Abstract
Splicing of pre-mRNA is a critical step in mRNA maturation and disturbances cause several genetic disorders. We apply the synthetic tetracycline (tc)-binding riboswitch to establish a gene expression system for conditional tc-dependent control of pre-mRNA splicing in yeast. Efficient regulation is obtained when the aptamer is inserted close to the 5′splice site (SS) with the consensus sequence of the SS located within the aptamer stem. Structural probing indicates limited spontaneous cleavage within this stem in the absence of the ligand. Addition of tc leads to tightening of the stem and the whole aptamer structure which probably prevents recognition of the 5′SS. Combination of more then one aptamer-regulated intron increases the extent of regulation leading to highly efficient conditional gene expression systems. Our findings highlight the potential of direct RNA–ligand interaction for regulation of gene expression.
INTRODUCTION
Alternative splicing of pre-mRNAs is recognized as the most important source of protein diversity in vertebrates (1,2) and defective regulation of pre-mRNA splicing has been identified as cause of several genetic diseases and various forms of cancer (3–6). Alternative splicing is regulated by the presence of enhancer/silencer elements, the strength of splicing signals and additional presence of protein factors. In addition, the structure and conformation of the pre-mRNA also has an influence on the efficiency of splicing (7). This is demonstrated by the recently identified riboswitches in the genomes of eukaryotes (8). Riboswitches are regulatory elements which can adopt a defined structure to directly sense a metabolite. Ligand-binding then leads to changes in the conformation which influences gene expression. In contrast to bacterial riboswitches, which mainly interfere with transcription termination or translation initiation, eukaryotic riboswitches preferentially seem to tackle mRNA processing steps (8–11).
A recent report unravels the mechanisms of riboswitch-controlled alternative splicing in the filamentous fungi Neurospora. The expression of genes involved in thiamine pyrophosphate metabolism is regulated by riboswitches which are located in introns in the 5′untranslated region (5′UTR) (11). They restructure upon metabolite-binding therewith forcing alternative splice site (SS) usage. These findings not only expand the scope of gene regulation by direct RNA ligand interaction, but also demonstrate that eukaryotic cells also employ riboswitches to control certain metabolic pathways by targeting pre-mRNA splicing.
These findings prompted us to develop a synthetic riboswitch able to control pre-mRNA splicing in yeast. For that, a small molecule-binding, in vitro selected RNA aptamer has been used. Aptamers display high binding affinity and specificity and adopt a unique conformation only upon ligand-binding with the ligand becoming an integral part of the complex (12). This inducible conformational change has already been used to develop conditional gene regulation systems. Inserting an aptamer into the 5′UTR of a eukaryotic mRNA led to interference of the aptamer-ligand complex with initial stages of translation initiation (13). We have identified a tetracycline (tc)-binding aptamer which confers tc-dependent gene regulation in yeast (14). The tc-aptamer complex inhibits the small ribosomal subunit joining when the aptamer is placed close to the cap structure and interferes with formation of the 80S ribosome when inserted directly in front of the start codon, probably by blocking scanning (15).
We describe here a conditional gene expression system in vivo, which exploits tc-aptamer mediated inhibition of pre-mRNA splicing (Figure 1). This expands the applicability of the tc-binding aptamer to regulate gene expression. In addition, we significantly enhanced the efficiency of tc-aptamer based regulation by combining the regulatory elements that influence both translation initiation and pre-mRNA splicing.
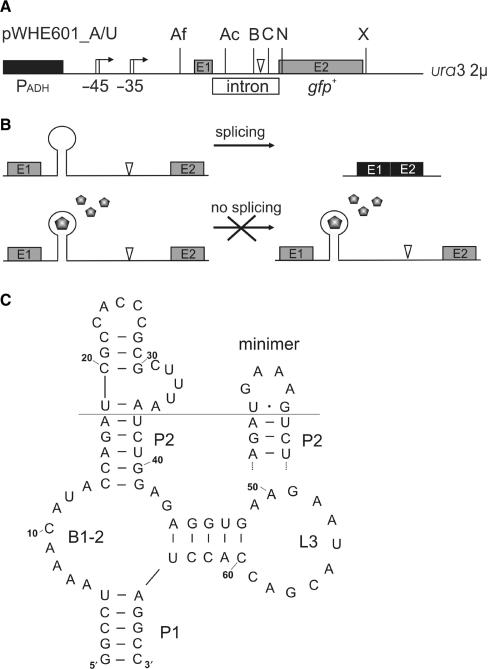
Figure 1.
Tc-aptamer regulated pre-mRNA splicing. (A) Schematic view of the vectors pWHE601_A/U. Shown is the adh promoter (PADH, black box), the gfp open reading frame with both exons E1 and E2 as gray boxes and the intron as open box. Unique restriction sites are indicated (AflII - Af, Acc65I - Ac, Bsu36I - B, ClaI - C, NheI - N, XbaI - X). The arrows indicate the transcriptional start sites with their distances to the start codon. The branch point of the intron is marked by an open triangle. (B) Model to explain tc-aptamer mediated control of splicing. The aptamer is inserted close to the 5′SS, the addition of tc facilitates the formation of a tc-aptamer complex which interferes with splicing. (C) The predicted secondary structure of the tc-aptamer is supported by structural probing (25). Important elements are indicated as stem = pedestal (P), bulge (B) and loop (L). The stem-loop 2 region of the minimer, where a GAAA tetraloop replaces nucleotides 19–36 of the aptamer, is shown top right.
MATERIAL AND METHODS
Plasmid constructions
We used the yeast 2 µ plasmid pWHE601 to constitutively express the gfp gene from an adh promoter (14). The actin- and the U3-intron were PCR amplified and inserted into a NheI restriction site directly downstream of the start codon. The resulting vectors were named pWH601_A and _U, respectively. To allow insertion of aptamer sequences at different intron positions, unique restriction sites for Acc65I and Bsu36I were introduced by PCR mutagenesis, thereby deleting the first NheI restriction site. Figure 1A schematically displays the positions of the respective restriction sites. For aptamer insertion, vectors were digested either with AflII/Acc65I or Bsu36I/ClaI. Double-aptamer constructs were generated by inserting the complete minimer-containing actin-intron either into the NheI restriction site or next to nucleotide 142 into either pWHE601AN32 (14) or pWHE601_A e min via PCR mutagenesis. Primer and vector sequences are available upon request.
GFP measurements
For all measurements, Saccharomyces cerevisiae strain RS453α was transformed according to the protocol supplied with the frozen EASY yeast transformation II kit (Zymo Research, Orange, CA, USA). Yeast cells transformed with the respective constructs were grown at 28°C for 48 h in minimal medium [0.2% (w/v) yeast nitrogen base, 0.55% ammonium sulfate, 2% (w/v) glucose, 12 µg/ml adenine, MEM amino acids, Gibco BRL] in the absence or presence of 250 µM tc in a final volume of 5 ml. Cells were harvested by centrifugation and re-suspended in 2 ml phosphate-buffered saline (PBS). For each construct, three independently grown cultures were analyzed. Fluorescence measurements were carried out at 25°C on a Fluorolog FL3-22 (SPEX. Industries Inc.) with the excitation wavelength set to 484 nm and an emission wavelength of 510 nm. Additionally, we determined the optical density (oD) at 600 nm to ensure homogeneous cell growth. The vector pVTU102 without gfp gene was analyzed in parallel as a blank and its value subtracted from all data. All measurements were repeated at least twice.
RT–PCR
Yeast cells transformed with the respective constructs were grown overnight at 28°C in minimal medium in the absence or presence of 250 µM tc. Total RNA was isolated according to the protocol supplied with the NucleoSpin RNA II kit (Macherey-Nagel, Düren, Germany). RT–PCR was performed according to manufacturer′s protocol using the SuperScript ™ III One-Step RT–PCR kit (Invitrogen, Karlsruhe, Germany) and analyzed on 3% agarose gels. gfp-cDNA was specifically amplified using primers adh_in (CAACTCCAAGCTAGATCTC) and GFP_rev_Julia (CCACTGACAGAAAATTTGTGC). ura3-cDNA, which is encoded as auxotrophy marker on all plasmids, was used as internal control and amplified using primers URA3_in (CAGCCTGCTTTTCTGTAACG) and URA3_out (GGAAGAGATGAAGGTTACG).
In-line probing
An RNA construct used for in-line probing was transcribed in vitro from PCR-amplified DNA, dephosphorylated, and 5′-32P-labeled as previously described (16). In-line probing was performed as described (17). In detail, 5′-32P-labeled RNA (100 pM) was incubated for 70 h at 25°C in 50 mM Tris–HCl pH 8.3, 20 mM MgCl2 and 100 mM KCl containing a tc concentration defined for each reaction. After incubation, 10 µl of 10 M urea was added and the products were separated using denaturating 10% polyacrylamide gel electrophoresis. Gels were dried and analyzed using a Storm Phosphoimager (GE Healthcare). The concentration of ligand needed to cause half-maximal modulation of spontaneous cleavage yields the KD for the RNA-ligand interaction.
RESULTS
Tc-aptamer mediated splice regulation is dependent on position and stability of the aptamer
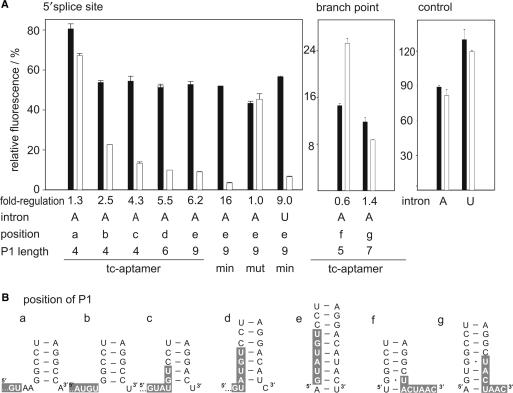
The introns of the actin (A) and U3 (U) genes from yeast were inserted directly downstream of the start codon of a constitutively expressed gfp gene resulting in the plasmids pWH601_A and _U, respectively (Figure 1A). GFP fluorescence only occurs when the mRNA has been correctly spliced. Unspliced mRNA should not be exported. If it does escape from the nucleus, several premature stop codons present in the intron sequence would either lead to rapid mRNA degradation via the nonsense-mediated decay pathway or to translation of a truncated, non-functional protein. We then introduced a set of unique restriction sites into the introns (Figure 1A) to allow easy insertion of the tc-aptamer next to the 5′ SS and the branch point (BP). Furthermore, we varied the distance between the aptamer and the 5′SS (shown in Figure 2 as positions a–e) or to the BP (positions f, g). Inhibition of splicing by the tc-aptamer complex can be monitored by a decrease in GFP fluorescence. Yeast cells were transformed with the respective constructs and the expression of GFP was measured in the presence and absence of 250 µM tc (Table 1 and Figure 2). The aptamer responds in a dose-dependent manner to tc with maximum repression level of 250 µM (14). Insertion of the tc-aptamer two nucleotides downstream of the 5′ SS (position a) leads to a negligible 1.3-fold decrease in GFP expression. Shifting the aptamer closer towards the 5′SS increases regulation to 2.5- and 4.3-fold for positions b and c, respectively.
Figure 2.
Dependence of GFP expression on tc-aptamer position and stability. (A) Constructs with the aptamer or the minimer variant (min) inserted near the 5′SS or the BP of the actin (A) or U3 (U) intron or without aptamer. The plots display the relative GFP activity measured in the absence (closed) and presence (open bars) of 250 µM tc with the activity of the construct without aptamer and without the intron, respectively, set to 100%. The regulation efficiency is given as factor determined as the ratio without and with tc. The mutant aptamer A9U which is unable to bind tc is denoted with mut. (B) Schematic view of the sequence environment of the aptamer inserted at several positions, the 5′SS and the BP, respectively, are shown with white bold letters highlighted with a gray box.
Table 1.
Regulatory properties of aptamer insertion
| Construct | GFP fluorescence − tc | GFP fluorescence + 250 µM tc | Regulatory factor | ||
|---|---|---|---|---|---|
| × 103 c.p.s.a | SD/% | × 103 c.p.s.a | SD/% | ||
| pWHE601 | 1780 | ±7.4 | 1830 | ±2.7 | 1.0 |
| pWHE601_A | 1590 | ±1.3 | 1500 | ±6.4 | 1.1 |
| pWHE601_A a | 1280 | ±3.1 | 1010 | ±1.6 | 1.3 |
| pWHE601_A b | 852 | ±1.7 | 339 | ±0.1 | 2.5 |
| pWHE601_A c | 863 | ±4.5 | 199 | ±5.5 | 4.3 |
| pWHE601_A d | 814 | ±3.1 | 149 | ±5.0 | 5.5 |
| pWHE601_A e | 836 | ±2.9 | 135 | ±1.7 | 6.2 |
| pWHE601_A e min | 822 | ±0.2 | 53 | ±8.7 | 15.7 |
| pWHE601_A e min (UUCG) | 751 | ±3.7 | 62 | ±4.5 | 12.2 |
| pWHE601_A e mut | 687 | ±2.3 | 679 | ±6.1 | 1.0 |
| pWHE601_U | 2310 | ±6.8 | 2200 | ±0.9 | 1.1 |
| pWHE601_U e min | 1310 | ±0.7 | 145 | ±4.2 | 9.0 |
| pWHE601_A f | 231 | ±2.6 | 379 | ±3.3 | 0.6 |
| pWHE601_A g | 187 | ±6.0 | 131 | ±0.2 | 1.4 |
| pWHE601 D1 | 41 | ±6.0 | 4 | ±5.0 | 11.4 |
| pWHE601 D2 | 89 | ±0.8 | 3 | ±4.3 | 28.6 |
| pWHE601 D3 | 134 | ±8.4 | 7 | ±4.1 | 19.7 |
| pWHE601 D4 | 235 | ±0.9 | 7 | ±2.5 | 32.5 |
aGFP fluorescence values are expressed as counts per second (c.p.s.) minus background levels (8000 c.p.s. -tc, 16 000 c.p.s. +250 µM tc).
Next we attempted to increase the efficiency of regulation by varying aptamer stability as it had already been shown for translational regulation (14). Therefore, we constructed P1 stems with six and nine base pairs. This was accompanied by further decreasing the distance between aptamer and 5′SS to include the complete 5′consensus sequence into P1 (Figure 2, positions d and e). Both constructs increase regulation to 5.5- and 6.2-fold, respectively. To further analyze the effect of stability on regulation, we replaced the aptamer by a minimer variant in which its complete head region is replaced by a GAAA tetraloop [Figure 1C (18)]. The minimer exhibits the same KD as the full-length aptamer and is proposed to adopt a more compact and stable conformation (18) and indeed, regulation was enhanced to 16-fold. Aptamer insertion close to the BP leads to negligible regulation, but caused a drastic decrease in fluorescence, already in the absence of tc.
Regulation was also detected with a minimer inserted into the U3 intron at the 5′SS. We observed only slightly changed regulatory properties, the aptamer sequence itself exerted less influence on basal expression of GFP in the absence of tc, however, regulation was also reduced slightly. This indicates that the aptamer-mediated regulation is universal and is not dependent on a specific intron. Control constructs either lacking an aptamer or containing an aptamer mutant A9U unable to bind tc show no regulation at all [Figure 2A: mut, (18)]. These findings indicate that the regulatory effect we observe is indeed tc-aptamer mediated.
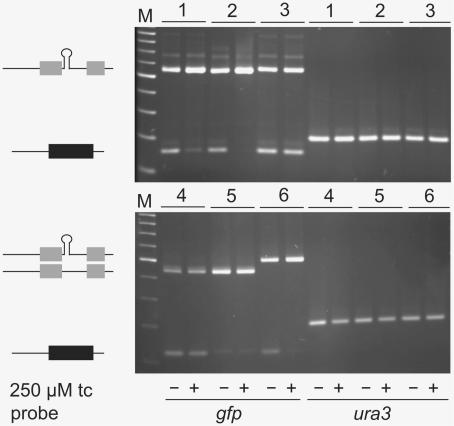
To verify that the tc-mediated decrease in fluorescence is due to altered pre-mRNA splicing we performed RT–PCR. Figure 3 shows that the amount of spliced gfp mRNA decreases in the presence of tc [exemplarily shown for an aptamer at position a (6), position c (1) and for the minimer at position e (2)], whereas control constructs show the same ratio of spliced and unspliced mRNA in the absence and presence of tc [pWH601_A without inserted aptamer (4) or with the mutant aptamer A9U (3)]. A mutation of the consensus sequence CCAUGU leads to tc-independent inhibition of splicing. In addition, ura3 mRNA as internal control shows no influence of tc on its expression for all constructs tested (Figure 3, six lanes at the right side of both blots).
Figure 3.
RT–PCR proves influence of tc on splicing. Tc-aptamer at position a (6) and c (1), the minimer (2) and the mutant variant A9U (3) at position e grown in the absence (–) and presence (+) of tc. As controls the actin–intron without the aptamer (4) or with a mutated 5′SS (5) were analyzed. Total RNA was isolated from yeast transformed with the respective constructs, RNA of gfp and ura3 (internal control) was reverse transcribed, amplified via PCR and analyzed on a 3% agarose gel. M = DNA size marker (see Material and Methods for experimental details).
Taken together, our data demonstrate that the tc-aptamer is able to regulate pre-mRNA splicing when inserted close to the 5′SS. This expands the application range of the aptamer as artificial tc-dependent riboswitch because it not only interferes with translation initiation when inserted into the 5′UTR but is also able to target mRNA processing.
Multiple aptamer insertion increases tc-mediated regulation
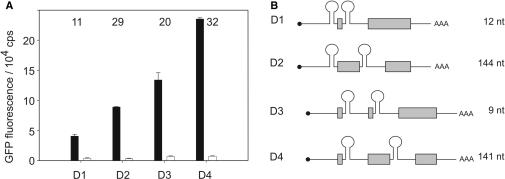
Previous studies showed that riboswitch activity of the tc-aptamer can inhibit different steps of translation initiation. It interferes with initial binding of the 43S subunit when located close to the cap structure but also inhibits 80S formation when inserted at a more downstream position, probably by impeding the scanning ribosome (15). Thereby, a 6-fold regulation was obtained when inserting the aptamer directly adjacent to the start codon. We speculated that combining translation with splicing control might further increase regulation. We used the intron construct with the highest regulation factor (minimer at position e with the 9 nt long stem) and combined it with an aptamer inserted directly in front of the start codon resulting in D1 (Figure 4). The construct shows a slightly reduced regulation compared to the single intron construct, however the two aptamers are separated by only 12 nt so that an interaction cannot be excluded. Therefore, we constructed a gfp gene with an intron at a position further downstream (at amino acid 47) resulting in D2. Both double constructs result in a remarkable increase in regulation (11- and 29-fold, respectively), however, regulation is accompanied by a more pronounced drop in basal activity.
Figure 4.
Double-intron constructs enhance regulatory efficiency. (A) Relative GFP activity measured in the absence (closed) and presence (open bars) of 250 µM tc. Regulation efficiency determined as the ratio of fluorescence without and with tc is given by the numbers above the bars within the blot. (B) Schematic view of the various constructs. The mRNA is shown as black line, the cap structure as black dot and the exons of the gfp open reading frame as gray boxes. The distance between two aptamers is given in nt.
We then inserted a second aptamer-containing intron at distances of either 9 or 141 nt downstream of the 3′SS of the first intron resulting in the double-intron constructs D3 and D4 (Figure 4). Both constructs lead to an increase in regulation with factors of 20- and 32-fold. Interestingly, the construct with the widely separated introns shows a less decrease in overall activity. Taken together, construct D4 containing two independently regulated introns has a regulatory efficiency of 32-fold which is the highest reported so far for aptamer-mediated conditional regulation of gene expression.
Structural probing indicates ligand-dependent changes in the accessibility of the 5′ SS
Previous studies indicated that the tc-aptamer has a pre-formed structure with the two single-stranded regions (B1-2 and L3) responsible for ligand-binding and the stem structures P1 and P2 forming the scaffold of the aptamer and proposed to be already present in the absence of tc. Since the 5′SS has to be accessible for efficient splicing, it was astonishing that the construct with the SS completely buried within P1 is still active. To analyze tc-mediated changes in the aptamer structure with a special focus on P1, we performed in-line probing of the minimer within the mRNA context. This method allows analysis of changes in the pattern of spontaneous RNA cleavage that occur upon ligand association (19). The aptamer exhibits substantial changes in spontaneous cleavage at several locations in response to increasing tc concentrations (Figure 5). These tc-induced structural modulations mainly occur in regions proposed to be involved in ligand-binding (B1-2 and L3) and fit the probing data of the aptamer in the context of the 5′UTR (20). We quantified the level of spontaneous cleavage at five positions (Figure 5, blue arrowheads) and used it to estimate the apparent dissociation constant (KD) for tc-binding (Figure 5B). The KD value of 800 pM determined independently at all five positions is in agreement with the value obtained by fluorescence titration spectroscopy and isothermal calorimetry [(18) and unpublished data] and represents one of the tightest binding of a small molecule by an aptamer.
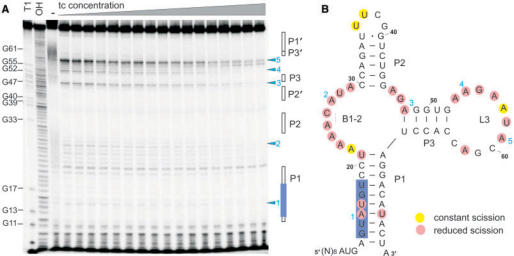
Figure 5.
Structural probing of the minimer in the context of the intron (A) Representative in-line probing data for the minimer. 5′-32P labelled RNA was examined after partial digestion with RNase T1 (T1) or alkali (OH), or after incubation for 70 h with increasing concentration of tc (lane 3–20 correspond to 0, 0.01, 0.06, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1, 1.5, 2, 3, 6, 10, 100, 1000 nM). Selected bands in the T1 lane (G-specific cleavage) are identified by nucleotide position, altered major sites are marked by blue arrowheads and denoted with 1–5, proposed stems are marked by open boxes, the 5′SS by a blue box. (B) Secondary structure model and tc-induced structural modulation of the tc-aptamer. Structural modulated sites (red-encircled nucleotides) were identified by monitoring spontaneous cleavage; the 5′SS is marked by a blue box.
In addition to the expected signals at the tc-binding sites within B1-2 and L3 (18,20), spontaneous cleavage also occurs in the lower part of stem P1 which resembles the 5′ SS. The signal intensity is not as strong as in the region proposed to be single stranded and involved in ligand binding. However, clear spontaneous cleavage can be observed at positions A15 and U16 which belong to the conserved nucleotides of the 5′SS (GUAUGU). For both positions, a pronounced tc-dependent decrease is seen concomitant with occurring changes in the tc-binding pocket. This clearly indicates that stem P1 is not completely formed in the absence of tc, so that the 5′SS can yet be recognized. Addition of tc then leads to a tightening of the complete aptamer structure including P1 which then efficiently interferes with recognition.
Taken together, our current findings expand the scope of conditional gene control by artificial riboswitches by showing that regulation of pre-mRNA splicing is feasible and represents a highly efficient aptamer-based conditional gene regulation system.
DISCUSSION
Pre-mRNA splicing is recognized as a critical step in mRNA maturation. Disturbances in this highly regulated process can lead to various severe diseases. Therefore, several systems which aim to prevent missplicing have been developed which either target the protein components involved (21,22) or the corresponding pre-mRNA (23). One approach makes use of antisense oligonucleotides which hybridize to distinct regions of the pre-mRNA and restore correct splicing. Antisense oligonucleotides have been employed to alternate splicing patterns for β-thalassemia, Duchenne muscular dystrophy and cancer (24). However, susceptibility to nuclease digestion, off-target effects and delivery problems prompted the development of cis active switches controllable by non-toxic small molecules with good cell permeability.
We have shown in the current report that in vivo control of pre-mRNA splicing using a synthetic small molecule-dependent riboswitch is possible. Regulation is efficient when the regulatory element is inserted near the 5′SS. This is in agreement with recently discovered TPP riboswitches in the filamentous fungus Neurospora crassa involved in regulation of alternative splicing. All riboswitches identified in this organism are exclusively located within the intron close to the 5′SS (11).
Insertion of the tc-aptamer close to the BP did not result in remarkable regulation. No aptamer was inserted between BP and 3′SS since the aptamer would introduce several alternative 3′SS. In addition, it has been shown that a 66 nt insertion at this position leads to a >70% drop in splicing efficiency of the actin intron (25). Gaur and co-workers (26) targeted this region by inserting the theophylline aptamer previously shown to be active as artificial riboswitch for translational regulation (27). In their report, the 3′SS is located within the theophylline aptamer sequence and a theophylline-dependent regulation of pre-mRNA splicing is shown using an in vitro splicing assay. However a transfer to any given mRNA would be difficult since parts of the theophylline aptamer are located within the coding region of the 3′exon.
The influence on splicing is position-dependent. Best regulation is obtained when the 5′SS is completely buried within the aptamer stem. This is surprising since previous data indicated that the P1 stem of the aptamer is already formed in the absence of tc (20). However, in-line probing revealed that P1 is not completely structured without ligand and tc-binding forces tightening of the stem. Thereby, the 5′SS is masked for recognition by U1 snRNP-binding which is discussed as the prime step of spliceosome formation. Further on, our data demonstrate that the efficiency of splice regulation is dependent on aptamer stability with stable constructs showing higher regulatory efficiency. In contrast to previous findings (28), in which gradually stabilized secondary structures next to the 5′SS lead to more pronounced splicing inhibition, stabilization of the tc-aptamer has no influence on splicing efficiency in the absence of ligand. First, upon tc addition the aptamer stabilization results in a marked decrease in splicing efficiency. A remarkable enhancement of regulation was achieved by combining more then one regulatable aptamer leading to the highest reported efficiency for aptamer-mediated gene regulation so far.
In comparison, the tc-regulated activator/repressor-mediated gene expression system in yeast shows an at least 10-fold higher dynamic range of the protein-based system (29). The disadvantage, however, of these systems is their complexity since they depend on the heterologous expression of the respective activator and repressor proteins and the manipulation of the target promoter. A further limitation of the system is that in many cases tight regulation is not possible due to leaky expression caused by not always predictable enhancer elements (30). Therefore, an individual choice between the two systems highly depends on the requirements of the specific experiment.
Taken together, our data demonstrate that the artificial tc-riboswitch is not only capable of regulating translation initiation, but can also modulate gene expression in eukaryotic organisms by facilitating ligand-dependent splicing of pre-mRNA. We therefore provide a valuable tool for alternative splicing which is a highly important step in regulation of gene expression. This work further highlights the importance of RNA structures and their capacity to influence alternative splicing. It underlines the need for simple model systems to study how secondary structures affect SS choice and opens up a wide field of applications for conditional gene expression systems based on regulatory active RNAs.
ACKNOWLEDGEMENTS
The studies were carried out in the laboratories of Wolfgang Hillen and Ronald R. Breaker, whose support is greatly appreciated. We thank Reinhard Lührmann for plasmids containing the yeast introns and Christian Berens and Andreas Wachter for discussions and critically reading the manuscript. We are grateful to the Volkswagenstiftung (I/79 950) and the Deutsche Forschungsgemeinschaft (SU 402/1-1) for financial support. Funding to pay the Open Access publication charges for this article was provided by Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
REFERENCES
- 1.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 2.Thanaraj TA, Stamm S, Clark F, Riethoven JJ, Le Texier V, Muilu J. ASD: the Alternative Splicing Database. Nucleic Acids Res. 2004;32:64–69. doi: 10.1093/nar/gkh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 4.Venables JP. Unbalanced alternative splicing and its significance in cancer. Bioessays. 2006;28:378–386. doi: 10.1002/bies.20390. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat. Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 6.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 7.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubodera T, Watanabe M, Yoshiuchi K, Yamashita N, Nishimura A, Nakai S, Gomi K, Hanamoto H. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003;555:516–520. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- 10.Borsuk P, Przykorska A, Blachnio K, Koper M, Pawlowicz JM, Pekala M, Weglenski P. L-arginine influences the structure and function of arginase mRNA in Aspergillus nidulans. Biol. Chem. 2007;388:135–144. doi: 10.1515/BC.2007.015. [DOI] [PubMed] [Google Scholar]
- 11.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 12.Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 13.Werstuck G, Green MR. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 14.Suess B, Hanson S, Berens C, Fink B, Schroeder R, Hillen W. Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucleic Acids Res. 2003;31:1853–1858. doi: 10.1093/nar/gkg285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson S, Berthelot K, Fink B, McCarthy JE, Suess B. Tetracycline-aptamer-mediated translational regulation in yeast. Mol. Microbiol. 2003;49:1627–1637. doi: 10.1046/j.1365-2958.2003.03656.x. [DOI] [PubMed] [Google Scholar]
- 16.Seetharaman S, Zivarts M, Sudarsan N, Breaker RR. Immobilized RNA switches for the analysis of complex chemical and biological mixtures. Nat. Biotechnol. 2001;19:336–341. doi: 10.1038/86723. [DOI] [PubMed] [Google Scholar]
- 17.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 18.Muller M, Weigand JE, Weichenrieder O, Suess B. Thermodynamic characterization of an engineered tetracycline-binding riboswitch. Nucleic Acids Res. 2006;34:2607–2617. doi: 10.1093/nar/gkl347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soukup GA, Breaker RR. Engineering precision RNA molecular switches. Proc. Natl Acad. Sci. USA. 1999;96:3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanson S, Bauer G, Fink B, Suess B. Molecular analysis of a synthetic tetracycline-binding riboswitch. RNA. 2005;11:503–511. doi: 10.1261/rna.7251305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagiwara M. Alternative splicing: a new drug target of the post-genome era. Biochim. Biophys. Acta. 2005;1754:324–331. doi: 10.1016/j.bbapap.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Soret J, Gabut M, Tazi J. SR proteins as potential targets for therapy. Prog. Mol. Subcell. Biol. 2006;44:65–87. doi: 10.1007/978-3-540-34449-0_4. [DOI] [PubMed] [Google Scholar]
- 23.Dominski Z, Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl Acad. Sci. USA. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilton SD, Fletcher S. RNA splicing manipulation: strategies to modify gene expression for a variety of therapeutic outcomes. Curr. Gene. Ther. 2005;5:467–483. doi: 10.2174/156652305774329249. [DOI] [PubMed] [Google Scholar]
- 25.Fouser LA, Friesen JD. Effects on mRNA splicing of mutations in the 3′ region of the Saccharomyces cerevisiae actin intron. Mol. Cell. Biol. 1987;7:225–230. doi: 10.1128/mcb.7.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim DS, Gusti V, Pillai SG, Gaur RK. An artificial riboswitch for controlling pre-mRNA splicing. RNA. 2005;11:1667–1677. doi: 10.1261/rna.2162205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harvey I, Garneau P, Pelletier J. Inhibition of translation by RNA-small molecule interactions. RNA. 2002;8:452–463. doi: 10.1017/s135583820202633x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goguel V, Wang Y, Rosbash M. Short artificial hairpins sequester splicing signals and inhibit yeast pre-mRNA splicing. Mol. Cell. Biol. 1993;13:6841–6848. doi: 10.1128/mcb.13.11.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belli G, Gari E, Pietrafita L, Aldea M, Herrero E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998;26:942–947. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogt K, Bhabhra R, Rhodes JC, Askew DS. Doxycycline-regulated gene expression in the opportunistic fungal pathogen. Aspergillus fumigatus. BMC Microbiol. 2005;5:1–11. doi: 10.1186/1471-2180-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]