay M, Bernard D, Cardin-Girard JF, Aubry S, Larouche L, Rousseau S, Huot J, Landry J, et al. Embryonic death of Mek1-deficient mice reveals a role for this kinase in angiogenesis in the labyrinthine region of the placenta. Curr. Biol. 1999;9:369–372. doi: 10.1016/s0960-9822(99)80164-x. [DOI] [PubMed] [Google Scholar]
- 37.Hemann MT, Fridman JS, Zilfou JT, Hernando E, Paddison PJ, Cordon-Cardo C, Hannon GJ, Lowe SW. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat. Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- 38.Seibler J, Kleinridders A, Kuter-Luks B, Niehaves S, Bruning JC, Schwenk F. Reversible gene knockdown in mice using a tight, inducible shRNA expression system. Nucleic Acids Res. 2007;35:e54. doi: 10.1093/nar/gkm122. [DOI] [PMC free article] [PubMed] [Google Scholar]