Abstract
Alternative splicing of competing 5′ splice sites is regulated by enhancers and silencers in the spliced exon. We have characterized sequences and splicing factors that regulate alternative splicing of PLP and DM20, myelin proteins produced by oligodendrocytes (OLs) by selection of 5′ splice sites in exon 3. We identify a G-rich enhancer (M2) of DM20 5′ splice site in exon 3B and show that individual G triplets forming M2 are functionally distinct and the distal group plays a dominant role. G-rich M2 and a G-rich splicing enhancer (ISE) in intron 3 share similarities in function and protein binding. The G-rich sequences are necessary for binding of hnRNPs to both enhancers. Reduction in hnRNPH and F expression in differentiated OLs correlates temporally with increased PLP/DM20 ratio. Knock down of hnRNPH increased PLP/DM20 ratio, while hnRNPF did not. Silencing hnRNPH and F increased the PLP/DM20 ratio more than hnRNPH alone, demonstrating a novel synergistic effect. Mutation of M2, but not ISE reduced the synergistic effect. Replacement of M2 and all G runs in exon 3B abolished it almost completely. We conclude that developmental changes in hnRNPH/F associated with OLs differentiation synergistically regulate PLP alternative splicing and a G-rich enhancer participates in the regulation.
INTRODUCTION
Alternative RNA splicing of a single gene transcript is a common strategy to generate multiple protein isoforms with different functional properties (1). A type of alternative splicing is the inclusion/exclusion of exon between competing 5′ splice sites (2). The regulation of this type of alternative splicing has been shown to be quite complex in genes expressed in a variety of tissues and in artificial chimeric genes (3,4). In the central nervous system (CNS), much of the transcript and protein complexity is generated by alternative splicing, which is regulated in a cell- and development-specific manner (5,6). Proteolipid protein (PLP), a major CNS myelin protein comprises two protein products, PLP and DM20, which are generated by alternative splicing of two competing 5′ donor sites resulting in either inclusion or exclusion of exon 3B (7).
In oligodendrocytes (OLs), the myelin-producing cells of the CNS, the PLP 5′ splice site is preferentially utilized, while in OLs progenitor cells (OPCs) and in other cell types, DM20 is the preferred site (8,9). Although DM20 is the ancestral gene and PLP did not appear until relatively recently in evolution (10,11), PLP is the more abundant of the two protein isoforms in the mammalian post-natal brain and is uniquely expressed in myelin. The inclusion of exon 3B confers PLP unique signaling functions in axo-glial interactions that maintain axonal integrity (12,13). In differentiated OLs and in the post-natal brain, the ratio of PLP to DM20 transcripts is 3 : 1 and accounts for the preponderance of the PLP protein isoform. Tight control of the PLP/DM20 ratio is critical for normal brain development and function. Mutations that impair the PLP/DM20 ratio cause a spectrum of developmental and degenerative disorders in humans (14–16). Thus, the identification of regulatory sequences and factors that control the PLP/DM20 ratio is of critical importance for the understanding of brain development and human disease and may also provide insight into the evolutionary history of alternative splicing.
A PLP splicing construct, in which the alternative inclusion of PLP exon 3B is reconstituted was used to characterize the effect of disease-causing mutations on exon 3B splicing (14–16). An analysis of mutations at and around the PLP and DM20 splice site showed that the intrinsic strength of the competing 5′ splice sites contributes to the final PLP/DM20 ratio (14). Although the DM20 5′ site is weaker than the PLP 5′ site (14), it is the preferred site in OPCs and non-glial cells, suggesting that either an enhancer of DM20 or a silencer of PLP 5′ splice site may regulate PLP alternative splicing in these cells. Analysis of mutations occurring in patients have identified enhancers of PLP 5′ splice site: a G-rich intronic splicing enhancer (ISE) (15) and an exonic splicing enhancer (ESE) containing an ASF/SF2 motif in exon 3B (16). In other genes, the exon between competing 5′ splice sites has been shown to contain enhancers and silencers that regulate the alternative splicing selection (4,17–19).
In this study, we investigate the role of regulatory sequences in PLP exon 3B by systematically mutating exon 3B. We characterize a novel G-rich enhancer of DM20 5′ splice site and its relationship with a G-rich ISE of PLP 5′ splice selection using functional and biochemical approaches. The hnRNP H and F bind to both enhancers. A reduction in the hnRNPH/F expression levels correlates temporally with the inclusion of PLP exon 3B in differentiated OLs. We investigate the role of hnRNPH and F in the regulation of PLP/DM20 ratio by knocking down their expression. This study defines a novel synergistic regulation of PLP/DM20 mediated by hnRNPH and F and establishes a role of a novel G-rich enhancer in PLP alternative splicing.
MATERIALS AND METHODS
Cell cultures and transfections
Immunoselected OPCs were cultured in B104-conditioned medium and differentiated into OLs in T3 medium (40 ng/ml) for 72 h (16,20). Oli-neu cells (kind gift of Dr Trotter) were cultured in SATO medium with 1% horse serum and differentiated in dbcAMP, 1 mM for 3–10 days (21). L cells were grown in DMEM containing 10% FBS. OPCs plated at 7 × 105 cells/well, Oli-neu cells at 5 × 105 cells/well and L cells at 3 × 105 cells/well were transfected for 6 h with 0.5 µg plasmid DNA using Qiagen Effectene Transfection Reagents (Qiagen, Valencia, CA, USA). After 18 h in growth medium, the cells were cultured in differentiation medium and harvested 72 h after transfection.
Plasmid constructs
Mutant PLP-neo constructs were generated by site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis Kit and the QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) (Figures 1–5).
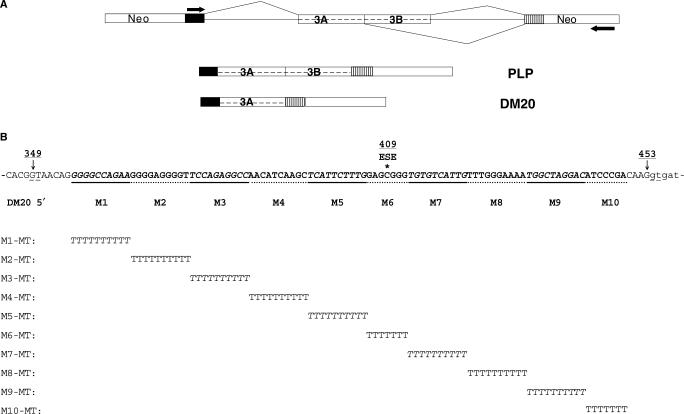
Figure 1.
PLP-neo construct and mutations in PLP exon 3B. (A) Schematic representation of the PLP-neo splicing construct. The arrows indicate the position of the PCR primers. The PLP and DM20 PCR products are shown. (B) Exon 3B: 349 indicates G at the invariant GT of DM20 5′ splice site, 453 is the last base of exon 3B at the PLP 5′ splice site, 409 is the position of a human mutation occurring in an ASF/SF2 motif and ESE indicates the ASF/SF2 binding motif (16). The linker scan sequence substitutions are aligned below the wild-type sequences. All mutant constructs replace 10 nt, except for M6 and M10 (see text), and are of the same length as the wild-type construct.
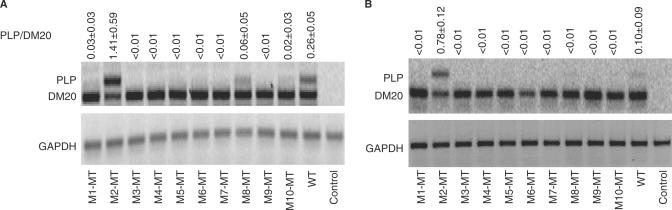
Figure 2.
Mutation of a G-rich sequence activates inclusion of PLP exon 3B. Results of RT-PCR assay of PLP and DM20 from total RNA isolated from Oli-neu cells (30 PCR cycles) (A) and L cells (35 PCR cycles) (B) transfected with wild-type PLP-neo (WT) and M1-MT to M10-MT. The PLP/DM20 ratios ± SD are shown (n = 3). GAPDH is used for accuracy of RNA loading (25 PCR cycles). Control represents the untransfected cells. The increase in PLP/DM20 ratio with the M2-MT construct is statistically significant (P = 0.017 for Oli-neu cells and P < 0.002 for L cells).
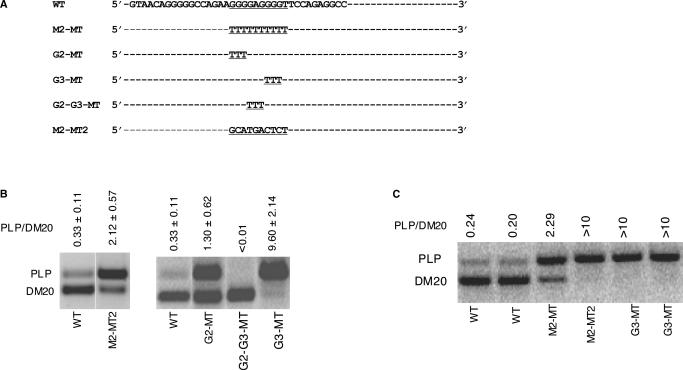
Figure 3.
Analysis of G triplets within M2 in Oli-neu cells and primary oligodendrocytes. (A) Partial wild-type sequences of exon 3B are shown and M2 is underlined. Linker scan mutations of M2 and G triplets within M2 are shown. (B) Results of RT-PCR assay of PLP and DM20 from RNA isolated from differentiated Oli-neu cells transfected with wild-type PLP-neo (WT) and mutated constructs, M2-MT, M2-MT2, G2-MT, G3-MT and G2-G3-MT. The PLP/DM20 ratios ± SD are shown (n = 3). The upper and lower limits of accurate quantification of the PLP/DM20 ratio are 10 and 0.01. (C) PLP and DM20 PCR products amplified in RNA extracted from differentiated OLs transfected with WT and M2-MT, M2-MT2 and G3-MT, duplicate transfections are shown.
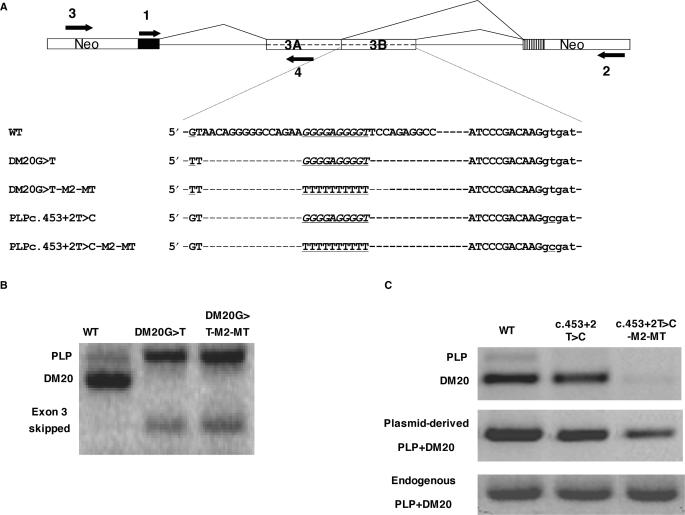
Figure 4.
M2 is an enhancer of DM20 5′ splice site selection. (A) PLP-neo construct and primers used for PCR amplification. Partial sequences of the natural exon 3B (WT) and mutated constructs are shown, underlined are mutations at the DM20 and PLP 5′ splice site and the M2-MT. (B) PLP and DM20 PCR products (35 PCR cycles) from WT and DM20 G>T and DM20 G>T-M2-MT amplified with primers 1 and 2 in RNA extracted from transfected Oli-neu cells. (C) PLP and DM20 PCR products (35 PCR cycles) derived from WT, c.453+2T>C and c.453T>C-M2-MT amplified with primers 1 and 2 in RNA extracted from Oli-neu cells. Plasmid-derived PLP+DM20 PCR product was amplified with primers 3 and 4 and represents the total plasmid-derived PLP/DM20 transcript. Endogenous PLP+DM20 PCR product amplified with primers 1 and 4 is the control for RNA loading.
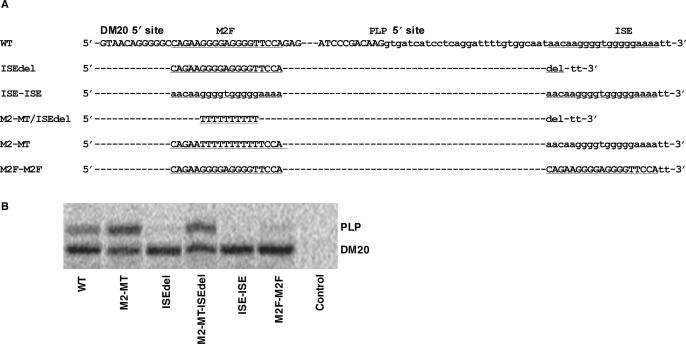
Figure 5.
Functional analysis of M2 and ISE. (A) Partial sequences of exon 3B: M2 and ISE (small cases) are underlined. The mutations and the name of the constructs are shown (see text for details). (B) Results of RT-PCR assay of PLP and DM20 from differentiated Oli-neu cells transfected with wild-type PLP-neo (WT) and mutant constructs, M2-MT, ISEdel, M2-MT-ISEdel, ISE-ISE and M2F-M2F. This is a representative experiment (n = 2).
siRNA and Western blot
Pre-designed double-stranded (ds) siRNAs targeting hnRNPF [175720 (siF1), 175721 (siF2) and 175722 (siF3); H [182015 (siH1), 182016 (siH2) and 75775 (siH3)], a custom-made ds siRNA, siF/H [target sequence 5′-GAA TAG GGC ACA GGT ATAT-3′, 524–542 nt of mouse hnRNP F cDNA (Accession# NM-133834), hnRNP H cDNA (Accession# NM-021510)] and Silencer® Negative Control #1 siRNA were purchased from Ambion (Austin, TX, USA).
Undifferentiated Oli-neu cells were transfected with siRNA (50–100 nM) alone or in combination with reporter plasmids (0.5 μg) in siPORT amine transfection reagent using the neofection protocol (Ambion, Austin, TX, USA). To keep constant the amount of siRNA, Silencer® Negative Control #1 siRNA was added to transfections in which a single siRNA was used. Western blots of cell lysates (10 μg proteins) were performed using the One-Step ™ Complete Western Kit (Genescript, Piscataway, NJ, USA). Blots were probed with 10 μg primary antibody to hnRNP F [rabbit polyclonal, generous gift of Dr Milcarek, (22,23)] and hnRNP H (Bethyl Laboratories). Bands were quantitated with Kodak 440CF Digital Image station using 1D analysis software with actin as a standard for loading.
RNA extraction and RT-PCR
Total RNA was extracted with RNeasy Mini Kit (Qiagen, Valencia, CA, USA) and treated with the DNA-free Kit (Ambion, Austin, TX, USA). Total RNA (0.5 μg) was reverse transcribed using random hexamer primers (BD Biosciences, Palo Alto, CA, USA). The PLP and DM20 PCR products and PLP+DM20 product derived from PLP-neo were amplified and quantitated as described (16) (Figures 1A and 4A).
Nuclear extracts and Western blot analysis
Nine micrograms of proteins of nuclear extracts (NEP, Pierce) were separated by 10% SDS-PAGE, blotted and reacted with MAb104 (ATCC), antibodies to β-tubulin (Sigma, T 4026), PCNA (Santa Cruz), hnRNPH and F [rabbit polyclonal, generous gift of Dr Milcarek, (22,23)], A1 (MAb9H10, generous gift of Dr Dreyfuss) and L (AbCam) diluted 1 : 2000 and QKI5 (Bethyl Laboratories) diluted 1 : 1000, HRP-conjugated secondary antibody (Jackson ImmunoResearch Laboratories) diluted 1 : 5000 and developed with enhanced chemiluminescence (ECL, Amersham) (16). Blots were quantified with Kodak 440CF Digital Image Station using 1D analysis software with tubulin as a standard for loading and quantification.
RNA-affinity precipitations
Biotinylated RNA oligonucleotides (500 pmol) (Integrated DNA Technologies, Inc) were incubated with 200 μg proteins of nuclear extracts in buffer containing 4 mM creatine, 2 mM ATP, 1.5 mM MgCl2, 1.2 mM DTT for 20 min at 30°C, heparin was added and the reactions were exposed to 254 nm UV light with a Spectronics XL-1000 UV crosslinker at a setting of 1.8 J/cm2 on ice (about 10 min). RNA-binding proteins were precipitated with streptavidin beads (Pierce, Rockford, IL, USA). Fifty microliters of prewashed immobilized streptavidin beads, 800 μl binding buffer and 10 μl Halt Protease Inhibitor Cocktail (Pierce, Rockford, IL, USA) were added to the UV-crosslinked reactions, incubated at 4°C in a rocking platform, and washed at least eight times with binding buffer prior to gel electrophoresis. Proteins from half of the entire mixture were separated by 10% SDS-PAGE and either visualized by silver staining (Bio-Rad) or detected by Western blot.
Protein isolation and identification by liquid chromatography–mass spectroscopy (LC–MS/MS)
Bands separated in 1D gel stained with silver stain were excised, destained and digested with trypsin. Samples were prepared and loaded on the MALDI plates and analyzed by Qstar XL with the MALDI source (Proteomics and Mass Spectroscopy Core Facility, University of Kentucky). The three most abundant peptide ions were automatically selected to perform MS/MS to obtain sequence information and help increase the confidence of a match with a known protein. Measured peptide masses obtained with MALDI–MS/MS and LC–MS/MS were compared with peptide masses from an in silico digestion of the protein database using MASCOT search engine for protein identification and modification detection.
RESULTS
PLP exon 3B contains sequences that regulate the PLP/DM20 ratio
To identify novel regulatory sequences that control the PLP/DM20 ratio, we have systematically mutated PLP exon 3B in a PLP splicing construct and assessed in vivo splicing in transfected Oli-neu cells (Figure 1A). In published work, we transfected primary OLs (14–16). In the present study, we have used Oli-neu cells, immortalized OPCs that are induced to differentiate by dbcAMP (21,24). Because the efficiency of transfection in Oli-neu cells is 36% compared with 6% in primary OLs (data not shown), changes in plasmid-derived PLP/DM20 ratio are more easily quantified. We have validated the use of Oli-neu cells as a model for the OLs in splicing studies. We have established that various PLP-neo constructs previously tested in primary OLs expressed the same PLP and DM20 products when transfected into Oli-neu cells (data not shown). Expression results for some of the critical mutations characterized in the present study in Oli-neu cells were confirmed in primary OLs (Figure 3). Finally, we show that differentiated Oli-neu cells replicate the trend of changes in hnRNP's expression that we detect in differentiated OLs (Figure 7).
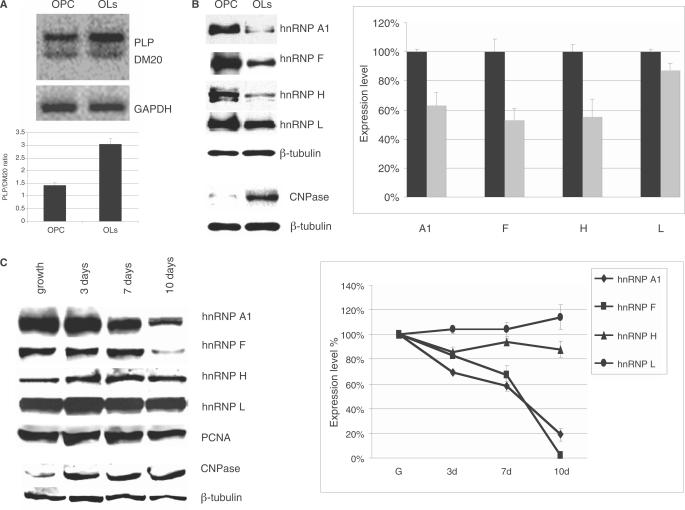
Figure 7.
The hnRNPs expression in primary OLs and Oli-neu cells. (A) Representative RT-PCR products of the endogenous PLP and DM20 transcripts in RNA isolated from OPC and OLs differentiated for 72 h. The bar graph represents the mean ± SD (n = 3). (B) Representative Western blot of nuclear extracts prepared from OPC and OLs differentiated for 72 h and probed with antibodies specific for hnRNPA1, H, F and L (see Materials and Methods section). CNPase expression was assessed in cytoplasmic extracts. Tubulin is a control for loading accuracy. The data were reproduced in five separate primary OLs preparations. Bands were quantified by densitometry and the value was corrected by the internal control tubulin. The bar graph represents the percent expression ± SD of each protein in OLs nuclear extracts relative to the level detected in OPC nuclear extracts, which is set at 100 (n = 5). Reduction of H (P = 0.006), F (P = 0.003) and A1 (P = 0.02) in differentiated OLs versus OPC were all significant. (C) Representative Western blot of nuclear extracts prepared from undifferentiated Oli-neu cells (growth) and differentiated for 3, 7 and 10 days and probed with antibodies specific for hnRNPA1, H, F and L. Tubulin is control for loading accuracy. The data were reproduced in three separate experiments. Bands were quantified by densitometry and the values were corrected by the internal control tubulin. CNPase expression was assessed in cytoplasmic extracts. The bar graph represents the percent expression ± SD of each protein in the Oli-neu nuclear extracts relative to the level detected in undifferentiated cells, which is set at 100 (n = 3). The reduction of F and A1 expression in differentiated Oli-neu cells was significant at 7 and 10 days versus undifferentiated Oli-neu cells (P = 0.0017 for F and P = 0.001 for A1). The decrease of H in differentiated Oli-neu cells versus undifferentiated Oli-neu cells did not reach statistical significance. Levels of hnRNPL did not change.
We made 10 PLP-neo constructs in which sequences of exon 3B (M1-M10) were replaced by a poly-T linker (25). In order to not affect sequences that are necessary for recognition of the 5′ splice site by the spliceosome, we began replacing bases after position +8 relative to the DM20 5′ splice site (349, Figure 1A) up to the fourth base preceding the PLP 5′ splice site (453, Figure 1A) (26). Ten bases were replaced except for M6 and M10, which both contain seven base changes. M6 spans an ASF/SF2 binding motif that regulates PLP alternative splicing (16) (Figure 1B).
Following transfection and expression of PLP-neo constructs carrying the mutated M1–M10 (M1-MT–M10-MT) in Oli-neu cells differentiated in medium containing dbcAMP for 72 h, the PLP and DM20 products were simultaneously amplified by RT-PCR (Figure 2A). Replacement of a G-rich sequence in the 5′ end of exon 3B (M2) resulted in 5.4-fold increase in PLP/DM20 ratio compared with the WT (Figure 2A). In contrast, replacement of the other exon 3B sequences caused a reduction in the PLP/DM20 ratio, with sequences replaced in M1, M8 and M10 having less of an effect (Figure 2A). Similar changes in the PLP/DM20 ratio were detected in Oli-neu cells transfected with M1-MT/M10-MT and differentiated for 6 days (data not shown).
To determine whether replacing M2 affects the PLP/DM20 ratio in non-glial cells, we transfected L cells with M1-MT through M10-MT and quantified the plasmid-derived PLP and DM20 products by RT-PCR. Although the PLP/DM20 ratio is low in L cells consistent with the cell-specific regulation of PLP 5′ splice site recognition (16), replacement of M2 resulted in 7.8-fold increase in the PLP/DM20 ratio, suggesting that M2 is active in L cells (Figure 2B). To confirm the functional relevance of M2, we replaced M2 with an IgM sequence (-GCATGACTCT-) (M2-MT2) (Figure 3A) (27). In M2-MT2-transfected Oli-neu cells, the PLP/DM20 ratio was 6.4-fold higher than in WT-transfected cells (Figure 3B), confirming that M2 regulates the PLP/DM20 ratio.
These data show that G-rich exonic sequences (M2) regulate the PLP/DM20 ratio in favor of the DM20 5′ splice site in both glial and non-glial cells, although the efficiency of exon 3B inclusion is greater in Oli-neu cells, suggesting a contribution of cell-specific context in the regulation of the PLP/DM20 ratio. The remainder of exon 3B favors PLP 5′ splice site selection. Interestingly, every substitution appears to affect the PLP/DM20 ratio, suggesting that multiple regulatory elements may be present in exon 3B. In support of this interpretation is the finding that one of these sequences M6 contains a previously characterized ASF/SF2 motif (Figure 1B, ESE) and M6-MT reduces the PLP/DM20 ratio consistent with results obtained by introducing patients’ mutations at this ASF/SF2 motif (16).
In an artificial α-globin minigene construct containing G triplets downstream of duplicated 5′ splice sites, the G triplet was the basic functional unit (28). To determine the functional relevance of G triplets within M2, PLP-neo constructs in which either the G triplets at the 5′ end (G2) or 3′ end (G3) or the internal GAG triplet (G2–G3) were replaced with Ts were transiently expressed in Oli-neu cells (Figure 3A). The PLP/DM20 ratio was increased 4-fold by mutation of G2 (G2–MT) and 29-fold by mutation of G3 (G3-MT) compared with WT (Figure 3B). In contrast, the PLP/DM20 ratio was greatly reduced by mutation of G2–G3 (G2–G3-MT) and the PLP product was not detectable (Figure 3B). The data suggest that M2 is functionally complex and is composed of G triplets of different strengths separated by a triplet that has an opposite effect.
We next sought to assess the function of M2 in primary OLs because these cells more closely replicate the developmental increase in the endogenous PLP/DM20 ratio observed in vivo (Figure 7A). OPCs were transfected with WT, M2-MT, M2-MT2 and G3-MT and differentiated into OLs for 72 h. The PLP/DM20 ratio was 9.4-fold higher in M2-MT-transfected OLs than in WT-transfected OLs and in M2-MT2- and G3-MT-transfected OLs only the PLP product was amplified. The results replicate those obtained with Oli-neu cells (compare Figure 3B with 3C), however, they also show that the increase in PLP/DM20 ratio in OLs is greater than in Oli-neu cells in keeping with the higher degree of differentiation reached by OLs versus Oli-neu cells. The data suggest that differentiation may affect the efficiency of PLP alternative splicing derived from the construct.
M2 is an enhancer of DM20 5′ splice site
We next sought to determine whether M2 functions as a silencer of PLP 5′ splice site or an enhancer of DM20 5′ splice site. We inactivated the DM20 5′ splice site by changing the invariant G (349 in exon 3B) to a T (Figure 4A). If M2 suppresses PLP 5′ splice site, inactivating the DM20 5′ splice site should not increase the PLP/DM20 ratio. The G>T change caused a complete loss of the DM20 product and a corresponding increase in the PLP product in differentiated Oli-neu cells (Figure 4B). When both the G>T and the M2-MT mutations were introduced in PLP-neo, the results were similar to those obtained with the G>T mutation alone (Figure 4B). A PCR product of ∼100 bp, which was consistently amplified from the G>T and G>T-M2-MT-transfected cells (Figure 4B) was sequenced and it is the spliced product in which exon 3 is completely skipped. These data suggest that M2 is an enhancer of DM20 5′ splice site.
We next inactivated the PLP 5′ splice site by mutating the invariant +2T (c.453+2T>C) in the M2-MT construct. This mutation was identified in humans and abolishes the PLP 5′ splice site (14). If M2 is an enhancer of DM20 5′ splice site, we would expect to detect a reduction of the DM20 product derived from c.453+2T>C-M2-MT compared with the WT and c.453+2T>C. The DM20 product is greatly reduced in c.453+2T>C-M2-MT-transfected Oli-neu cells, while it is amplified in approximately the same amount in c.453+2T>C as in WT-transfected Oli-neu (Figure 4C). No PLP product is detected in either construct. The plasmid-derived PLP+DM20 PCR product amplified with a forward primer in the neo gene and reverse primer in PLP exon 3A is drastically reduced in c.453+2T>C-M2-MT compared with WT and c.453+2T>C (Figure 4C). This reduction could be explained by skipping of exon 3 due to inefficient recognition of the DM20 5′ splice site caused by the M2-MT or less likely inefficient 3′ splice site utilization. Alternatively, the reduction in the combined PLP+DM20 product may be explained by retention of intron 3 resulting in an unstable transcript. However, PCR products of the predicted size for exon 3 skipping and intron 3 retention were not detected. Collectively, these results support the interpretation that M2 is an enhancer of DM20 5′ splice site.
We next tested the function of M2 in a globin-PLP construct in which PLP exon 3B and 100 nt of intron 3 spanning the G-rich ISE were fused to the β–globin gene (GloPLP, Supplementary Figure S1A and Supplementary Data). In transfected Oli-neu cells, the ratio of product including exon 3B to the product excluding exon 3B was increased 6.3-fold in Glo-PLPM2-MT compared with Glo-PLP (Supplementary Figure S1B). The data show that M2 exerts its function in a minimal construct and suggest that exon 3B and the first 100 nt of intron 3 are sufficient to regulate the PLP/DM20 ratio. Collectively, the data show that M2 is an enhancer of DM20 5′ site selection and its function is replicated in a minimal PLP-globin construct.
Functional analysis of M2 and ISE in the regulation of PLP/DM20 ratio
M2 and ISE contain G-rich sequences that have nearly identical composition, raising the possibility that a balance between these enhancers regulates PLP/DM20 ratio (Figure 5A). To evaluate their functional relationship, we asked whether M2 and ISE enhance the upstream 5′ splice site when they are exchanged. The G-rich core and flanking sequences of the 19-bp ISE contribute to the enhancer's function (15). In order to move the ISE and not change the distance of G sequences from the 5′ splice site, we replaced M2 and the five bases 5′ and four bases 3′ of M2 (19 nt, M2F) with the ISE and vice versa. We have made the following constructs: ISE-ISE, M2F-M2F and M2-MT/ISEdel (Figure 5A). The naturally occurring ISE deletion (ISEdel) reduces PLP 5′ splice site selection (15). PLP and DM20 PCR products were amplified in Oli-neu cells transfected with these constructs and with PLP-neo, M2-MT and ISEdel (Figure 5B). The PLP/DM20 ratio derived from ISE-ISE and M2F-M2F was lower than the ratio derived from the WT, suggesting that the ISE is a strong enhancer of DM20 5′ splice site and M2F is a weak enhancer of PLP 5′ splice site. The PLP/DM20 ratio derived from M2-MT/ISEdel was increased compared with WT, although it was not as high as that derived from M2-MT (Figures 5B and 9C). These data suggest that although the ISE and M2F can replace each other, they differ in the strength by which they enhance the 5′ splice site when removed from their natural position.
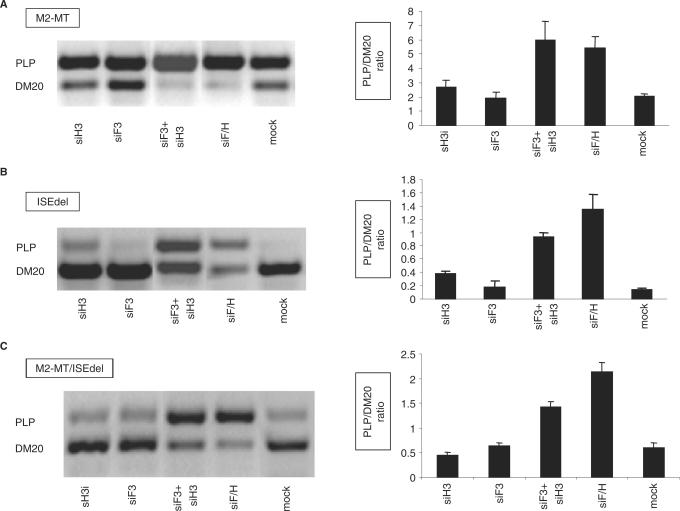
Figure 9.
The effect of mutation of M2 and ISE on the hnRNPH and F-mediated regulation of PLP/DM20. (A) Representative RT-PCR analysis of the M2-MT derived PLP and DM20 products amplified from RNA isolated from Oli-neu cells treated with siH3, siF3, siF3+H3, siF/H (35 PCR cycles). Mock are cells treated with control siRNA. The bar graph shows the PLP/DM20 ratios ± SD (n = 3). The increase in PLP/DM20 ratio induced by siF3+H3 and siF/H are statistically significant (P < 0.01). (B) Representative RT-PCR analysis of the ISEdel derived PLP and DM20 products amplified from RNA prepared from Oli-neu cells treated with siH3, siF3, siF3+H3, siF/H (35 PCR cycles). Mock are cells treated with control siRNA. The bar graph shows the PLP/DM20 ratios ± SD (n = 3). The increase in PLP/DM20 ratio induced by siH3, siF3+H3 and siF/H is statistically significant (P < 0.01). (C) Representative RT-PCR analysis of the M2-MT/ISEdel derived PLP and DM20 products amplified from RNA prepared from Oli-neu cells treated with siH3, siF3, siF3+H3, siF/H (35 PCR cycles). Mock are cells treated with control siRNA. The bar graph shows the PLP/DM20 ratios ± SD (n = 3). The changes in PLP/DM20 ratio induced by siF3+H3 and siF/H are statistically significant (P < 0.05).
Biochemical analysis of proteins that bind to M2F and ISE in Oli-neu extracts
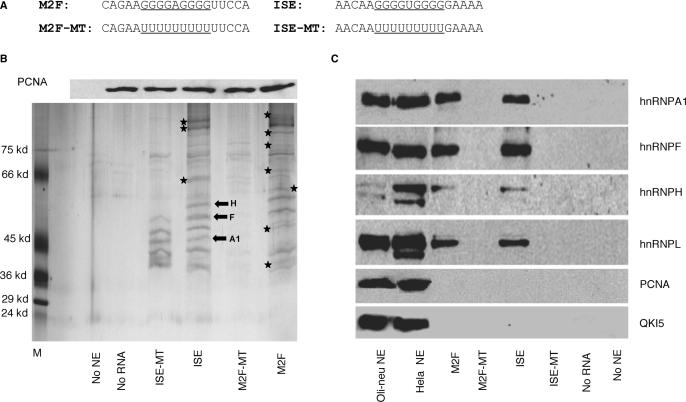
To further examine M2F and ISE, we analyzed proteins that bind to M2F and ISE in differentiated Oli-neu nuclear extracts by RNA-affinity precipitations. Biotinylated 19 bp RNA oligoribonucleotides containing the M2F and ISE sequences and templates in which the Gs were replaced by poly-U, M2F-MT and ISE-MT (Figure 6A) were incubated with nuclear extracts. A reaction without the oligoribonucleotide was used as control for non-specific binding to the matrix. The UV-crosslinked protein–RNA complexes were precipitated with streptavidin–agarose beads and the proteins were separated by 10% SDS-PAGE and visualized by silver staining. Representative RNA-affinity precipitates (n = 2) are shown in Figure 6B.
Figure 6.
Biochemical analysis of proteins that bind to M2F and ISE in Oli-neu extracts. (A) RNA templates used in RNA affinity precipitations. The natural G-rich sequences and the mutated poly-U sequences are underlined. (B) Upper panel: Western blot analysis of PCNA levels in each RNA affinity precipitate. One-tenth of each RNA/protein mixture prior to streptavidin beads precipitation was separated by SDS-PAGE and probed with an antibody to PCNA. Lower panel: representative silver stained gel of RNA affinity precipitates with biotinylated RNA templates containing wild-type (M2F and ISE), poly-U (M2F-MT and ISE-MT) and Oli-neu extracts (n = 2) (see Materials and Methods section). Controls are precipitates without nuclear extracts (no NE) and without RNA template (no RNA). The asterisks indicate protein bands that are uniquely present in precipitates with either M2F or ISE. The block arrows indicate the protein bands that were analyzed by LC/MS/MS and their identity is shown. (C). Western blot analysis of hnRNPA1, F, H and L in the RNA affinity precipitates (see Materials and Methods Section). Precipitates without nuclear extracts (no NE) and without RNA template (no RNA) are used as controls. Western blot of Oli-neu and HeLa nuclear extracts (9 μg) were used as control for the reactivity of the antibody. PCNA and QKI5 antibodies, used as control of the specificity of RNA affinity precipitates, detect a band in the nuclear extracts, but not in the RNA affinity precipitates.
While a number of protein bands of similar molecular weights (MWs) in the >100 to 50–55 kDa and 45–37 kDa range were detected in both ISE and M2F precipitates, there were bands uniquely present in either precipitate (shown by asterisks, Figure 6B). The protein bands detected in non UV-crosslinked precipitates were similar in gel mobility and pattern to those detected in UV-crosslinked precipitates, suggesting that the differences in protein bands between ISE and M2F precipitates are likely to reflect real differences in protein binding rather than shifts in mobility caused by UV-crosslinking (n = 2, data not shown). The data suggest that proteins of similar and distinct MWs bind to M2F and ISE.
In precipitates with the ISE-MT, high MW bands were absent while protein bands in the ∼45–37 kDa range were still detected (Figure 6B). In contrast, nearly all protein bands were not detected in precipitates with M2F-MT (Figure 6B). The generalized reduction in protein binding to M2F-MT is not due to inefficient synthesis and/or amount of the M2F-MT template, since all RNA templates were quantified prior to the RNA-affinity precipitations (data not shown). In addition, the differences in binding are not due to unequal protein loading to each reaction. Similar levels of PCNA were detected by Western blot analysis of each RNA–protein mixture separated by SDS-PAGE prior to streptavidin-beads precipitation (Figure 6B, upper panel). These data support the interpretation that the presence of the G-rich sequences is necessary for binding of all proteins to M2F, whereas it is required for binding of some, but not all proteins to the ISE.
To identify proteins whose binding depends on the presence of G-rich sequences, we have analyzed the affinity-precipitated proteins by Western blot with antibodies to known RNA binding proteins. On the basis of MW, binding motifs in the G-rich sequences, and published data on proteins binding to G-rich elements (18,29,30), we anticipated that proteins in the MW range 37–65 kDa were likely to be hnRNPs. The RNA-affinity precipitates were separated by SDS-PAGE, blotted and probed with antibodies to hnRNPs. The hnRNPA1, H, F and L were detected in the precipitates with M2F and ISE, but were not detected in precipitates with M2F-MT and ISE-MT (Figure 6C). While very strong binding motifs are present in M2F and ISE, only partial motifs for hnRNPL are identified. To test the specificity of the RNA affinity precipitations, we have probed Western blots of the RNA affinity precipitates with an antibody to QKI5, an RNA binding protein that contains a heterogeneous nuclear ribonucleoprotein K homology (KH) domain (31) and is not expected to bind to M2F and ISE and an antibody to PCNA, an unrelated nuclear protein. We found that both QKI5 and PCNA do not bind to either the wild-type or mutated RNA templates (Figure 6C). These data demonstrate the specificity of the RNA affinity assay.
We next demonstrated that hnRNPH, F and A1 are present in the RNA–protein complexes by LC/MS/MS of three discrete bands in the 37–65 kDa MW range of the ISE precipitates. The peptide sequence data were matched with the protein database and confirmed to be hnRNPF, H and A1 (block arrows, Figure 6B). The results demonstrate that hnRNPs bind to the ISE and M2F in G sequence-dependent manner and suggest that these factors may participate in the enhancer's function.
Western blot analysis reveals differences in expression levels of hnRNPs in OLs compared with OPC
We next investigated whether hnRNP A1, H, F and L are differentially expressed in OLs versus OPC by Western blot analysis of nuclear extracts prepared from OPC and OLs differentiated for 72 h using antibodies specific for these hnRNPs (Figure 7B). Significant reductions in hnRNP F (47%), hnRNPH (45%) and hnRNPA1 (37%) levels were detected in OLs compared with OPC extracts (n = 5) (Figure 7B). No change in the hnRNPL level was detected (Figure 7B). Expression of the SR proteins detected with the MAb104 antibody did not change in OLs versus OPC (data not shown) and is in keeping with previous expression studies of ASF/SF2 in OLs (16). CNPase, a marker of OLs differentiation was expressed in cytoplasmic extracts of OLs, but not in OPC, indicating that the cells have differentiated (Figure 7B).
Changes in hnRNPs expression are associated with the developmental switch in the endogenous PLP/DM20 ratio. The PLP/DM20 ratio was 3 : 1 in OLs compared with 1.5 : 1 in OPC (n = 3) (Figure 7A) and replicates the developmental increase in PLP/DM20 ratio in the post-natal brain (32). The data show a temporal association between decrease in hnRNPs expression levels and increase in the PLP/DM20 ratio.
We next assessed whether changes in the expression of hnRNPs in differentiated versus undifferentiated Oli-neu cells replicate the pattern observed in primary OLs. Oli-neu cells were either grown in serum-containing medium or differentiated for 3, 7 and 10 days in dbcAMP-containing medium. Oli-neu cells reach a higher degree of differentiation after 7 and 10 days of culture in dbcAMP-containing medium (21). Significant reductions in hnRNP F and A1, but only modest decreases in hnRNPH were detected in Oli-neu differentiated for 7 and 10 days compared with undifferentiated and 3 day-differentiated Oli-neu cells (Figure 7C). CNPase expression was used as an internal marker of differentiation (Figure 7C). These results demonstrate that changes in hnRNPs associated with differentiation of Oli-neu cells generally replicate the results obtained with OLs. Together, the data show that a decrease in hnRNP H, F and A1 expression occurs in differentiated OLs and is temporally associated with differentiation-dependent increase in PLP/DM20 ratio in OLs.
Knock down of hnRNPH and F increases the PLP/DM20 ratio in undifferentiated OLs
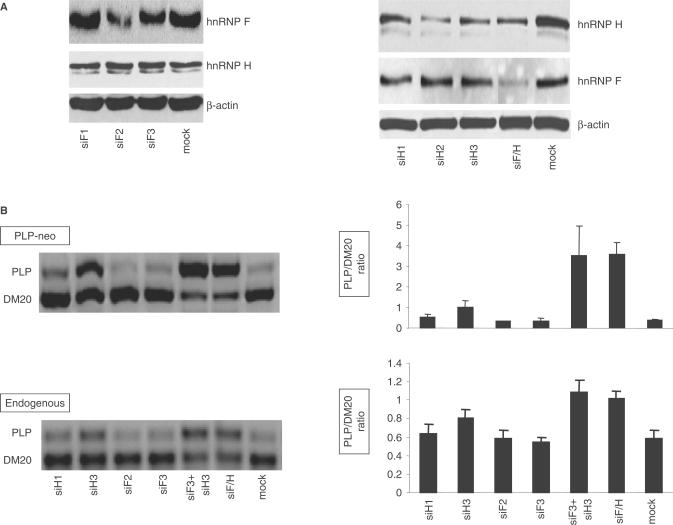
The coordinate decrease in the expression of hnRNP H, F and A1 in OLs suggests that these splicing factors may regulate the changes in the PLP/DM20 ratio. We have tested whether RNAi-mediated removal of either hnRNPH or F is sufficient to increase the PLP/DM20 ratio in undifferentiated Oli-neu cells in which the endogenous expression of H and F is high. Oli-neu cells were treated with siRNA that either target hnRNPH (siH1, H2, H3) or hnRNPF (siF1, F2 and F3) (Ambion, see Materials and Methods section) and hnRNPH and F levels were quantified by Western blot of cell lysates prepared 72 h after transfection. hnRNPH was reduced by 70% with siH2, 50% with siH3 and 40% with siH1 (Figure 8A). hnRNPF was reduced by 60% with siF2 and 40% with siF3, while was unaffected by siF1 (Figure 8A). Expression of hnRNP A1 and L was not changed by treatment with the siRNAs, confirming the specificity of the knock down (data not shown). Subsequent experiments were carried out with siH3, H1 and siF2, F3.
Figure 8.
RNAi-mediated knock down of hnRNPH and F increases the PLP/DM20 ratio in Oli-neu cells. (A) Representative Western blot of cell extracts prepared from Oli-neu cells treated with siRNAs that target hnRNPH (siH1, H2, H3), hnRNPF (siF1, F2 and F3) and both H and F (siF/H). Mock are cells treated with negative control siRNA. After quantification of the bands, the values were corrected by actin, used as control for loading accuracy. The hnRNPH was reduced by 40, 70 and 50% in cells treated with siH1, siH2 and siH3 and 50% in cells treated with siH/F versus control (n = 2). The hnRNPF was reduced by 60 and 40% in cells treated with siF2 and siF3 and by 60% in cells treated with siF/H versus controls. (B) Representative RT-PCR analysis of the PLP-neo derived PLP and DM20 spliced products and the endogenous PLP and DM20 transcripts amplified from RNA isolated from Oli-neu cells treated with siH1, siH3, siF2, siF3, siF3 + H3 and siF/H (35 PCR cycles). The bar graph shows the PLP/DM20 ratios ± SD (n = 3). Mock are cells treated with control siRNA. Increase in PLP/DM20 ratio is statistically significant for siH3 (P < 0.05) and for siF3+H3 and siF/H (P < 0.01)-treated cells compared with mock-treated cells.
To determine whether knock down of either hnRNPF or H affects the PLP/DM20 ratio, we transfected PLP-neo and siRNAs into Oli-neu cells. The plasmid-derived PCR products were amplified in RNA prepared from transfected Oli-neu cells cultured for 72 h in growth medium (Figure 8B). Knock down of hnRNPH resulted in >2-fold increase in the PLP/DM20 ratio in cells treated with siH3 compared with mock treated cells (0.98 versus 0.36) (similar results were obtained with H2, data not shown), a smaller increase was induced by siH1 (0.56 versus 0.36), consistent with lower silencing efficacy of siH1 on hnRNPH expression (Figure 8B, upper panel). The PLP/DM20 ratio was not increased by treatment with siF2 and siF3, although the levels of hnRNPF are significantly reduced by the treatment (Figure 8B, upper panel). These data show that reduction in hnRNPH is sufficient to increase the PLP/DM20 ratio, while knock down of hnRNPF does not have an effect on the PLP/DM20 ratio.
To determine whether knock down of both hnRNP H and F cooperatively regulates the PLP/DM20 ratio, we co-transfected Oli-neu cells with siF3+siH3 and PLP-neo. The simultaneous knock down of hnRNPF and H caused 10-fold increase in the PLP/DM20 ratio compared with mock treated cells (3.5 versus 0.36) (Figure 8B). siF2 combined with either siH2 or siH3 treatments gave similar results (data not shown). The data suggest that hnRNPH and F cooperatively regulate PLP/DM20 ratio and their knock down results in a synergistic effect. These findings were confirmed by knock down of hnRNPH and F with a custom-made siRNA that targets both hnRNPF and H (siF/H) (Ambion, see Methods and Materials section). siF/H reduced the expression of hnRNPF by 60% and hnRNPH by 50% (Figure 8A) and caused a 10-fold increase in the PLP/DM20 ratio (Figure 8B, upper panel).
We have also determined that knock down of hnRNPH and hnRNPH+F increased exon 3B inclusion in GloPLP-derived products similarly to PLP-neo (data not shown), suggesting that exon 3B and intron 3 contain regulatory elements that mediate the effects of hnRNPH and F. We next evaluated whether knock down of hnRNPH and F increases the endogenous PLP/DM20 ratio. Knock down of both hnRNPH and F caused 2-fold increase in the endogenous PLP/DM20 ratio compared with mock treated cells (1 versus 0.5), while removal of hnRNPH increased the PLP/DM20 ratio modestly (0.8 versus 0.5) (Figure 8B, lower panel). These results demonstrate a role of hnRNPH and F in the regulation of the endogenous and plasmid-derived PLP alternative splicing.
Because hnRNPH and F bind to M2 and ISE, we tested whether the presence of M2 and ISE is required for the hnRNPH and F-mediated effect on PLP/DM20 ratio. If either M2 or ISE is necessary for hnRNPH and F transactivation, mutation of these enhancers would reduce the increase in the PLP/DM20 ratio induced by removal of hnRNPH and F. Knock down of hnRNPH did not change the PLP/DM20 ratio derived from M2-MT compared with mock treated cells, while knock down of both hnRNPH and F caused <3-fold increase in PLP/DM20 ratio (5.5 versus 2), which is much lower than the 10-fold increase in PLP/DM20 ratio derived from the WT (Figure 9A). Although the PLP/DM20 ratio derived from M2-MT is relatively high (2), the smaller increase in PLP/DM20 ratio is not due to limitation in detection and quantification of the PCR products, as we can accurately quantify PLP/DM20 ratio of up to 10 (Figure 3).
The PLP/DM20 ratio derived from the ISEdel was increased 3-fold by removal of hnRNPH (0.37 versus 0.13) (Figure 9B). Knock down of hnRNPH and F with siF3+siH3 increased the PLP/DM20 ratio 7-fold (0.93 versus 0.13) and with siF/H increased the ratio 10-fold (1.32 versus 0.13) (Figure 9B). Although the efficiency of PLP 5′ splice site selection is low in the ISEdel, the fold increase in the PLP/DM20 ratio induced by knock down of hnRNPH and hnRNPH+F is similar to that derived from the PLP-neo. When both M2 and ISE are mutated (M2-MT/ISEdel) knock down of hnRNPH did not change the PLP/DM20 ratio (Figure 9C) similar to the results obtained with M2-MT (Figure 9A). The PLP/DM20 ratio derived from M2-MT/ISEdel was increased >2-fold by treatment with siF3+siH3 (1.43 versus 0.6) and >3-fold by siH/F (2.13 versus 0.6) (Figure 9C) similar to the results obtained with M2-MT (Figure 9A). These data suggest that disabling M2 interferes with hnRNPH-mediated effect on the PLP/DM20 and greatly reduces, although does not completely eliminate the synergistic effect induced by removal of hnRNPH and F. The latter result suggests that sequences other than M2 and ISE and contained in PLP exon 3B and intron 3, also participate in mediating the synergistic effect.
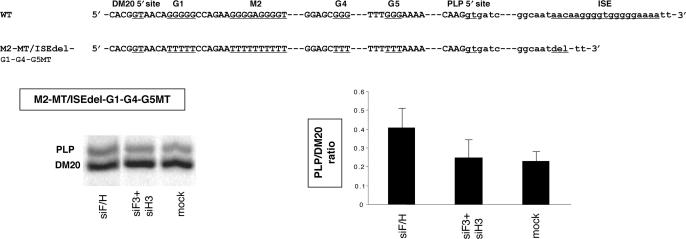
We next determined whether the other G-rich sequences present in exon 3B may mediate the increase in PLP/DM20 ratio derived from M2-MT/ISEdel after knock down of hnRNPH and F. We have replaced G runs in M1 (G1), M6 (G4) and M8 (G5) with T's in the M2-MT/ISEdel (Figure 10A) and determined the PLP/DM20 ratio derived from this construct in transfected Oli-neu cells (Figure 10B). The PLP/DM20 ratio derived from M2-MT/ISEdel/G1-G4-G5-MT was 0.22 which is similar to the ratio derived from the WT (Figure 2A). The PLP/DM20 ratio derived from M2-MT/ISEdel-G1-G4-G5MT in Oli-neu cells treated with siF3 + siH3 did not change compared with mock treated cells (0.27 versus 0.22), while ∼1.6-fold increase was detected after treatment with siF/H (0.4 versus 0.22) (Figure 10B). The data show that mutations of G1, G4 and G5 in addition to M2 almost completely abolish the PLP/DM20 increase induced by knock down of hnRNPH/F and suggest that some or all of these G runs participate in mediating the synergistic effect.
Figure 10.
The effect of mutation of G1, G4 and G5 on the hnRNPH and F-mediated regulation of PLP/DM20. (A) Partial sequences of PLP exon 3B/intron 3 (WT) and of mutations of M2/ISE and G1, G4 and G5 in exon 3B (M2-MT/ISEdel-G1-G4-G5MT) are shown. (B) Representative RT-PCR analysis of the M2-MT/ISEdel-G1-G4-G5MT derived PLP and DM20 products amplified from RNA isolated from Oli-neu cells treated with siF3+H3, siF/H (35 PCR cycles). Mock are cells transfected with M2-MT/ISEdel-G1-G4-G5MT and treated with control siRNA. The bar graph shows the PLP/DM20 ratios ± SD (n = 3).
DISCUSSION
Regulation of PLP alternative splicing and maintenance of the PLP/DM20 ratio are critical for brain function and are relevant to neurological disorders in humans (15). In this study, we have identified a novel G-rich enhancer (M2) of DM20 5′ splice site selection and show that it is active in OLs and non-glial cells. However, the overall efficiency of PLP alternative splicing and the impact that mutation of M2 has on the PLP/DM20 ratio is dependent on cell-specific (compare Oli-neu with L cells) and differentiation-dependent factors (compare OLs versus Oli-neu).
The presence of M2 and ISE downstream of DM20 and PLP 5′ splice sites suggests that the PLP/DM20 ratio may be regulated by a balance between these enhancers. We have addressed this question both functionally and biochemically. When switched, M2 and ISE differ in the strength with which they enhance the upstream 5′ splice site. Changes in the distance of the G-rich sequences from the 5′ splice site in the switched position and the presence of putative ESE in the exonic context and their potential interaction with the G-rich enhancer may account for the difference in strength. However, we favor the hypothesis that sequences immediately adjacent to the G runs may influence the enhancer's strength. In the ISE, the flanking sequences enhance the PLP 5′ site selection (15), while sequences adjacent to M2 reduce the DM20 5′ splice site selection (Figure 2, M1-MT and M3-MT). In addition, the biochemical studies show that some proteins exclusively bind to either M2 or ISE and mutations of the G-rich sequences have a different impact on the binding of proteins to M2F and ISE. Together, these data would support the interpretation that sequence-dependent differences in protein binding may play a role in defining the function of M2 and ISE. Identification and characterization of these proteins in future studies will allow to test this hypothesis.
In this report, we show that the G runs are necessary for binding of hnRNPs to both enhancers. Potential binding motifs for hnRNPH and F are poly G sequences in M2 and ISE (29,33) while hnRNPA1 may bind to a partial consensus sequence -AGGG- (Figure 6) (34) present in single copy in ISE and in double copy in M2. An unexpected finding is that hnRNPL binds to M2 and ISE, since the high affinity site for L is not present in M2 and ISE (35,36) and only partial motifs, CCA in M2F and ACA in the ISE are found. It is possible that hnRNPL does not bind directly to M2 and ISE, but is a part of a molecular complex formed by sequence-specific binding of hnRNPH, F and A1 to these enhancers.
Unlike previous findings that the G triplet is the basic functional unit of G-rich enhancers (28), in M2 the 3′ G triplet exerts a dominant enhancing effect, while the bases separating the G triplets reduce DM20 5′ splice site selection. The functional complexity of M2 may modulate the overall strength of the enhancer in response to changes in RNA-binding proteins during OLs differentiation.
The developmental decrease in hnRNPF, H and A1 in differentiated OLs would support the hypothesis. The hnRNPs are differentially expressed in various tissues (37) and hnRNPH is linked to differentiation of muscle cells (38), however, to our knowledge, coordinate decrease in hnRNPH, F and A1 associated with differentiation was not previously reported.
Removal of both hnRNPH and F has uncovered a novel synergistic effect on PLP alternative splicing regulation mediated by hnRNPH and F, and points to a critical role of hnRNPH in this process. The hnRNPH is sufficient to affect the PLP/DM20 ratio and is necessary for hnRNPF to affect the PLP/DM20 ratio. Although knock down of both hnRNPH and F reduces the shorter of the two products derived by alternative splicing of competing 5′ splice sites in the bclx gene more strongly than knock down of either hnRNPH or F alone, a synergistic effect was not identified in these studies (18). The broader significance of hnRNPH and F synergistic effect in regulating alternative 5′ splice site selection of other genes remains to be investigated. Our findings raise important questions as to whether the synergism of hnRNPF and H is cell-specific and limited to OLs or represents a more general way of regulating 5′ alternative splicing and whether the synergism is gene context specific.
We have started to investigate cis-acting elements that may participate in the hnRNP's mediated regulation and have assessed whether M2 and ISE are necessary for hnRNPH and F to affect the PLP/DM20 ratio. The presence of M2 appears to be required for the effect of hnRNPH, since removal of hnRNPH does not increase the PLP/DM20 ratio when M2 is mutated. In contrast, the synergistic effect of hnRNPH+F knock down depends only in part on the presence of M2, since mutation of M2 either alone or in combination with ISEdel greatly reduces the fold increase in the PLP/DM20 ratio, but does not completely abolish it. However, when the other G runs in exon 3B are mutated the synergistic effect is almost completely eliminated. We interpret these results to indicate that M2 in part contributes to the effect of hnRNPH+F on the PLP/DM20 ratio and that G runs in exon 3 B sequences other than M2 and ISE also play a role. The finding that replacement of G runs in different positions in exon 3B affects the hnRNPH/F-mediated regulation of PLP/DM20 ratio corroborates the data obtained with the M1–M10 substitutions and support that multiple regulatory sequences are present in PLP exon 3B. The role and relative contribution of each G run to the synergistic effect remain to be elucidated.
How M2 may participate in hnRNPH- and F-dependent regulation of PLP alternative splicing remains to be determined. Our data do not demonstrate that hnRNPH and F act directly through M2. For instance, M2 may enhance DM20 5′ splice site selection through a mechanism that is independent from hnRNPH and F such as formation of stacked G structures (39). Loss of the stacked G structure may impair DM20 5′ splice site recognition reducing the efficiency of splicing rather than affecting hnRNPH- and F-mediated trans-activation directly. Alternatively, loss of G stacked structure may indirectly reduce the efficiency of hnRNPH and F binding to other cognate sequences.
Our data suggest that ISE does not participate in the hnRNPF- and H-dependent regulation of the PLP/DM20 ratio at least in undifferentiated OLs. However, it is possible that a role of ISE is overlooked in our studies if reduction of hnRNPH and F needs to be coupled to differentiation-induced changes in other cell-specific and ubiquitous trans-acting splicing factors (40,41).
In summary, our studies have identified a novel G-rich enhancer and a synergistic effect of hnRNPF and H in regulating the PLP/DM20 ratio and suggest that these splicing factors, the G-rich enhancer and other G runs may together regulate PLP/DM20 ratio in OLs. Elucidation of the mechanism that mediates the synergistic interactions of hnRNPH and F may reveal a novel role of these splicing factors in 5′ alternative splicing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
ACKNOWLEDGEMENTS
The authors thank Dr Martha Peterson for helpful discussion and critical reading of the manuscript, Dr Doug Black for critical reading of the manuscript and helpful suggestions, Dr Milcarek for hnRNPF and H antibodies, Dr Dreyfuss for hnRNPA1 antibody and Dr Black for the DUP4-1 construct. This work was supported in part by the European Leukodystrophy Association and by NIH/NINDS (RO1NS053905). Funding to pay the Open Access publication charges for this article was provided by NIH/NINDS (RO1NS053905) and European Leukodystrophy Association (ELA Fondation).
Conflict of interest statement. None declared.
REFERENCES
- 1.Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 2.Sugnet CW, Kent WJ, Ares M, Jr, Haussler D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac. Symp. Biocomput. 2004:66–77. doi: 10.1142/9789812704856_0007. [DOI] [PubMed] [Google Scholar]
- 3.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Xiao X, Van Nostrand E, Burge CB. General and specific functions of exonic splicing silencers in splicing control. Mol. Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black DL, Grabowski PJ. Alternative pre-mRNA splicing and neuronal function. Prog. Mol. Subcell. Biol. 2003;31:187–216. doi: 10.1007/978-3-662-09728-1_7. [DOI] [PubMed] [Google Scholar]
- 6.Grabowski PJ, Black DL. Alternative RNA splicing in the nervous system. Prog. Neurobiol. 2001;65:289–308. doi: 10.1016/s0301-0082(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 7.Nave KA, Lai C, Bloom FE, Milner RJ. Splice site selection in the proteolipid protein (PLP) gene transcript and primary structure of the DM-20 protein of central nervous system myelin. Proc. Natl Acad. Sci. USA. 1987;84:5665–5669. doi: 10.1073/pnas.84.16.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campagnoni AT, Macklin WB. Cellular and molecular aspects of myelin protein gene expression. Mol. Neurobiol. 1988;2:41–89. doi: 10.1007/BF02935632. [DOI] [PubMed] [Google Scholar]
- 9.Timsit SG, Bally-Cuif L, Colman DR, Zalc B. DM-20 mRNA is expressed during the embryonic development of the nervous system of the mouse. ([A-Z]) J. Neurochem. 1992;58:1172–1175. doi: 10.1111/j.1471-4159.1992.tb09378.x. [DOI] [PubMed] [Google Scholar]
- 10.Venkatesh B, Erdmann MV, Brenner S. Molecular synapomorphies resolve evolutionary relationships of extant jawed vertebrates. Proc. Natl Acad. Sci. USA. 2001;98:11382–11387. doi: 10.1073/pnas.201415598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida M, Colman DR. Parallel evolution and coexpression of the proteolipid proteins and protein zero in vertebrate myelin. Neuron. 1996;16:1115–1126. doi: 10.1016/s0896-6273(00)80138-5. [DOI] [PubMed] [Google Scholar]
- 12.Gudz TI, Schneider TE, Haas TA, Macklin WB. Myelin proteolipid protein forms a complex with integrins and may participate in integrin receptor signaling in oligodendrocytes. J. Neurosci. 2002;22:7398–7407. doi: 10.1523/JNEUROSCI.22-17-07398.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stecca B, Southwood CM, Gragerov A, Kelley KA, Friedrich VL, Jr, Gow A. The evolution of lipophilin genes from invertebrates to tetrapods: DM-20 cannot replace proteolipid protein in CNS myelin. J. Neurosci. 2000;20:4002–4010. doi: 10.1523/JNEUROSCI.20-11-04002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hobson GM, Huang Z, Sperle K, Sistermans E, Rogan PK, Garbern JY, Kolodny E, Naidu S, Cambi F. Splice-site contribution in alternative splicing of PLP1 and DM20: molecular studies in oligodendrocytes. Hum. Mutat. 2006;27:69–77. doi: 10.1002/humu.20276. [DOI] [PubMed] [Google Scholar]
- 15.Hobson GM, Huang Z, Sperle K, Stabley DL, Marks HG, Cambi F. A PLP splicing abnormality is associated with an unusual presentation of PMD. Ann. Neurol. 2002;52:477–488. doi: 10.1002/ana.10320. [DOI] [PubMed] [Google Scholar]
- 16.Wang E, Huang Z, Hobson GM, Dimova N, Sperle K, McCullough A, Cambi F. PLP1 alternative splicing in differentiating oligodendrocytes: characterization of an exonic splicing enhancer. J. Cell Biochem. 2006;97:999–1016. doi: 10.1002/jcb.20692. [DOI] [PubMed] [Google Scholar]
- 17.Elrick LL, Humphrey MB, Cooper TA, Berget SM. A short sequence within two purine-rich enhancers determines 5′ splice site specificity. Mol. Cell Biol. 1998;18:343–352. doi: 10.1128/mcb.18.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garneau D, Revil T, Fisette JF, Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 2005;280:22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- 19.Humphrey MB, Bryan J, Cooper TA, Berget SM. A 32-nucleotide exon-splicing enhancer regulates usage of competing 5′ splice sites in a differential internal exon. Mol. Cell Biol. 1995;15:3979–3988. doi: 10.1128/mcb.15.8.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Z, Tang XM, Cambi F. Down-regulation of the retinoblastoma protein (rb) is associated with rat oligodendrocyte differentiation. Mol. Cell Neurosci. 2002;19:250–262. doi: 10.1006/mcne.2001.1077. [DOI] [PubMed] [Google Scholar]
- 21.Jung M, Kramer E, Grzenkowski M, Tang K, Blakemore W, Aguzzi A, Khazaie K, Chlichlia K, von Blankenfeld G, et al. Lines of murine oligodendroglial precursor cells immortalized by an activated neu tyrosine kinase show distinct degrees of interaction with axons in vitro and in vivo. Eur. J. Neurosci. 1995;7:1245–1265. doi: 10.1111/j.1460-9568.1995.tb01115.x. [DOI] [PubMed] [Google Scholar]
- 22.Alkan SA, Martincic K, Milcarek C. The hnRNPs F and H2 bind to similar sequences to influence gene expression. Biochem. J. 2006;393:361–371. doi: 10.1042/BJ20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veraldi KL, Arhin GK, Martincic K, Chung-Ganster LH, Wilusz J, Milcarek C. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell Biol. 2001;21:1228–1238. doi: 10.1128/MCB.21.4.1228-1238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simons M, Kramer EM, Macchi P, Rathke-Hartlieb S, Trotter J, Nave KA, Schulz JB. Overexpression of the myelin proteolipid protein leads to accumulation of cholesterol and proteolipid protein in endosomes/lysosomes: implications for Pelizaeus-Merzbacher disease. J. Cell Biol. 2002;157:327–336. doi: 10.1083/jcb.200110138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang G, Huang SC, Wu JY, Benz EJ. An erythroid differentiation-specific splicing switch in protein 4.1R mediated by the interaction of SF2/ASF with an exonic splicing enhancer. Blood. 2005;105:2146–2153. doi: 10.1182/blood-2004-05-1757. [DOI] [PubMed] [Google Scholar]
- 26.Nalla VK, Rogan PK. Automated splicing mutation analysis by information theory. Hum. Mutat. 2005;25:334–342. doi: 10.1002/humu.20151. [DOI] [PubMed] [Google Scholar]
- 27.Lynch KW, Weiss A. A CD45 polymorphism associated with multiple sclerosis disrupts an exonic splicing silencer. J. Biol. Chem. 2001;276:24341–24347. doi: 10.1074/jbc.M102175200. [DOI] [PubMed] [Google Scholar]
- 28.McCullough AJ, Berget SM. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol. Cell Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caputi M, Zahler AM. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H'/F/2H9 family. J. Biol. Chem. 2001;276:43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- 30.Grabowski PJ. A molecular code for splicing silencing: configurations of guanosine-rich motifs. Biochem. Soc. Trans. 2004;32:924–927. doi: 10.1042/BST0320924. [DOI] [PubMed] [Google Scholar]
- 31.Galarneau A, Richard S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat. Struct. Mol. Biol. 2005;12:691–698. doi: 10.1038/nsmb963. [DOI] [PubMed] [Google Scholar]
- 32.Sporkel O, Uschkureit T, Bussow H, Stoffel W. Oligodendrocytes expressing exclusively the DM20 isoform of the proteolipid protein gene: myelination and development. GLIA. 2002;37:19–30. doi: 10.1002/glia.10014. [DOI] [PubMed] [Google Scholar]
- 33.Matunis MJ, Xing J, Dreyfuss G. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 1994;22:1059–1067. doi: 10.1093/nar/22.6.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanchette M, Chabot B. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 1999;18:1939–1952. doi: 10.1093/emboj/18.7.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hui J, Hung LH, Heiner M, Schreiner S, Neumuller N, Reither G, Haas SA, Bindereif A. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 2005;24:1988–1998. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hui J, Stangl K, Lane WS, Bindereif A. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat. Struct. Biol. 2003;10:33–37. doi: 10.1038/nsb875. [DOI] [PubMed] [Google Scholar]
- 37.Kamma H, Portman DS, Dreyfuss G. Cell type-specific expression of hnRNP proteins. Exp. Cell Res. 1995;221:187–196. doi: 10.1006/excr.1995.1366. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Beqaj S, Yang Y, Honore B, Schuger L. Heterogeneous nuclear ribonucleoprotein-H plays a suppressive role in visceral myogenesis. Mech. Dev. 2001;104:79–87. doi: 10.1016/s0925-4773(01)00377-x. [DOI] [PubMed] [Google Scholar]
- 39.Kostadinov R, Malhotra N, Viotti M, Shine R, D’Antonio L, Bagga P. GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006;34:D119–D124. doi: 10.1093/nar/gkj073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chou MY, Rooke N, Turck CW, Black DL. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.