Abstract
Selenoprotein P (Sel P) is a selenium-rich glycoprotein believed to play a key role in selenium (Se) transport throughout the body. Development of a Sel P knockout mouse model has supported this notion and initial studies have indicated that selenium supply to various tissues is differentially affected by genetic deletion of Sel P. Se in the form of the amino acid, selenocysteine, is incorporated into selenoproteins at UGA codons. Thus, Se availability affects not only selenoprotein levels, but also the turnover of selenoprotein mRNAs via the nonsense-mediated decay pathway. We investigated how genetic deletion of Sel P in mice affected levels of the mRNAs encoding all known members of the murine selenoprotein family, as well as three non-selenoprotein factors involved in their synthesis, selenophosphate synthetase 1 (SPS1), SECIS-binding protein 2 (SBP2) and SECp43. Our findings present a comprehensive description of selenoprotein mRNA expression in the following murine tissues: brain, heart, intestine, kidney, liver, lung, spleen and testes. We also describe how abundance of selenoproteins and selenoprotein-synthesis factors are affected by genetic deletion of Sel P in some of these tissues, providing insight into how the presence of this selenoprotein influences selenoprotein mRNA levels, and thus, the selenoproteome.
INTRODUCTION
Selenium (Se) is an essential dietary trace element that benefits many aspects of human health (1–3). Se is believed to exert its biological effects through its incorporation into selenoproteins as the amino acid, selenocysteine (Sec). To date, 25 selenoproteins have been identified in humans and all but one of these exist as selenocysteine-containing proteins in mice (4). The functions of members of the selenoprotein family elucidated to date include roles in thyroid hormone metabolism, intra- and extra-cellular antioxidation, redox regulation, glucose metabolism, and sperm maturation and protection. However, more comprehensive details regarding tissue distribution and physiological roles for the entire selenoproteome are necessary for a better understanding of the mechanisms by which Se affects human health.
Selenoprotein P (Sel P) is a unique member of the selenoprotein family in that it contains multiple Sec residues per protein molecule. In particular, human and mouse Sel P both contain 10 Sec residues. Culturing cells in Sel P-depleted media was found to reduce activity of the glutathione peroxidase (GPX) family of selenoproteins, and activity was restored following reconstitution of the media with Sel P (5). These facts combined with the finding that a majority of plasma Se is contained within Sel P suggests that this protein plays an important role in transporting Se throughout the body for use in synthesis of other selenoproteins. This notion was supported by two independently developed Sel P-knockout mouse models, both of which exhibited altered tissue distribution of Se (6,7). Hill et al. found that, in terms of Se content, the tissues most affected by Sel P-deficiency were testes and brain, while kidney and heart were less affected. The phenotype of the knockout mice was consistent with these findings and included neurological problems and male sterility. Sel P-knockout mice injected with 75Se had lower levels of isotope in brain and testes, but higher accumulation in livers compared to controls. These findings suggested that the liver is a tissue that readily takes up Se and incorporates it into Sel P, which is then secreted into the plasma for transport of Se to other tissues. This was supported by studies of Schweitzer et al. demonstrating that liver-specific inactivation of the gene encoding Sec tRNA (trsp) resulted in decreased plasma and kidney GPX activity (8). In contrast, GPX activity in brain tissue in these mice remained unaffected, suggesting that liver Sel P is not required to the same extent for expression of different selenoproteins in different tissues.
Synthesis of selenoproteins is regulated at the levels of mRNA transcription and stability, and protein translation (9–12). Turnover of mRNA is a particularly important point of regulation in that selenoprotein mRNAs require recoding of UGAs within the coding regions from stop codons to Sec-insertion sites. Under circumstances of low selenium, some selenoprotein mRNAs are degraded through nonsense-mediated decay (NMD), a pathway that degrades mRNAs containing premature stop codons (13). Se-deficiency decreases the efficiency with which Sec incorporation is directed by the Sec incorporation machinery and increases the efficiency with which the codons are recognized as nonsense, thus eliciting NMD (13,14). Sensitivity to NMD is determined by the location of termination codons in the mRNA in relation to exon-junction complexes, which are deposited upstream of exon–exon boundaries during mRNA splicing and export. In higher eukaryotes, a nonsense codon is usually recognized as premature if it is located more than 50–55 nt upstream of the last intron in the pre-mRNA (13,15).
Interestingly, Se-deficient conditions result in degradation of different selenoprotein mRNAs to different extents. Several studies have shown that, within the GPX family of proteins (GPX1 through 4), Se-deficiency leads to degradation of GPX1 and GPX3 mRNAs to lower levels than GPX2 and GPX4 mRNAs (16–18). Other studies have supported a similar notion that GPX1 mRNA levels are decreased by Se-deficiency while other GPX mRNAs are relatively unchanged (19,20). However, how expression of other members of the selenoprotein family or the protein factors involved in their synthesis is affected by Se availability in different tissues remains unknown.
Given the important roles that Se transport and metabolism play in selenoprotein expression, we investigated the relative levels of mRNA abundance in eight different mouse tissues and how these levels are affected by genetic deletion of Sel P. Our results describe mRNA levels for selenoproteins in different mouse tissues including brain, heart, intestine, kidney, liver, lung, spleen and testes. In addition, deletion of Sel P resulted in lower levels of many selenoprotein mRNAs and some proteins in brain and testes, but not in heart and lung. Taken together, the results provide insight into how the presence of Sel P influences the different selenoproteins in different tissues.
MATERIALS AND METHODS
Mice and tissue collection
Sel P−/+ mice on a C57Bl/6 background were kindly provided by Drs Raymond Burk and Kristina Hill and were used to generate a colony of mice here at the University of Hawaii. Genotyping of the mice was carried out using methods previously described (6) and heterozygous breeding pairs were used to produce litters consisting of pups homozygous for Sel P deletion (Sel P−/−) as well as wild-type littermate controls (Sel P+/+). Male weanlings were fed standard mouse chow until 8 to 10 weeks of age, when they were sacrificed and eight tissues quickly harvested. This was carried out prior to onset of any apparent neurological disorders. Each tissue was immediately washed in PBS and frozen in liquid nitrogen. Tissues were ground into powder on dry ice, which was then divided into separate tubes for RNA or protein extraction.
RNA extraction and real-time qPCR
Frozen, powdered tissue samples were thawed and RNA extracted using RNeasy Mini kit and RNase-free DNase I (all from Qiagen, Valencia, CA, USA). Concentration and purity of extracted RNA was determined using A260/A280 measured on an ND1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Synthesis of cDNA was carried out using Superscript III (Invitrogen, Carlsbad, CA, USA) and oligo dT primer, with 2 μg RNA per 50 μl reaction. For real-time PCR, 1 μl of the cDNA was used in 10 μl reactions with Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen). Reactions were carried out in a LightCycler 2.0 thermal cycler (Roche Applied Biosystems, Indianapolis, IN, USA). Oligonucleotides used for qPCR are listed in Table 1. Cycling conditions were used as suggested in the SYBR Green kit instructions and results analyzed using Relative Quantification Software (Roche).
Table 1.
Oligonucleotide primers used for real-time PCR
| Name of target sequence | GenBank accession number | PCR product length | ||
|---|---|---|---|---|
| hprt | NM_013556.2 | fwd | TCCTCCTCAGACCGCTTTT | 90 |
| rev | CCTGGTTCATCGCTAATC | |||
| gapdh | NM_001001303 | fwd | TGACATCAAGAAGGTGGTGAAGC | 203 |
| rev | CCCTGTTGCTGTAGCCGTATTC | |||
| actin | NM_007393.1 | fwd | TGACAGGATGCAGAAGGAGA | 75 |
| rev | CGCTCAGGAGGAGCAATG | |||
| ubc | NM_019639.3 | fwd | GAGGGCATCTCCCCTGAC | 60 |
| rev | GCCATCTTCCAGCTGCTT | |||
| gpx1 | NM_008160.2 | fwd | ACAGTCCACCGTGTATGCCTTC | 238 |
| rev | CTCTTCATTCTTGCCATTCTCCTG | |||
| gpx2 | NM_030677.1 | fwd | GTTCTCGGCTTCCCTTGC | 64 |
| rev | TCAGGATCTCCTCGTTCTGAC | |||
| gpx3 | NM_008161.1 | fwd | ATTTGGCTTGGTCATTCTGG | 105 |
| rev | CCACCTGGTCGAACATACTTG | |||
| gpx4 | NM_001037741.1 | fwd | TCTGTGTAAATGGGGACGATGC | 174 |
| rev | TCTCTATCACCTGGGGCTCCTC | |||
| sel H | NM_001033166.2 | fwd | GGAAGAAAGCGTAAGGCGGG | 191 |
| rev | GGTTTGGACGGGTTCACTTGC | |||
| sel I | NM_027652.1 | fwd | CCTGACATACTTCGACCCTGA | 63 |
| rev | CAGTCAGGCACATGCTTATGA | |||
| sel K | NM_019979.1 | fwd | TCCACGAAGAATGGGTAGGA | 94 |
| rev | GCTTCTCAGAGCAGACATTTACCT | |||
| sel M | NM_053267 | fwd | TGACAGTTGAATCGCCTAAAGGAG | 113 |
| rev | AACAGCACGAGTTCGGGGTC | |||
| sel N | NM_029100.1 | fwd | ACCTGGTCCCTGGTAAAGGAGC | 177 |
| rev | GGTGATGTCAAGGAAGTAGTTGGC | |||
| sel O | NM_027905.2 | fwd | GGCTGCCCATACCTGTGA | 240 |
| rev | CGTGCTGTTCGCTGTGTC | |||
| sel P | NM_009155.2 | fwd | CCTTGGTTTGCCTTACTCCTTCC | 199 |
| rev | TTTGTTGTGGTGTTTGTGGTGG | |||
| sel R | NM_013759.1 | fwd | TTCGTCCCTAAAGGCAAAGA | 62 |
| rev | CATTCGCAGTCCATGTCCTA | |||
| sel S | NM_024439.2 | fwd | CAGAAGATTGAAATGTGGGACAGC | 116 |
| rev | CCTTTGGGGATGACAGATGAAGTAG | |||
| sel T | XR_004549.1 | fwd | TGAGGCTCCTGCTGCTTC | 110 |
| rev | GGTGGCGTACTGCATCTTTAAT | |||
| sel V | NM_175033.3 | fwd | CAGGAGGAAGCATGCAAGA | 62 |
| rev | TGGTTTGGCAGCATTTCAG | |||
| sel W | NM_009156.2 | fwd | GGTGCCTCCCCAGAATCTAC | 136 |
| rev | TGGGGGAATTCAGAGAGAGA | |||
| sps1 | NM_175400.5 | fwd | TGAACTGAAAGGCACAGGCTGC | 143 |
| rev | CGCAAGTATCCATCCCAATGC | |||
| sps2 | NM_009266.2 | fwd | ACCGACTTCTTTTACCCCTTGG | 166 |
| rev | TCACCTTCTCTCGTTCCTTTTCAC | |||
| tr1 | NM_015762.1 | fwd | CCTATGTCGCCTTGGAATGTGC | 244 |
| rev | ATGGTCTCCTCGCTGTTTGTGG | |||
| tr2 | NM_013711.1 | fwd | GTTCCCCACATCTATGCCATTG | 114 |
| rev | GGTTGAGGATTTCCCAAAGAGC | |||
| tr3 | NM_153162.2 | fwd | CTTTGCAAGATGCCAAGAAA | 69 |
| rev | TCATGGCCTCCCAGTTGT | |||
| dio1 | NM_007860.2 | fwd | GCCTCTCAGGACAGAAGTGC | 73 |
| rev | CTGCCAAAGTTCAACACCAG | |||
| dio2 | NM_010050.1 | fwd | CTGCGCTGTGTCTGGAAC | 67 |
| rev | GGAGCATCTTCACCCAGTTT | |||
| dio3 | NM_172119.1 | fwd | GGAGGTTGTCCGACCTGAT | 60 |
| rev | GTCCCTTGTGCGTAGTCGAG | |||
| sep15 | NM_053102.1 | fwd | GCTGTCAGGAAGAAGCACAA | 74 |
| rev | TTTTCATCCGCAGACTTCAA | |||
| sbp2 | XM_127336.5 | fwd | GAAGCAGAGGGAGATACCTAAGGC | 110 |
| rev | GGCTCACAGCACTTTCTTGGAG | |||
| secp43 | NM_027925.2 | fwd | ACCGTGATGAGCGTCAAAAT | 74 |
| rev | GCGAATTCCACAAAGCAGTAG |
Protein extraction and western blots
Protein was extracted from tissue by homogenizing 0.5 g of tissue on ice in 10 ml of CellLytic MT buffer (Sigma) containing 1 mM DTT, 1X protease inhibitor cocktail (Calbiochem, San Diego, CA, USA), and 5 mM EDTA. Homogenate was centrifuged at 12 000 r.p.m. (13 000 g; Beckman Coulter microcentrifuge) for 10 min and supernatant removed and stored at −70°C. Bradford assay was carried out using Bradford Reagent (Bio-Rad, Hercules, CA, USA) and 30 μg total protein was combined with reduced Laemeli buffer, boiled at 95°C for 10 min, cooled on ice, and loaded into wells of 10–14.5% polyacrylamide gels (Bio-Rad). Protein was transferred to PVDF membranes, which were blocked for 1 h with 5% BSA and then probed for 1 h with primary antibodies, including rabbit polyclonal anti-GPX1 and anti-GPX4 (Lab Frontier, Seoul, Korea), rabbit anti-Sel W raised against the peptide: SKKRGDGYVDTESKFRK (custom synthesized and affinity purified by ProSci, Inc., Poway, CA, USA), mouse monoclonal IgG anti-β-actin (Sigma) and anti-α-tubulin (Novus Biologicals, Littleton, CO, USA). Appropriate HRP-conjugated secondary antibodies were purchased from Jackson Immunolabs (West Grove, PA), incubated with the membranes for 45 min and detected using ECL Plus (GE Healthcare). In further attempts to detect Sel W in heart and lung, immunoprecipitation reactions were carried out to concentrate any Sel W that might be present in protein extracts from these tissues, followed by electrophoresis and western blotting. However, these and other methods did not result in detection of Sel W in these two tissues except for questionable bands upon overexposure of the film. For densitometry, digital images of autoradiographic film were captured using Gel Logic 200 and Kodak MI software (Kodak Scientific Imaging Systems, Rochester, NY, USA). This software was used to measure mean intensity from regions of interest (ROI) that corresponded to bands to be measured. The intensity of the target bands (e.g. GPX1 band) was normalized to that of the loading control band (e.g. β-actin band) to obtain normalized levels of target proteins.
Statistical analyses
All statistical tests were performed using GraphPad Prism version 4.0 for Windows (GraphPad Software, San Diego, CA, USA). Means of two groups were compared using a student's t-test and significance was considered at P < 0.05.
RESULTS
Evaluation of mRNA levels for selenoproteins in various mouse tissues
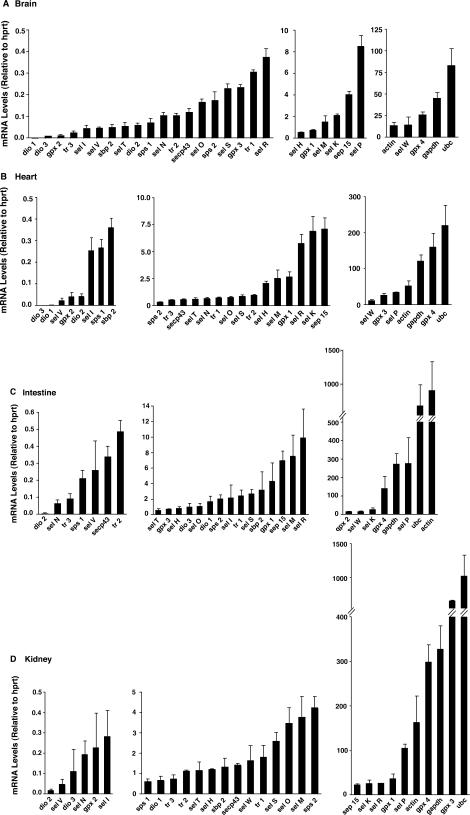
We first set out to evaluate mRNA levels for the entire murine selenoproteome in eight different mouse tissues: brain, testes, liver, kidney, lung, heart, intestine and spleen. To achieve this goal, we utilized real-time PCR and included oligonucleotides specific for all 24 murine selenoproteins, three non-selenoprotein factors involved in their synthesis (sbp2, secp43 and sps1), and four housekeeping genes: hypoxanthinephosphoribosyltransferase (hprt), ubiquitin c (UBC), β-actin (actin) and glyceraldehyde-3-phosphate dehydrogenase (gapdh) (Table 1). Because levels of hprt were most consistent between different tissues and different mice, this housekeeping gene product was used to normalize the abundance of other mRNAs. Results for the complete list of mRNA levels in the eight different tissues are shown in Figure 1. The mRNAs for each tissue were divided into low (<0.5), medium (0.5 − 10) and high (>10) copy groups, based on levels of mRNAs relative to the stable hprt housekeeping mRNA. Use of these cutoff values facilitated comparative analyses of selenoproteome and housekeeping mRNA levels that ranged from rare to highly abundant transcripts.
Figure 1.
Abundance of mRNA for selenoproteins, selenoprotein-synthesis factors, and housekeeping genes. Total RNA was extracted from brain (A), heart (B), intestine (C), kidney (D), liver (E), lung (F), spleen (G), and testes (H), which were used for cDNA synthesis and real-time PCR analysis. Levels of each target mRNA relative to hprt are shown for each tissue and grouped into either low copy (average <0.5) on the left, medium copy (average 0.5–10) in the middle and high copy (average >10) on the right. Number of mice used for each tissue were 3 (intestine, kidney, liver and spleen), 4 (brain and testes), 5 (heart) or 6 (lung) and results represent mean +SE.
While the specific pattern of mRNA abundance differs between tissues, there are some common features of selenoproteome mRNAs when comparing the eight different tissues. First, gpx4 is among the mRNAs detected at the highest levels for nearly all tissues examined. This reinforces the notion that this antioxidant enzyme may be essential for nearly all cell types and tissues, consistent with the embryonic lethal phenotype displayed by the gpx4 knockout mouse model (21). Another mRNA near the top of the abundance hierarchy is sel P, except in the spleen and testes, which both contain sel P as a medium copy mRNA. Sel P mRNA is detected at particularly high levels in the liver. This finding is consistent with other studies demonstrating the liver as a source of a majority of sel P found in plasma (22). However, it is interesting to note the relatively high levels of sel P message in most tissues, including the kidney, which was shown to be a tissue particularly dependent on hepatically derived Sel P protein as a Se source (8).
Another common feature of mRNA abundance shared among the different tissues is the relatively low levels of all three deiodinase enzymes (dio1, 2 and 3). The lone exception to this is in liver, where dio1 is a medium copy mRNA. Also, sel V expression is low in most tissues, except in testes where it is detected at very high levels, a finding that is consistent with previous results (4). The selenoprotein-synthesis factors that we included in this study were detected at relatively low levels, suggesting high specific activity, low requirement or rapid mRNA turnover. The only notable exception was sps2, which is both a selenoprotein and a factor involved in synthesis of other selenoproteins. Sps2 mRNA was detected at relatively high levels in the liver, where it was the fifth most abundant selenoprotein mRNA.
After evaluating the results in Figure 1 for similarities, we next turned our attention to the differences found between the various tissues. One result that is apparent is the high abundance of gpx3 mRNA in the kidney, which is consistent with previously reported results (23). A surprising finding is that the heart also displays a relatively high level of gpx3 mRNA. The liver has high sel P mRNA levels compared to other tissues and confirms previous findings suggesting that this tissue produces high quantities of Sel P protein for secretion into blood plasma. Both liver and kidney are relatively high in abundance of mRNA for sel R, a selenoprotein that has been characterized as a zinc-containing stereo-specific methionine sulfoxide reductase (24). Two selenoprotein mRNAs present at high levels in the testes compared to other tissues are sel K and tr3.
Effects of genetic deletion of selenoprotein P on expression of selenoproteins in different tissues
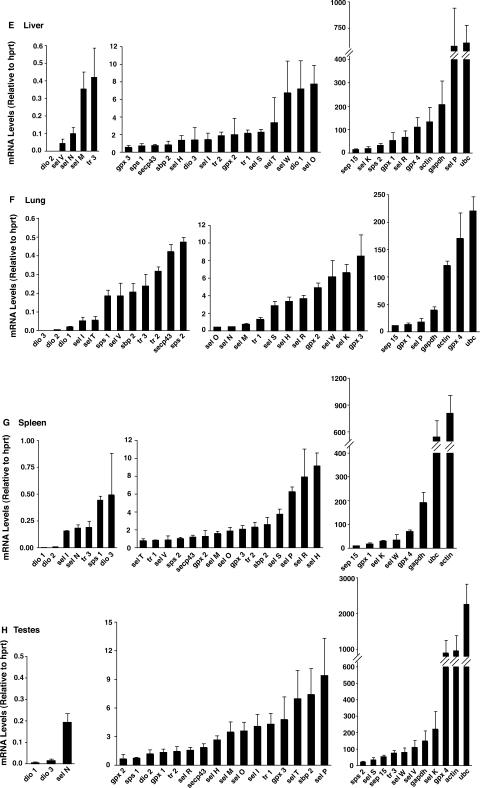
We next investigated how deletion of Sel P in mice affects the mRNA levels of the selenoproteins and non-selenoprotein synthesis factors in four different tissues: brain, testes, heart and lung. These particular tissues were chosen due to the phenotype of the Sel P−/− mice. These mice have been shown to demonstrate neurological disorders and sterility, but have no apparent physiological problems related to the lung or heart (6,7). In this manner, we set out to compare mRNA abundance for the selenoproteins in two tissues involved with the phenotypical problems (brain and testes) and two seemingly unaffected tissues (heart and lung).
Results from our comparisons are displayed in Figure 2 and reveal several interesting features regarding how genetic deletion of Sel P affects different tissues. First, both testes and brain displayed overall decreases in selenoprotein mRNA abundance as well as decreases in housekeeping mRNAs. Although statistical significance was not achieved for most of the mRNAs due to variability and low numbers of mice per group (N = 3 − 6), the brain and testes were clearly more affected than heart and lung. This was particularly evident for mRNAs for the medium and high copy genes and may reflect apoptotic or necrotic cellular damage, which may be involved in or lead to the physiological problems in these tissues. In contrast, in heart and lung mRNA abundance was decreased only in a small number of selenoproteins expressed in the low copy group. The medium and high copy selenoprotein mRNAs in heart and lung are much less affected by genetic deletion of Sel P.
Figure 2.
Comparison of Sel P knockout to wild-type mice for abundance of mRNA for selenoproteins, selenoprotein-synthesis factors and housekeeping genes. Total RNA was extracted from heart (A), lung (B), brain (C), and testes (D), which were used for cDNA synthesis and real-time PCR analysis. Levels of each target mRNA relative to hprt are shown for each tissue and grouped into either low copy (average <0.5) on the left, medium copy (average 0.5–10) in the middle and high copy (average >10) on the right. Black bars represent wild-type mice and white bars represent Sel P knockout mice. Number of mice used for each wild-type tissue were, 4 (brain and testes), 5 (heart) or 6 (lung) and for all 3 knockout tissues. Results represent mean +SE.
Other interesting features that emerge from these results include the changes in sel W and gpx4 mRNA levels. In brain and testes, the levels decreased dramatically in Sel P−/− compared to Sel P+/+ mice, while in lung and heart they increased. Other mRNA levels also increased in response to genetic deletion of Sel P. For example, sps2 increased in three tissues (brain, testes, and heart) in Sel P−/− versus Sel P+/+ mice. Both sps2 and sep15 are high copy mRNAs in testes, and both demonstrate high resistance to effects of Sel P knockout in this tissue.
Lastly, the brain and testes tissue from Sel P−/− mice appear to have low, but detectable levels of sel P mRNA (Figure 2C and D). Levels of sel P mRNA transcripts should be near zero considering the oligonucleotides used for real-time PCR (exons 4 and 5) are located downstream of the neo cassette insertion site (exon 2) (6). Insertion of the neo cassette results in a premature stop codon, which leads to degradation of the transcript. Thus, for some tissues the rate at which the transcript containing the neo cassette is degraded may be slower than others, resulting in detection of transcript en route to NMD degradation.
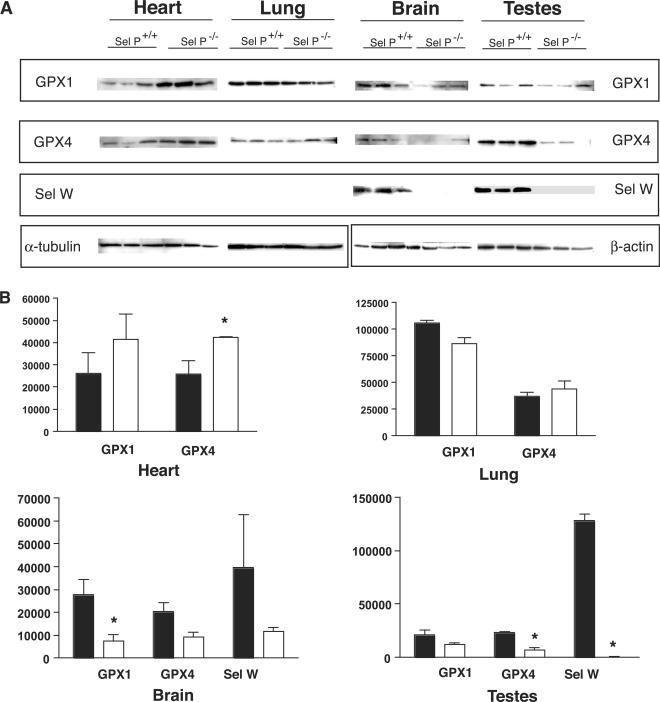
Analyses of protein levels in heart, lung, brain and testes
To further investigate the effects of genetic deletion of Sel P on the expression of selenoproteins in different tissues, we used western blot analyses to measure levels of three selenoproteins: GPX1, GPX4 and Sel W. As described above, we included two physiologically affected tissues (brain and testes) and two seemingly unaffected tissues (lung and heart) in these analyses.
As shown in Figure 3A, levels of all three proteins are decreased in brain and testes. Using densitometry to quantify the levels of proteins, we found significant reductions in GPX1 in brain and both GPX4 and Sel W in testes. These results are consistent with the mRNA results described above. The results from lung and heart tissue differ from brain and testes in several ways. First, the antibody that detects Sel W in brain and testes does not detect Sel W in lung and heart. This could be due to different isoforms of Sel W expressed in the different tissues, as mRNA levels for this selenoprotein were relatively high in heart and lung. A second feature that is evident from the western blot analyses is that the decreased levels of GPX1 and GPX4 detected in Sel P+/+ brain and testes tissues are not detected in lung and heart, which is consistent with the mRNA levels described above. In fact, GPX4 levels are significantly increased in heart tissue from Sel P−/− mice and GPX1 levels show a trend toward an increase in the knockout mice. These results are similar to those found at the mRNA level and raise the question of why these two members of the GPX family may increase in heart with the deletion of Sel P.
Figure 3.
Comparison of GPX1, GPX4, and Sel W levels in Sel P knockout versus wild-type mice. (A) Protein was extracted from heart, lung, brain and testes, which were then used for western blot analysis. Rabbit polyclonal antibodies were used to detect GPX1, GPX4 and Sel W for three Sel P knockout mice and three wild-type mice for each tissue. Equivalent protein loading was determined using α-tubulin for heart and lung and β-actin for brain and testes. (B) Densitometry was used to quantify western blot data as described in the Methods section. Results represent mean ±SE. Statistical significance was determined by student's t-test (*P < 0.05) Black and white bars represent wildtype (N=3) and knockout (N=3) mice, respectively.
DISCUSSION
The study presented herein provides a comprehensive description of selenoprotein mRNA abundance for eight murine tissues including brain, testes, kidney, liver, spleen, intestine, lung, and heart. This multi-tissue analysis allows comparisons to be made within and between tissues for relative levels of the different selenoprotein mRNAs. Our results suggest that certain selenoprotein mRNAs (e.g. gpx4 and sel P) are found at high levels in most of these tissues, while others are generally expressed at very low levels (e.g. deiodinase enzymes, dio1-3). Differences in mRNA levels may be due to multiple factors acting individually or in combination. Stability of mRNA has been shown to play a key role in regulation of selenoprotein synthesis (25). Selenoprotein mRNA stabilities are affected differentially by limiting availability of Se or selenoprotein synthesis factors (25–27). The degradation of these and other mRNAs is believed to be carried out by NMD (10,15,28). Differences in susceptibility to degradation may lie in the presence of cis-elements in the selenoprotein mRNA that either lead to recognition by the NMD machinery, affect their ability to compete for stabilizing factors such as SBP2, or alter subcellular localization (13,14).
In addition to mRNA stability, differential transcription may contribute to varying mRNA levels for the various selenoproteins. Preliminary analysis of the promoters of selenoprotein genes suggests that several of these may be under control of shared regulatory pathways, including those upregulated under conditions of stress (29–32). In particular, the genes for several selenoprotein mRNAs exhibiting increased expression in Sel P−/− tissues share several common transcription factor motifs that may result in their upregulation (manuscript in preparation). In addition, multiple transcripts are annotated in the mouse genome for many of the selenoprotein genes, and the tissue expression patterns of many of these transcripts are not well characterized. These are relatively unexplored but undoubtedly important areas for future investigation.
The main goal of this study was to determine the relationship between Sel P and the expression of other selenoprotein family members in different tissues. Because of the Se transport role that Sel P has been demonstrated to play, the Sel P knockout mouse model provides a useful tool to investigate how the absence of this protein affects expression of the different selenoprotein family members in various tissues. Our data suggest that brain and testes are similarly affected by genetic deletion of Sel P in mice. In both tissues, Sel P deletion leads to a generalized reduction in abundance of all medium and high copy selenoprotein mRNAs. The substantial decrease in levels of most selenoprotein mRNAs in these two tissues in Sel P−/− mice suggests that Sel P is important for transport of Se within or to these tissues and is consistent with the neurological and sterility problems observed in the Sel P−/− mice. Recent findings by the Burk lab further support the role of Sel P in transporting Se to the brain and testes (33,34). Olson et al. have proposed a mechanism in the testes by which Sel P is taken into cells of this Se-sensitive tissue via the apolipoprotein E receptor (35). In contrast to the brain and testes, heart and lung tissue from Sel P−/− mice did not demonstrate decreased selenoprotein mRNA levels. In fact, gpx4 and sel W mRNA levels were increased in heart and lung from Sel P−/− mice compared to Sel P+/+ controls. In heart, GPX4 protein was also found to be significantly increased in mice lacking Sel P. This may be due to a feedback mechanism by which transcription of these two family members is upregulated during increased oxidative stress in these two tissues.
When comparing our results involving Sel P deletion to those involving dietary Se-deficiency, some interesting differences are evident. For example, studies in rats have demonstrated that Se-deficiency results in decreased expression of GPX-1 in several tissues including liver, kidney, heart and colon (36–38). In contrast, GPX-4 levels were found to be unaffected or less affected by Se-deficiency in most of these tissues, except in colon where GPX-4 was decreased at nearly equivalent levels as GPX-1. Our results suggest that Sel P deletion in mice affected GPX-4 and GPX-1 similarly in that they were lowered in brain and testes, but increased in heart. Furthermore, Se-deficiency in rats has been shown to decrease mRNA levels for Sel W in colon, similar to our results showing a decrease in Sel W mRNA and protein levels in brain and testes from Sel P-deficient mice compared to wild-type controls. Differences between species may account for some of these differences and further investigation is needed to fully understand how deficiencies in dietary Se compare with decreases in Sel P levels.
An interesting finding of this study was the detection by western blot of Sel W in brain and testes, but the conspicuous absence of this selenoprotein in the heart and lung. The rabbit antibody used for this assay was specific for a 17 amino acid region in the C-terminal portion of the protein. Despite several different approaches including immunoprecipitation concentration of protein extracts prior to western blots, the antibody failed to detect Sel W in the heart and lung. This suggests that, despite high mRNA levels in heart and lung, the protein is not expressed at detectable levels in these tissues. This is consistent with previous findings that Sel W was detected by western blot in muscle, spleen, brain and testes only (39). Interestingly, we used this antibody raised against murine Sel W to detect human Sel W on a commercially purchased human tissue blot and found that it detected the protein in nearly all tissues except for ovaries and testes (data not shown). This occurred despite a two amino acid mismatch between the antigenic region in mouse and human Sel W. It remains unclear whether different isoforms of Sel W are expressed in different tissues, but differences between rodents and primates have been previously noted (40).
The selenoprotein-synthesis factors analyzed in this study included sbp2, secp43, sps1 and sps2. These factors were detected at relatively low to moderate levels, suggesting high specific activity, low requirement or rapid mRNA turnover. The only notable exception was sps2, which is both a selenoprotein and a factor involved in synthesis of other selenoproteins. Sps2 mRNA was detected at relatively high levels in the liver, where it was the fifth most abundant selenoprotein mRNA. This may be an adaptation to relatively high levels of selenoprotein synthesis occurring in this tissue. Given that testes would seemingly require high levels of selenoprotein synthesis, it was surprising that this tissue exhibited only low to moderate abundance for the synthesis factors.
Importantly, our findings are based on comparisons between target mRNAs and the most stable housekeeping mRNA, hprt. Using other housekeeping mRNAs as comparison transcripts produces different results. However, results from our protein analysis described above correlate best with the mRNA data obtained with the stable hprt as the comparison transcript.
Overall, our study provides an extensive comparison of the selenoprotein mRNA abundance in terms of tissue distribution in the mouse under normal conditions and, in four tissues, under conditions of sel P deletion. Determining exactly how tissues cope differentially with changes in Se availability, metabolism and transport will lead to a better understanding of how expression of the selenoproteome throughout the organism contributes to its overall health status.
ACKNOWLEDGEMENTS
We are grateful to Drs Raymond Burk and Kristina Hill for providing the SelP knockout mice. Also, we thank Dr Ronald J. Koenig for his help in developing reagents and protocols and Dr Zoia Stoytcheva for her assistance with genome database searches. This publication was made possible by NIH awards R01 NS40302, DK47320 and DK52963. Funding to pay the Open Access publication charges for this article was provided by the three NIH awards listed above.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rayman MP. The importance of selenium to human health. Lancet. 2000;356:233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 2.Driscoll DM, Copeland PR. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- 3.Hatfield DL, Carlson BA, Xu XM, Mix H, Gladyshev VN. Selenocysteine incorporation machinery and the role of selenoproteins in development and health. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:97–142. doi: 10.1016/S0079-6603(06)81003-2. [DOI] [PubMed] [Google Scholar]
- 4.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 5.Saito Y, Takahashi K. Characterization of selenoprotein P as a selenium supply protein. Eur. J. Biochem. 2002;269:5746–5751. doi: 10.1046/j.1432-1033.2002.03298.x. [DOI] [PubMed] [Google Scholar]
- 6.Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, Gesteland RF, Burk RF. Deletion of selenoprotein P alters distribution of selenium in the mouse. J. Biol. Chem. 2003;278:13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- 7.Schomburg L, Schweizer U, Holtmann B, Flohe L, Sendtner M, Kohrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem. J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schweizer U, Streckfuss F, Pelt P, Carlson BA, Hatfield DL, Kohrle J, Schomburg L. Hepatically derived selenoprotein P is a key factor for kidney but not for brain selenium supply. Biochem. J. 2005;386:221–226. doi: 10.1042/BJ20041973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dreher I, Jakobs TC, Kohrle J. Cloning and characterization of the human selenoprotein P promoter. Response of selenoprotein P expression to cytokines in liver cells. J. Biol. Chem. 1997;272:29364–29371. doi: 10.1074/jbc.272.46.29364. [DOI] [PubMed] [Google Scholar]
- 10.Copeland PR. Regulation of gene expression by stop codon recoding: selenocysteine. Gene. 2003;312:17–25. doi: 10.1016/s0378-1119(03)00588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Jesus LA, Hoffmann PR, Michaud T, Forry EP, Small-Howard A, Stillwell RJ, Morozova N, Harney JW, Berry MJ. Nuclear assembly of UGA decoding complexes on selenoprotein mRNAs: a mechanism for eluding nonsense-mediated decay? Mol. Cell. Biol. 2006;26:1795–1805. doi: 10.1128/MCB.26.5.1795-1805.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Small-Howard A, Morozova N, Stoytcheva Z, Forry EP, Mansell JB, Harney JW, Carlson BA, Xu XM, Hatfield DL, et al. Supramolecular complexes mediate selenocysteine incorporation in vivo. Mol. Cell. Biol. 2006;26:2337–2346. doi: 10.1128/MCB.26.6.2337-2346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maquat LE. Evidence that selenium deficiency results in the cytoplasmic decay of GPx1 mRNA dependent on pre-mRNA splicing proteins bound to the mRNA exon-exon junction. Biofactors. 2001;14:37–42. doi: 10.1002/biof.5520140106. [DOI] [PubMed] [Google Scholar]
- 14.Weiss Sachdev S, Sunde RA. Selenium regulation of transcript abundance and translational efficiency of glutathione peroxidase-1 and -4 in rat liver. Biochem. J. 2001;357:851–858. doi: 10.1042/0264-6021:3570851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 16.Weiss SL, Sunde RA. Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. RNA. 1998;4:816–827. doi: 10.1017/s1355838298971990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wingler K, Muller C, Brigelius-Flohe R. Stability of gastrointestinal glutathione peroxidase mRNA in selenium deficiency depends on its 3′UTR. Biofactors. 2001;14:43–50. doi: 10.1002/biof.5520140107. [DOI] [PubMed] [Google Scholar]
- 18.Muller C, Wingler K, Brigelius-Flohe R. 3′UTRs of glutathione peroxidases differentially affect selenium-dependent mRNA stability and selenocysteine incorporation efficiency. Biol. Chem. 2003;384:11–18. doi: 10.1515/BC.2003.002. [DOI] [PubMed] [Google Scholar]
- 19.Hill KE, Lyons PR, Burk RF. Differential regulation of rat liver selenoprotein mRNAs in selenium deficiency. Biochem. Biophys. Res. Commun. 1992;185:260–263. doi: 10.1016/s0006-291x(05)80984-2. [DOI] [PubMed] [Google Scholar]
- 20.Lei XG, Evenson JK, Thompson KM, Sunde RA. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J. Nutr. 1995;125:1438–1446. doi: 10.1093/jn/125.6.1438. [DOI] [PubMed] [Google Scholar]
- 21.Yant LJ, Ran Q, Rao L, Van Remmen H, Shibatani T, Belter JG, Motta L, Richardson A, Prolla TA. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free. Radic. Biol. Med. 2003;34:496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- 22.Burk RF, Hill KE. Selenoprotein P: an extracellular protein with unique physical characteristics and a role in selenium homeostasis. Annu. Rev. Nutr. 2005;25:215–235. doi: 10.1146/annurev.nutr.24.012003.132120. [DOI] [PubMed] [Google Scholar]
- 23.Avissar N, Ornt DB, Yagil Y, Horowitz S, Watkins RH, Kerl EA, Takahashi K, Palmer IS, Cohen HJ. Human kidney proximal tubules are the main source of plasma glutathione peroxidase. Am. J. Physiol. 1994;266:C367–C375. doi: 10.1152/ajpcell.1994.266.2.C367. [DOI] [PubMed] [Google Scholar]
- 24.Kryukov GV, Kumar RA, Koc A, Sun Z, Gladyshev VN. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc. Natl Acad. Sci. USA. 2002;99:4245–4250. doi: 10.1073/pnas.072603099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss SL, Sunde RA. Selenium regulation of classical glutathione peroxidase expression requires the 3′ untranslated region in Chinese hamster ovary cells. J. Nutr. 1997;127:1304–1310. doi: 10.1093/jn/127.7.1304. [DOI] [PubMed] [Google Scholar]
- 26.Christensen MJ, Burgener KW. Dietary selenium stabilizes glutathione peroxidase mRNA in rat liver. J. Nutr. 1992;122:1620–1626. doi: 10.1093/jn/122.8.1620. [DOI] [PubMed] [Google Scholar]
- 27.Moriarty PM, Reddy CC, Maquat LE. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol. Cell. Biol. 1998;18:2932–2939. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun X, Li X, Moriarty PM, Henics T, LaDuca JP, Maquat LE. Nonsense-mediated decay of mRNA for the selenoprotein phospholipid hydroperoxide glutathione peroxidase is detectable in cultured cells but masked or inhibited in rat tissues. Mol. Biol. Cell. 2001;12:1009–1017. doi: 10.1091/mbc.12.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mostert V, Wolff S, Dreher I, Kohrle J, Abel J. Identification of an element within the promoter of human selenoprotein P responsive to transforming growth factor-beta. Eur. J. Biochem. 2001;268:6176–6181. doi: 10.1046/j.0014-2956.2001.02565.x. [DOI] [PubMed] [Google Scholar]
- 30.Rundlof AK, Arner ES. Regulation of the mammalian selenoprotein thioredoxin reductase 1 in relation to cellular phenotype, growth, and signaling events. Antioxid. Redox. Signal. 2004;6:41–52. doi: 10.1089/152308604771978336. [DOI] [PubMed] [Google Scholar]
- 31.Kohrle J. Selenium and the control of thyroid hormone metabolism. Thyroid. 2005;15:841–853. doi: 10.1089/thy.2005.15.841. [DOI] [PubMed] [Google Scholar]
- 32.Riese C, Michaelis M, Mentrup B, Gotz F, Kohrle J, Schweizer U, Schomburg L. Selenium-dependent pre- and posttranscriptional mechanisms are responsible for sexual dimorphic expression of selenoproteins in murine tissues. Endocrinology. 2006;147:5883–5892. doi: 10.1210/en.2006-0689. [DOI] [PubMed] [Google Scholar]
- 33.Nakayama A, Hill KE, Austin LM, Motley AK, Burk RF. All regions of mouse brain are dependent on selenoprotein p for maintenance of selenium. J. Nutr. 2007;137:690–693. doi: 10.1093/jn/137.3.690. [DOI] [PubMed] [Google Scholar]
- 34.Hill KE, Zhou J, Austin LM, Motley AK, Ham AJ, Olson GE, Atkins JF, Gesteland RF, Burk RF. The selenium-rich C-terminal domain of mouse selenoprotein P is necessary for supply of selenium to brain and testis but not for maintenance of whole-body selenium. J. Biol. Chem. 2007;282:10972–10980. doi: 10.1074/jbc.M700436200. [DOI] [PubMed] [Google Scholar]
- 35.Olson GE, Winfrey VP, Nagdas SK, Hill KE, Burk RF. Apolipoprotein E receptor-2 (ApoER2) mediates selenium uptake from selenoprotein P by the mouse testis. J. Biol. Chem. 2007;10:1074. doi: 10.1074/jbc.M611403200. [DOI] [PubMed] [Google Scholar]
- 36.Lei XG, Evenson JK, Thompson KM, Sunde RA. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J. Nutr. 1995;125:1438–1446. doi: 10.1093/jn/125.6.1438. [DOI] [PubMed] [Google Scholar]
- 37.Bermano G, Nicol F, Dyer JA, Sunde RA, Beckett GJ, Arthur JR, Hesketh JE. Tissue-specific regulation of selenoenzyme gene expression during selenium deficiency in rats. Biochem. J. 1995;311:425–430. doi: 10.1042/bj3110425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pagmantidis V, Bermano G, Villette S, Broom I, Arthur JR, Hesketh JE. Effects of Se-depletion on glutathione peroxidase and selenoprotein W gene expression in the colon. FEBS Lett. 2005;579:792–796. doi: 10.1016/j.febslet.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 39.Yeh JY, Beilstein MA, Andrews JS, Whanger PD. Tissue distribution and influence of selenium status on levels of selenoprotein W. Faseb J. 1995;9:392–396. doi: 10.1096/fasebj.9.5.7896009. [DOI] [PubMed] [Google Scholar]
- 40.Gu QP, Sun Y, Ream LW, Whanger PD. Selenoprotein W accumulates primarily in primate skeletal muscle, heart, brain and tongue. Mol. Cell. Biochem. 2000;204:49–56. doi: 10.1023/a:1007065829068. [DOI] [PubMed] [Google Scholar]