Abstract
The eukaryotic mismatch repair protein Msh6 shares five domains in common with other MutS members. However, it also contains several hundred additional residues at its N-terminus. A few of these residues bind to PCNA, but the functions of the other amino acids in the N-terminal region (NTR) are unknown. Here we demonstrate that the Msh6 NTR binds to duplex DNA in a salt-sensitive, mismatch-independent manner. Partial proteolysis, DNA affinity chromatography and mass spectrometry identified a fragment comprised of residues 228–299 of yeast Msh6 that binds to DNA and is rich in positively charged residues. Deleting these residues, or replacing lysines and arginines with glutamate, reduces DNA binding in vitro and elevates spontaneous mutation rates and resistance to MNNG treatment in vivo. Similar in vivo defects are conferred by alanine substitutions in a highly conserved motif in the NTR that immediately precedes domain I of MutS proteins, the domain that interacts with mismatched DNA. These data suggest that, in addition to PCNA binding, DNA binding and possibly other functions in the amino terminal region of Msh6 are important for eukaryotic DNA mismatch repair and cellular response to alkylation damage.
INTRODUCTION
In eukaryotes, repair of mismatches generated during replication is initiated when a complex of either Msh2 and Msh3 or Msh2 and Msh6 binds to mismatched DNA. These MutS proteins also have other functions in cells (1–4), one of which is binding to damaged DNA to initiate events that result in damage-induced cytotoxicity (5). Msh2, Msh3 and Msh6 share five conserved protein domains (designated I–V) in common with bacterial MutS. The crystal structures of these five domains in Taq and Escherichia coli MutS are known (6,7), and their roles in binding to mismatched DNA, ADP/ATP binding and hydrolysis, heterodimerization, conformational changes and partnerships with proteins downstream in the mismatch repair pathway have been extensively studied and are partially understood (1–4). However, unlike bacterial MutS or Msh2, Msh6 and Msh3 have an additional evolutionarily conserved region preceding domain I comprised of from ∼100 to more than 600 amino acids, depending on the organism. These N-Terminal Regions (NTRs) of Msh3 and Msh6 contain short, conserved PIP (PCNA interacting protein) boxes near the N-terminus that interact with PCNA (8–10), the sliding clamp that participates in both DNA replication and DNA mismatch repair. Non-conservative amino acid replacement of residues in these PIP boxes partially reduces Msh2-Msh3-dependent and Msh2-Msh6-dependent mismatch repair in yeast (8,9), and deletion of the PIP box in human Msh6 partially inactivates mismatch repair in vitro (10).
In addition to residues important for binding to PCNA, other residues in the NTR, diagramed in Figure 1, could also be functionally important. This importance is suggested by the evolutionary conservation of NTRs in both Msh6 and Msh3 (albeit of different lengths and sequence), by the identification (11) in human Msh6 NTR of a PWWP domain characteristic of proteins associated with chromatin, and by the presence in the human Msh6 NTR of missense mutations that are associated with cancer (12–14). In the present study, we examine the possibility that residues in the Msh6 NTR other than those in the PIP box are functionally important. We demonstrate that recombinant yeast and human Msh6 NTRs bind to duplex DNA, identify amino acids in yeast Msh6 that contribute to this binding, and characterize msh6 mutants that concomitantly reduce DNA binding in vitro and reduce Msh6-dependent mismatch repair and sensitivity to killing by MNNG in vivo. We also show that substituting alanine for residues in a previously unrecognized, highly conserved motif at the extreme C-terminus of the Msh6 NTR also reduce Msh6-dependent mismatch repair and sensitivity to MNNG treatment. These results suggest that the Msh6 NTR has multiple roles in Msh6-dependent mismatch repair and in cellular response to alkylation damage.
Figure 1.
Residues of interest in the yeast and human Msh6 NTRs. The N-terminal 310 amino acids from S. cerevisiae are shown. Acidic residues are colored red and basic residues are colored blue. The green box highlights the PIP box, the red box highlights the region of acidic residues from 144 to 212 (a putative ‘DNA mimic’), the blue box highlights the DNA binding fragment identified by mass spectrometry and the black box outlines residues that are highly conserved in Msh6 homologs. See text for further descriptions.
EXPERIMENTAL PROCEDURES
Expression and purification of yeast and human Msh6 NTRs
The coding sequences of the NTRs of yeast and human Msh6 were amplified from plasmids pRDK439 (gift from Richard Kolodner) and pAC61.2 (15) using primers to create restriction sites at the 5′ and 3′ ends. The amplified fragments were digested and ligated into pET28a(+). The yMsh6 and human proteins contained a TAG stop codon immediately after codon 299 and 394, respectively (see description in Results). The yeast DNA fragment was ligated into an NdeI and HindIII digested vector placing a His tag at the N-terminus of the protein. The human Msh6 N-terminal fragment was ligated into the NcoI and XhoI sites of pET28a(+) to place the His tag at the C-terminus of the protein. The yeast and human NTRs were expressed in E. coli upon IPTG induction of a log phase culture followed by a 3-h incubation. Cells were collected from 12-l cultures, washed with 20 mM Tris pH 8, centrifuged, frozen in liquid nitrogen and stored at −80°C. Frozen cell pellets were thawed and re-suspended in an equal volume (milliliter) of 20 mM Tris pH 8, 150 mM NaCl, 1 mM β-mercaptoethanol (Ni-A buffer) per weight (gram) of pellet, and lysed either by sonication or with a French press. The lysate was applied to a 4-ml nickel NTA (Qiagen) column, washed with 40 ml of NiA buffer + 5 mM imidazole, 40 ml of NiA buffer + 30 mM imidazole and eluted with 25 ml of NiA buffer + 100 mM imidazole. The eluted fraction was diluted 2-fold with 20 mM Tris pH 8, 1 mM EDTA, 1 mM β-mercaptoethanol, 10% glycerol (HepQ-A buffer) and loaded onto a 5-ml HiTrap heparin column (GE). The NTR proteins were eluted by a 100 to 700 mM NaCl gradient over 150 ml. The yeast protein eluted near 400 mM NaCl and the human NTR eluted near 600 mM. The proteins were diluted to 100 mM NaCl with HepQ-A buffer, loaded onto a 1-ml HiTrap Q column (GE), and eluted with a 20-ml gradient of NaCl from 100 mM to 1 M. Both yeast and human proteins elute at about 400 mM NaCl concentration. The eluted proteins were dialyzed against 20 mM Tris pH 8, 150 mM NaCl, 1 mM DTT, 5% glycerol, frozen in liquid nitrogen and stored at −80°C.
DNA cellulose affinity chromatography
Fifty micrograms of yeast or human NTR was diluted to 1 ml with 20 mM Tris pH 8, 15 mM NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, 10% glycerol (column buffer) and applied to a 0.1-ml volume of dsDNA cellulose (Sigma) packed into Poly-Prep chromatography columns (Bio-Rad). Columns were washed with 1 ml of column buffer and step eluted with 0.2 ml of column buffer with NaCl concentrations from 25 to 300 mM.
DNA binding assays
DNA filter binding assays were performed as previously described (16). DNA electrophorectic mobility shift assays were performed as previously described (15).
Identification of DNA binding region in the yMsh6 NTR
The yeast NTR was diluted to 50 µg/ml in 1 ml of 20 mM Tris pH 8, 100 mM NaCl and 1 mM β-mercaptoethanol and digested with 200 µl of 1 µg/ml chymotrypsin for 15 min at 25°C. This condition was selected because it produced a partial proteolysis of the protein resulting in a wide range of fragments. After digestion, a 100 µl aliquot was removed and the remainder of the reaction was diluted 10-fold with 20 mM Tris pH 8, 1 mM EDTA, 1 mM β-mercaptoethanol and 10% glycerol and loaded onto a 0.1-ml dsDNA cellulose column. After loading, the column was washed with loading buffer containing 10 mM NaCl and step eluted with 0.2 ml of loading buffer containing a range of NaCl concentrations from 25mM to 300 mM. A 25 µl sample from each elution was loaded on a 4–12% SDS polyacrylamide gel and fragments were resolved by electrophoresis. Two bands from the 200 mM NaCl elution were excised from the gel, cut into small pieces, and transferred into a 96-well microtiter plate. Gel pieces were subjected to automatic tryptic digestion using an Investigator™ Progest protein digestion station (Genomic Solutions, Ann Arbor, MI) as previously described (17). Briefly, gels were sequentially washed twice with 25 mM ammonium bicarbonate buffer (pH 7) and acetonitrile, dehydrated, rehydrated with 25 ml of the enzyme solution and digested at 37°C for 8 h. The enzyme solution used was sequencing grade, modified trypsin (Promega Corporation, Madison, WI) at a concentration of 0.01 mg/ml in 25 mM ammonium bicarbonate buffer (pH 7). Resulting tryptic peptides were extracted from the gel, lyophilized and stored at −80°C.
Mass spectrometry
Prior to matrix assisted laser desorption ionization mass spectrometry (MALDI/MS) analysis, the peptides were reconstituted in 10 ml of a 50:50 solution of acetonitrile:water (0.1% formic acid). MALDI analyses were performed on the digested fragments using a Voyager-DE STR (Applied Biosystems, Framingham, MA) delayed-extraction time-of-flight (TOF) mass spectrometer in the positive ion reflector and/or linear modes. The instrument is equipped with a nitrogen laser (337 nm) to desorb and ionize the samples. A close external calibration, using two points to bracket the mass range of interest, was used. A 0.5 µl aliquot of the tryptic peptide solution was spotted with 0.5 µl MALDI matrix on a stainless steel sample target and allowed to dry at room temperature. A saturated solution of α-cyano-4-hydroxycinnamic acid in 45:45:10 ethanol:water:formic acid (v/v) was used as the MALDI matrix. Spectra were obtained over the mass range of 800–4000 Da in the reflector mode and 1000–25 000 in the linear mode with 50–100 laser shots per spectrum. For the in-gel digest analyses, ions corresponding in mass to trypsin autolysis products were used to internally calibrate mass spectra when possible.
Construction of truncations, in-frame deletion and missense mutants of Msh6 NTR
Msh6 NTR truncations were made in the pET28a construct expressing the 299-amino acid yMsh6 NTR. Truncations were constructed by designing a phosphorylated primer that included a termination codon to anneal to vector sequence at the 3′ end of the truncation and paired with phosphorylated primers annealing to the site of the desired truncation. The phosphorylated primer p-TAG AAG CTT GCG GCC GCA CTC GAG CAC CAC CAC CAC was paired with p-TGA AGA ATG CGA AGT GTT GTA TGA AAA TTT TTT CTT AG for truncation N227 (after S227), p-TCT ACT TGG TGC CTG ATT TGG CCT GCT TTT CTT TTT TG for N251 (after R251), p-TTT GCT AGA CTT AGA AGT CGC TGA TGG TTG ACT ATG for N268 (after K268), p-ACG GCG CTG AGC ATC TCG TTC ATC CAC TAA CCA TTG for N289 (R289). pET-28a yMsh6NTR plasmid was amplified by Pfu turbo polymerase (Stratagene) in 20 cycles of a 3-step amplification reaction (95° × 30 s/55° × 1 min/68° × 10 min). After amplification, products were digested with DpnI at 37° for 1 h. Digested products were dialyzed against TE buffer 2x using a microcon (Amicon). During the final centrifugation, the sample was concentrated to a 10–20 µl volume. An aliquot was ligated and used to transform E. coli. Plasmid DNA was isolated and constructs were verified by DNA sequencing.
Msh6 internal in-frame deletions were constructed by designing phosphorylated primers to anneal at each end of the desired deletion. For the 3′ end of yMsh6 in-frame deletions, either p-ACA CTG TAC ATC CCA TCT TCT GCA TGG AAC AAG TTT ACT C for deletions including R299 or p-CCC AAG AGT GAT CCA GAG TAC GAT CCA AGA for deletions including R289 were paired with phosphorylated primers at the 5′ end. The 5′ primers included p-GGC CAT TTT GGA GTT CAA ATT GGC TTT GTC AAA ATT AA for deletions including P3, p-TGA AGA ATG CGA AGT GTT GTA TGA AAA TTT TTT CTT AG for deletions including S228, p-TCT ACT TGG TGC CTG ATT TGG CCT GCT TTT CTT TTT TG for deletions including S252, p-TTT GCT AGA CTT AGA AGT CGC TGA TGG TTG ACT ATG for deletions including F269, p-ACG GCG CTG AGC ATC TCG TTC ATC CAC TAA CCA TTG for deletions including P290. A centromeric plasmid (pRDK439) that expresses Msh6 from its natural promoter was amplified by Pfu turbo polymerase (Stratagene) using reaction conditions similar to the truncation reactions above but with longer extension times. Plasmid DNA was isolated from the transformants and msh6 internal in-frame deletions were confirmed by DNA sequencing.
Construction of the strain for measuring sensitivity to killing by MNNG
Sensitivity to MNNG was measured in AC711a (mgt1Δ rad52Δ msh6::kanMX) that was derived from E203 (18). MGT1 was deleted by delito perfetto. KlURA3 and hyg was amplified from pGSHU with MGT1 ends for recombination using amplification primers 5′yMGT1-P.IIS: ACAAAAAAAAAATTGAAAACGGTCGCATTTTTGATCTAAATGGACCAACG tagggataacagggtaatccgcgcgttggccgattcat and 3′yMGT1-P.I: ATACATAACTATTTCTTATGTTTATTTTCCTAAAATCCTTTATCCAACTAttcgtacgctgcaggtcgac. Capitalized letters represent MGT1 sequence. Italicized letters represent an SceI unique restriction endonuclease site. Lower case letters represent either hyg or KlURA3 sequence. E203 was transformed with amplified DNA and Ura+ and Hygr colonies were selected for transformation with IRO oligos. MGT1 IRO oligos were yMGT1-IRO-S: ACAAAAAAAAAATTGAAAACGGTCGCATTTTTGATCTAAATGGACCAACGTAGTTGGATA AAGGATTTTAGGAAAATAAACATAAGAAATAGTTATGTAT and yMGT1-IRO-A: ATACATAACTATTTCTTATGTTTATTTTCCTAAAATCCTTTATCCAACTACGTTGGTCCATT TAGATCAAAAATGCGACCGTTTTCAATTTTTTTTTTGT. FOAr and Hygs IRO transformed colonies were selected, screened for mgt1Δ by PCR and confirmed by sequencing (19). Colonies deleted for MGT1 were selected for deleting RAD52 by transformation with SalI digested pΔRAD52blast and selecting on plates lacking uracil (20). Ura+ rad52Δ transformants were screened for pΔRAD52blast containing sequences by PCR and sensitivity to MMS. Ura+ and MMSs transformants confirmed by PCR were patched onto FOA plates for deleting URA3 sequence. Deletions of RAD52 were confirmed by DNA sequencing. These rad52Δ strains contained a copy of hisG sequence from pΔRAD52blast.
Measuring mutation rates
Mutation rates and 95% confidence intervals were determined as described (21), using 9–16 individual cultures.
Sensitivity to killing by MNNG
The strain AC711a was transformed with wild-type and mutant Msh6 alleles to measure sensitivity to MNNG. Overnight cultures were grown at 30°C from single cell isolates. The cultures were diluted 10-fold and grown for three additional hours. Each culture was divided and either a 100x stock of MNNG in DMSO or DMSO alone was added. The cultures were incubated for an additional hour, washed with water, diluted and plated to determine survival.
RESULTS
The Msh6 NTR binds to DNA
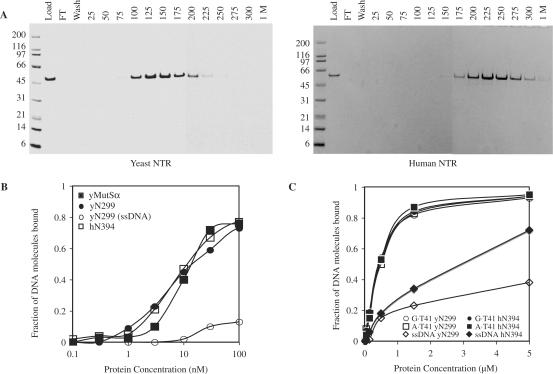
When the structure of Thermus aquaticus MutS protein was solved, a structure-based amino acid sequence alignment was provided [Figure 5 in (6)]. This alignment suggested that domain I of Msh6 may begin at approximately residue 300 in yeast Msh6 and at approximately residue 395 in human MSH6. On that basis, and absent structural information on Msh6 proteins per se, here we studied and refer to the preceding residues as Msh6 NTRs. As a first step towards determining if these Msh6 NTRs interact with macromolecules other than PCNA, we expressed and purified the yeast Msh6 NTR comprised of residues 1–299 and the human Msh6 NTR comprised of residues 1–394. Both proteins were expressed in E. coli with a 6-His tag. This tag was placed at the N-terminus of the yeast NTR, but at the C-terminus of the human NTR to avoid perturbing the PIP box that is at the extreme N-terminus. Both NTRs were purified using three chromatographic steps, one of which involved binding to a heparin column. Both NTRs were obtained in highly purified form (see lanes labeled ‘load’ in Figure 2A). Because heparin is a negatively charged resin to which many DNA binding proteins bind, we tested whether the Msh6 NTRs could bind to a dsDNA cellulose column. Indeed, the NTR of yeast Msh6 bound, and peak fractions eluted from the column at 125–150 mM NaCl (Figure 2A). The NTR of human Msh6 also bound, and the peak fraction eluted from the column at 225 mM NaCl (Figure 2A). Thus both proteins can bind to dsDNA via ionic interactions, and the NTR of human Msh6 appears to bind more tightly than the NTR of yeast Msh6. When DNA binding capacity was measured using a filter-binding assay (16), both the yeast and human NTRs bound to double-stranded plasmid DNA with an affinity similar to yMutSα (Figure 2B) The yMsh6 NTR also bound to single-stranded M13 DNA (open circles in Figure 2B), but with lower affinity. Using an electrophoretic mobility shift assay (EMSA), both yeast and human Msh6 NTRs were observed to bind similarly to homoduplex DNA and to heteroduplex DNA containing a G–T mismatch (Figure 2C).
Figure 5.
A conserved motif near the C-terminus of Msh6 NTRs. The Msh6 NTRs from a variety of eukaryote species were compared by Clustal alignment. A conserved 41 amino acid region at the C-terminus of the NTRs is shown. The numbering indicates the amino acid positions. The consensus sequence is shown below the alignment. Amino acids that are similar to the consensus are shaded and invariant residues are highlighted by bold type. Lower case (h) represents a hydrophobic residue, (+) and (−) represent positive and negative charged residues, respectively. Asterisks above the alignment indicate positions of the Tyr296, Asp297, Thr300 and Leu301 that were changed to alanine.
Figure 2.
DNA binding by yeast and human Msh6 NTRs. A. Binding of the yeast NTR (left panel) and human NTR (right panel) to dsDNA cellulose. Purified NTR protein (50 µg) was diluted in 1 ml of 20 mM Tris pH 8, 15 mM NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, 10% glycerol (column buffer) and applied to a 0.1-ml volume of dsDNA cellulose (Sigma) packed into Poly-Prep chromatography columns (Bio-Rad). Columns were washed with 1 ml of column buffer and step eluted with 0.2 ml of column buffer containing NaCl concentrations from 25 to 300 mM. B. Nitrocellulose filter binding assays were performed as described in Materials and Methods. Twenty-microliter reactions containing between 2 fmoles and 2 pmoles of purified yeast N-terminal 299 (filled circle) or human N-terminal 394 (open square) residues were incubated with 1 nmole of nucleotide of 3H labeled pGBT9. DNA bound to protein is retained during filtration through nitrocellulose membranes. For comparison, pGBT9 binding by identical concentrations of yMutSα is shown (filled square). Binding of yN299 to 1 nmole of 3H labeled M13mp2 phage DNA (open circle) shows reduced affinity for ssDNA. C. The binding of 0.3 to 100 pmol of either the yeast N-terminal 299 residues (open symbols) and human N-terminal 394 residues (closed symbols) to 0.1 pmole of heteroduplex (G·T41), homoduplex (A·T41) or ssDNA oligonucleotide substrates in 20 µl reactions was monitored by electrophoretic mobility shift assays as described in Materials and Methods. The fraction of substrate bound is the ratio of substrate migrating with a slower mobility compared with the total amount of substrate in the lane in comparison with a reference mock lane lacking NTR protein.
Identifying a polypeptide in the Msh6 NTR that binds DNA
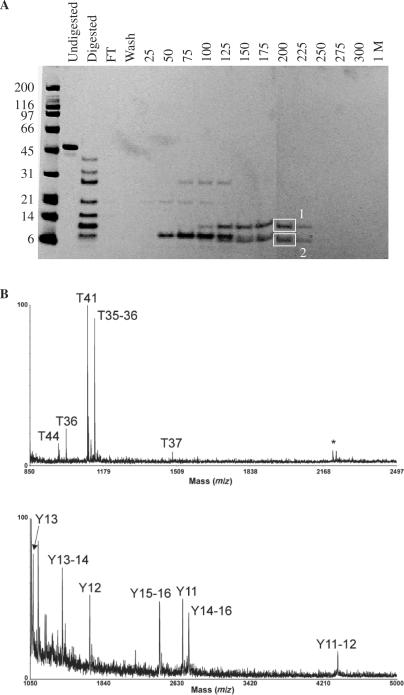
As a step towards determining if DNA binding by the NTR is important for Msh6 functions in vivo, we partially proteolyzed the yeast Msh6 NTR and tested the ability of the resulting polypeptide fragments to bind to the dsDNA cellulose column. Limited digestion with chymotrypsin generated a ladder of polypeptides (lanes labeled ‘Digested’ in Figure 3A), several of which bound to the dsDNA cellulose column and eluted at high NaCl concentrations (Figure 3A). To determine the identity of strongly bound chymotryptic fragments, two bands (boxed in Figure 3A) were excised from the SDS–PAGE gel, digested and analyzed by MALDI-TOF mass spectrometry. In the MALDI mass spectra of the tryptic digestion of band 1 (Figure 3B, upper panel), several ions corresponding in mass to predicted theoretical tryptic peptides of yeast Msh6 are observed (peptides labeled T44, T36, T41, T35–36 and T37). In addition, an autolysis product of trypsin (m/z 2211.11) was observed (labeled with an asterisk in Figure 3B). The corresponding residues for these peptides are amino acids 292–299, 243–251, 277–284, 242–251 and 252–265, respectively. Similar data were obtained for band 2 except that peptide T44 was not observed (data not shown). To establish the amino termini of the chymotryptic fragments eluted from the dsDNA column, the column eluents were also analyzed by MALDI/MS analysis in the positive ion linear mode (Figure 3B lower panel). Ions were observed corresponding in mass to theoretical chymotryptic peptides consisting of amino acids 270–277, 270–279, 254–269, 280–299, 231–253, 278–299 and 231–269. These ions are labeled as Y13, Y13–14, Y12, Y15–16, Y11, Y14–16 and Y11–12, respectively. Collectively, these data indicate that the chymotryptic fragments that bound strongly to the dsDNA cellulose column contain residues 231 through 299.
Figure 3.
Identifying the DNA binding region in the yeast Msh6 NTRs. A. Proteolysis–DNA cellulose chromatography. Proteolysis with chymotrypsin was performed as described in Materials and Methods, and DNA cellulose chromatography was performed as described in the legend to Figure 1. Boxes 1 and 2 designate peptide bands excised from gel for analysis by mass spectrometry. B. Upper panel, MALDI/mass spectrum for the in-gel tryptic digestion of the top band boxed in panel A. The ions corresponding in mass to theoretical tryptic peptides of yMSH6 are labeled as T44, T36, T41, T35–36 and T37. The ion labeled with an asterisk (*) corresponds in mass to an autolysis product of trypsin. Lower panel, MALDI mass spectrum of the 200-mM elution from the dsDNA column. The ions corresponding in mass to theoretical chymotryptic peptides of yMSH6 are labeled as Y13, Y13–14, Y12, Y15–16, Y11, Y14–16 and Y11–12.
DNA binding properties of deletion mutants of the yeast Msh6 NTR
Next, we expressed, purified and examined the DNA binding properties of yeast NTRs containing C-terminal truncations of residues in the DNA binding region of the yeast Msh6 NTR. Deletion of residues 290–299 did not significantly reduce DNA binding (Figure 4, closed boxes), while deletion of more residues progressively diminished DNA binding (269–299, closed triangles; 252–299, closed diamonds). Deletion of all residues of the DNA binding fragment (228–299, open circles) completely eliminated DNA binding (Figure 4).
Figure 4.
DNA binding by mutant yMsh6 NTRs. Twenty-microliter reactions containing between 2 fmol and 2 pmol of purified yeast Msh6 NTR, or truncated or mutant derivative NTRs, were mixed with 1 nmole (nucleotide) of 3H labeled pGBT9 in 20 µl reactions and filtered through nitrocellulose membranes. NTR proteins used included NTRs with C-terminal truncations. N299 represents the binding of pGBT9 by the N-terminal 299 amino acids, N289 represents binding by the N-terminal 289 residues, N268 the N-terminal 268 residues, etc. N299-R232E K271E R289E protein represents the N-terminal 299 amino acids with glutamate substitutions at Arg232, Lys271 and Arg289.
Reduced mismatch repair capacity of in-frame deletion mutants
Next, we tested whether the reduced DNA binding capacity of the mutant Msh6 NTR proteins correlated with reduced DNA mismatch repair activity in vivo, as measured by elevated spontaneous mutation rates in haploid yeast strains. We constructed Msh6 alleles with in-frame deletions and measured mutation rates for resistance to canavanine, which results from a wide variety of mutations that inactivate the CAN1 gene encoding arginine permease, and for lysine prototrophy resulting from deletion of a single base pair from a run of 14 A-T base pairs in the Lys2::A14 gene. An msh6 yeast strain lacking functional Msh6 has mutation rates at these two loci that are elevated by 10- and 400-fold, respectively (Table 1, line 2), in comparison with the rates in this same strain into which we introduced an ARS-CEN vector expressing Msh6 from its natural promoter (line 1). When we examined an msh6 mutant with an in-frame deletion of residues 3–289 of yeast Msh6, mutation rates remained as high (line 3) as for the complete absence of msh6 (line 2). This complete lack of complementation is consistent with one or more functions for the yeast NTR in addition to its interactions with PCNA. This interpretation is based on the fact that an msh6 derivative with alanine replacing two residues in the PIP box strongly reduced Msh2–Msh6 interactions with PCNA (8), but nonetheless exhibit partial complementation (Table 1, line 4 and (8)). Moreover, several different msh6 mutants with in-frame deletion of residues that reduce DNA binding (Figure 4) but do not change the PIP box all have mutation rates that are significantly elevated, and combining a PIP box mutation with an in-frame deletion results in a higher mutation rate (Table 1, last line) than observed in either single mutant alone. The correlations between loss of DNA binding in vitro (Figure 4) and elevated mutation rates in yeast (Table 1) are consistent with the interpretation that DNA binding by the yeast Msh6 NTR contributes to mismatch repair activity in vivo.
Table 1.
Complementation by msh6 alleles with in-frame deletions in the NTR
| MSH6 allele | Canavaniner | Lysine+ | ||||
|---|---|---|---|---|---|---|
| Mut. ratea (× 10−6) | Conf. limitb (× 10−6) | Relative rate | Mut. ratea (× 10−6) | Conf. limitb (× 10−6) | Relative rate | |
| MSH6 | 0.8 | 0.6–1.2 | 1 | 3 | 2–4 | 1 |
| none (empty vector) | 7.9 | 5.7–14 | 10 | 1200 | 930–2600 | 400 |
| msh6Δ3-289 | 10.3 | 7.5–20 | 13 | 1600 | 1280–2900 | 530 |
| msh6FF33,34AA | 1.9 | 0.9–3.1 | 2 | 20 | 14–37 | 7 |
| msh6Δ228-289 | 3.8 | 1.6–10 | 5 | 125 | 100–183 | 42 |
| msh6Δ269-299 | 2.0 | 0.9–3.4 | 3 | 190 | 130–240 | 60 |
| msh6Δ252-299 | 3.9 | 1.5–7.2 | 5 | 380 | 270–650 | 130 |
| msh6Δ228-299 | 4.1 | 1.3–4.4 | 5 | 480 | 290–574 | 160 |
| msh6Δ290-299 | 2.0 | 1.2–5.6 | 3 | 83 | 59–128 | 30 |
| msh6Δ228-289 + FF33,34AA | 6.8 | 5.8–10 | 8 | 950 | 700–1200 | 320 |
aMutation rates were determined using 9–12 individual cultures.
b95% confidence interval for mutation rate.
Reduced mismatch repair and reduced DNA binding capacity of Msh6 NTR missense mutants
As a further test of the importance to mismatch repair of DNA binding by the Msh6 NTR, we examined complementation of the msh6 mutant strain with full-length Msh6 containing negatively charged glutamate substituted for positively charged lysines and arginines within the DNA binding peptide. For this purpose, we measured mutation rates at the lys2::A14 locus, which gives a greater mutator response when mismatch repair is defective. Three individual single residue replacements (R232E, K271E, R289E) had no apparent effect on mutation rates (Table 2, lines 3–5). However, mutator effects of 2–5-fold were detected in mutants containing either two or three replacements (lines 6–8), mutator effects of 7–12-fold were detected in mutants containing four replacements (lines 9–11), and five replacements yields 16-fold increases in mutation rate at the lys2::A14 locus (lines 12 and13). We then expressed and purified one NTR derivative that contained glutamates replacing Arg232, Lys271 and Arg289. Compared with the normal yeast Msh6 NTR, the DNA binding affinity of this mutant protein was somewhat reduced (open squares in Figure 4). These observations further support the interpretation that DNA binding by the yeast Msh6 NTR contributes to mismatch repair activity.
Table 2.
Complementation by msh6 alleles with charge-reversal substitutions in the NTR
| msh6 allele | Lysine+ | ||
|---|---|---|---|
| Mut. ratea (× 10−6) | Conf. limitb (× 10−6) | Relative rate | |
| MSH6 | 3 | 2–4 | 1 |
| None (empty vector) | 1200 | 934–2600 | 400 |
| msh6R232E | 4 | 2–4 | 1 |
| msh6K271E | 4 | 2–7 | 1 |
| msh6R289E | 5 | 4–6 | 2 |
| msh6R232E R289E | 10 | 8–33 | 3 |
| msh6K271E R289E | 11 | 7–15 | 4 |
| msh6R232E K271E R289E | 15 | 12–21 | 5 |
| msh6R232E R236E K271E R289E | 22 | 17–31 | 7 |
| msh6R232E R268E K271E R289E | 24 | 16–29 | 8 |
| msh6R232E K271E R288E R289E | 36 | 20–122 | 12 |
| msh6R232E R236E K268E K271E R289E | 48 | 25–53 | 16 |
| msh6R232E R236E K271E R288E R289E | 49 | 31–59 | 16 |
aMutation rates were determined using 9–12 individual cultures.
b95% confidence interval for mutation rate.
Identification of a conserved motif near the C-terminus of the Msh6 NTR
The DNA binding polypeptides identified by mass spectrometry are in a region of Msh6 that is not present in bacterial MutS or eukaryotic Msh2 proteins. To determine if residues in this region are conserved in multiple Msh6 proteins, a sequence alignment was performed that excluded other MutS proteins and focused only on residues in Msh6, specifically those residues within the yeast Msh6 DNA binding fragment through amino acid Phe311. Phe311 was used as an endpoint for this alignment because Thr312 was previously aligned with the first residue in helix 1a of domain 1 of Thermus aquaticus MutS [Figure 5 in (6)]. This focused alignment reveals that the amino acid sequence in this region is conserved (Figure 5). Within a span of nine residues, two residues, corresponding to yMsh6 T300 and P304, are invariant and six others are highly conserved, yielding a consensus sequence of ‘YDPxTLYI/V/LP’. Additional residues proximal to these are also conserved, through an invariant residue corresponding to E275 in yMsh6.
In vivo consequences of altering residues in the conserved motif
Because this conservation suggests functional significance, we examined the in vivo consequences of substituting alanine for one, two or four of the conserved residues in this region. Each of these msh6 mutants was partially defective in complementing the mutator phenotype of an msh6Δ strain (Table 3). These partial defects were similar to that resulting from alanine substitutions in the PIP box (line 3). Moreover, combining the alanine substitutions in the PIP box with a quadruple YDTL-AAAA mutant resulted in a much stronger defect (line 9), approaching that of the msh6Δ mutant. These results are consistent with the interpretation that the conserved residues at the C-terminus of the yeast NTR contribute to mismatch repair function in a manner different from PCNA binding.
Table 3.
Complementation by msh6 alleles with amino acid substitutions in the conserved motif at the C-terminal end of the N-terminal region
| msh6 allele | Canavaniner | Lysine+ | ||||
|---|---|---|---|---|---|---|
| Mut. ratea (× 10−6) | Conf. limitb (× 10−6) | Relative rate | Mut. ratea (× 10−6) | Conf. limitb (× 10−6) | Relative rate | |
| MSH6 | 0.8 | 0.6–1.0 | 1 | 3 | 2–4 | 1 |
| None (empty vector) | 7.9 | 7.6–10 | 10 | 1200 | 1100–1500 | 400 |
| msh6F33A,F34A (PIP) | 1.9 | 0.9–3.1 | 2 | 20 | 14–37 | 7 |
| msh6Y296A | 1.8 | 0.9–3.7 | 2 | 6 | 3–8 | 2 |
| msh6T300A | 1.5 | 0.7–2.2 | 2 | 7 | 5–9 | 2 |
| msh6Y296A D297A | 2.5 | 2.1–3.3 | 3 | 78 | 60–111 | 25 |
| msh6T300A L301A | 2.4 | 2.3–2.7 | 3 | 66 | 27–80 | 20 |
| msh6Y296A,D297A,T300A,L301A | 1.8 | 1.2–3.4 | 2 | 120 | 97–140 | 40 |
| msh6Y296A,D297A,T300A,L301A + PIP | 8.5 | 6.2–15 | 9 | 950 | 680–1780 | 320 |
aMutation rates were determined using 9–12 individual cultures.
b95% confidence interval for mutation rate.
Increased resistance to MNNG of mutant derivatives of the yeast Msh6 NTR
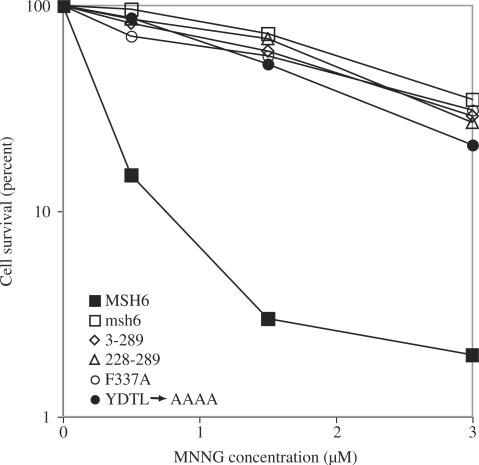
In addition to DNA mismatch repair, Msh6 also participates in cellular processes that determine sensitivity to killing upon exposure to agents that damage DNA (5), and reviewed in references 2–4. Yeast strains defective in Rad52-dependent homologous recombination are highly sensitive to killing by treatment with the methylating agent MNNG, and resistance to MNNG-induced killing is conferred by a loss of DNA mismatch repair (22). An mgt1Δ rad52Δ msh6::kanMX strain lacking Msh6 function is resistant to MNNG-induced killing (Figure 6) and complementation with Msh6 transforms it into an MNNG-sensitive strain. A similar degree of resistance was observed with msh6 mutants that lack residues 3–289 or residues 228–289 and with msh6 mutants that have alanine substituted for Phe337 in domain I that binds to the mismatched base, or for conserved residues in the consensus motif at the C-terminus of the NTR (Figure 6). Thus, in addition to contributing to DNA mismatch repair, NTR residues involved in DNA binding and residues in the conserved motif near the C-terminus of the NTR all contribute to cellular response to MNNG treatment in vivo. This contribution to cellular response to DNA damage is not by specific recognition of MNNG induced O6-methyl guanine (O6-meG) base pairs in DNA by the NTR. The yeast NTR displayed similar binding affinities for O6-meG-T, O6-meG-C, G-T and G-C containing duplex oligonucleotide substrates (EMSA data not shown).
Figure 6.
MNNG sensitivity data. An mgt1Δ rad52Δ msh6::kanMX strain was transformed by yeast centromeric plasmids expressing Msh6 or the indicated mutant derivative from the Msh6 promoter. Log phase cultures were exposed to the indicated concentration of MNNG for 1 h, washed and plated as described in Materials and Methods.
DISCUSSION
In conjunction with previous studies, the results presented here suggest that the NTR of Msh6 makes several contributions to DNA mismatch repair and cellular response to alkylation damage to DNA. The first role to be identified for the Msh6 NTR was in mismatch repair (8,9), and involved PCNA binding via the PIP box at the N-terminus (boxed in green in Figure 1). The DNA binding data in Figures 2 and 3, the elevated mutation rates in Tables 1 and 2 and the resistance to MNNG in Figure 6 suggest that DNA binding by the Msh6 NTR also contributes to mismatch repair, as well as to cellular response to alkylation damage. Residues near the C-terminus of the NTR (boxed in blue in Figure 1) contribute to this DNA binding, and at least some of these residues are likely to be lysines and arginines that may interact with the phosphate backbone of DNA (Table 2). A conserved motif (Figure 5) near the C-terminus of the NTR (boxed in black in Figure 1) also appears to be functionally important for mismatch repair and response to MNNG (Table 3, Figure 6). Residues in this conserved region could contribute to DNA binding, especially since they are located immediately proximal to core domain I that is already known to interact with mismatched DNA [reviewed in (2–4)]. However, it cannot be excluded that this conserved motif may have a role other than or in addition to DNA binding. This possibility is consistent with the fact that an in-frame deletion of residues 290–299 had no detectable affect on DNA binding (Figure 4), but did result in a mutator phenotype (Table 1). Double mutants that concomitantly reduce PCNA binding and DNA binding (Table 1), or double mutants that concomitantly reduce PCNA binding and alter the consensus motif at the C-terminus of the NTR (Table 3), yield mutator effects that are greater than observed with single mutants, suggesting that these multiple functions are combined for ensuring the full efficiency of mismatch repair. However, their relative contribution to cellular response to MNNG treatment is somewhat different. For example, in considering the contribution of DNA binding, in-frame deletion of residues 228–299 yields a modest MMR defect (Table 1), but results in resistance to MNNG similar to that of an msh6 null mutant (Figure 6). Likewise, whatever the function of the consensus motif at the C-terminus of the NTR, a quadruple alanine mutant only partially inactivates MMR (Table 3) but strongly inactivates the damage response (Figure 6).
The evidence that the Msh6 NTR has multiple functions leads one to wonder how many of these functions might be shared by Msh3. One shared function is PCNA binding via a PIP box near the N-terminus (8,9). Initial attempts to express the yeast Msh3 NTR in E. coli resulted in cell lysis, precluding an examination of DNA binding capacity. The NTRs of Msh3 proteins are generally shorter than those of Msh6, and initial alignments have not suggested the presence of a PWWP domain. As mentioned in the introduction, the human Msh6 NTR does contain a PWWP sequence motif located distal to the PIP box, and Slater and colleagues (11) have predicted that this is part of a PWWP domain, a module of unknown function that is sometimes found in proteins associated with chromatin. While the yeast Msh6 NTR lacks a PWWP motif per se, certain Msh6 NTRs have been suggested to contain a structurally related Tudor domain, again just distal to the PIP box. Just beyond this, we further note that 45% (31 of 69) of residues between amino acids 144 and 212 of yeast Msh6 are negatively charged (boxed in red in Figure 1). Similarly, 49% (19/39) of residues from 192 through 230 in human Msh6 are negatively charged. This feature is reminiscent of proteins that act as DNA mimics [reviewed in (23)], wherein side chain carboxylates of aspartates and glutamates mimic the charge pattern of phosphates in a DNA backbone. Putnam and Tainer (23) have suggested that DNA mimics provide ‘an elegant mechanism by which interfaces can be reused to force sequential rather than simultaneous complex formations such as seen in systems involving polar protein assemblies and DNA repair machinery.’ If the highly negatively charged regions of yeast and human Msh6 NTRs are indeed DNA mimics, their location (boxed in red in Figure 1) immediately adjacent to the region of the Msh6 NTR that binds to DNA is intriguing. This juxtaposition suggests a model wherein the putative DNA mimic (red in Figure 1) and the DNA binding region (blue in Figure 1) might cooperate to regulate sequential steps in mismatch repair. Such regulation could involve interactions with other proteins such as PCNA or MutLα, or perhaps the proposed transition from an initial mismatch recognition complex containing bent DNA to an ultimate recognition complex in which the DNA is unbent (24).
ACKNOWLEDGEMENTS
The authors wish to thank Shannon F. Holmes and Jana E. Stone for thoughtful comments on the manuscript. The authors also wish to thank John Tainer for pointing out that the negatively charged region in the Msh6 NTR could be a DNA mimic. This research was supported by the Intramural Research Program of the NIH, NIEHS. Funding to pay the Open Access publication charges for this article was provided by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences to T.A.K.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 2.Kunkel TA, Erie DA. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 3.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 4.Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell. Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 5.Kat A, Thilly WG, Fang WH, Longley MJ, Li GM, Modrich P. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc. Natl Acad. Sci. USA. 1993;90:6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 7.Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 8.Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. J. Biol. Chem. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- 9.Flores-Rozas H, Clark D, Kolodner RD. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat. Genet. 2000;26:375–378. doi: 10.1038/81708. [DOI] [PubMed] [Google Scholar]
- 10.Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slater LM, Allen MD, Bycroft M. Structural variation in PWWP domains. J. Mol. Biol. 2003;330:571–576. doi: 10.1016/s0022-2836(03)00470-4. [DOI] [PubMed] [Google Scholar]
- 12.Kolodner RD, Tytell JD, Schmeits JL, Kane MF, Gupta RD, Weger J, Wahlberg S, Fox EA, Peel D, et al. Germ-line msh6 mutations in colorectal cancer families. Cancer Res. 1999;59:5068–5074. [PubMed] [Google Scholar]
- 13.Wu Y, Berends MJ, Mensink RG, Kempinga C, Sijmons RH, van Der Zee AG, Hollema H, Kleibeuker JH, Buys CH, et al. Association of hereditary nonpolyposis colorectal cancer-related tumors displaying low microsatellite instability with MSH6 germline mutations. Am. J. Hum. Genet. 1999;65:1291–1298. doi: 10.1086/302612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plaschke J, Kruppa C, Tischler R, Bocker T, Pistorius S, Dralle H, Ruschoff J, Saeger HD, Fishel R, et al. Sequence analysis of the mismatch repair gene hMSH6 in the germline of patients with familial and sporadic colorectal cancer. Int. J. Cancer. 2000;85:606–613. doi: 10.1002/(sici)1097-0215(20000301)85:5<606::aid-ijc2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 15.Clark AB, Cook ME, Tran HT, Gordenin DA, Resnick MA, Kunkel TA. Functional analysis of human MutSalpha and MutSbeta complexes in yeast. Nucleic Acids Res. 1999;27:736–742. doi: 10.1093/nar/27.3.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall MC, Wang H, Erie DA, Kunkel TA. High affinity cooperative DNA binding by the yeast Mlh1-Pms1 heterodimer. J. Mol. Biol. 2001;312:637–647. doi: 10.1006/jmbi.2001.4958. [DOI] [PubMed] [Google Scholar]
- 17.Detweiler CD, Deterding LJ, Tomer KB, Chignell CF, Germolec D, Mason RP. Immunological identification of the heart myoglobin radical formed by hydrogen peroxide. Free Radic. Biol. Med. 2002;33:364–369. doi: 10.1016/s0891-5849(02)00895-x. [DOI] [PubMed] [Google Scholar]
- 18.Tran HT, Gordenin DA, Resnick MA. The 3′→5′ exonucleases of DNA polymerases delta and epsilon and the 5′→3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storici F, Resnick MA. Delitto perfetto targeted mutagenesis in yeast with oligonucleotides. Genet. Eng. (NY) 2003;25:189–207. [PubMed] [Google Scholar]
- 20.Lewis LK, Kirchner JM, Resnick MA. Requirement for end-joining and checkpoint functions, but not RAD52-mediated recombination, after EcoRI endonuclease cleavage of Saccharomyces cerevisiae DNA. Mol. Cell. Biol. 1998;18:1891–1902. doi: 10.1128/mcb.18.4.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drotschmann K, Clark AB, Tran HT, Resnick MA, Gordenin DA, Kunkel TA. Mutator phenotypes of yeast strains heterozygous for mutations in the MSH2 gene. Proc. Natl Acad. Sci. USA. 1999;96:2970–2975. doi: 10.1073/pnas.96.6.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cejka P, Mojas N, Gillet L, Schar P, Jiricny J. Homologous recombination rescues mismatch-repair-dependent cytotoxicity of S(N)1-type methylating agents in S. cerevisiae. Curr.Biol. 2005;15:1395–1400. doi: 10.1016/j.cub.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 23.Putnam CD, Tainer JA. Protein mimicry of DNA and pathway regulation. DNA Repair (Amst.) 2005;4:1410–1420. doi: 10.1016/j.dnarep.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Yang Y, Schofield MJ, Du C, Fridman Y, Lee SD, Larson ED, Drummond JT, Alani E, et al. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proc. Natl Acad. Sci. USA. 2003;100:14822–14827. doi: 10.1073/pnas.2433654100. [DOI] [PMC free article] [PubMed] [Google Scholar]