Abstract
Here, we examined the effects of molecular crowding on the function, structure and stability of nucleases. We found that the hydrolysis of a 29-mer double-stranded DNA by the endonucleases DNase I and S1 nuclease was substantially enhanced by molecular crowding using polyethylene glycol (PEG); however, molecular crowding had little effect on hydrolysis by exo III and exo I exonucleases. Moreover, kinetic analysis showed that the maximum velocity for the reaction of DNase I at 25°C was increased from 0.1 to 2.7 μM/min by molecular crowding with 20% (w/v) PEG, whereas that of exonuclease I at 37°C decreased from 2.2 to 0.4 μM/min. In contrast, molecular crowding did not significantly affect the Michaelis constant of DNase I or exonuclease I. These results indicate that molecular crowding has different effects on the catalytic activities of exonucleases and endonucleases.
INTRODUCTION
Nucleases, which cleave the phosphodiester bonds of nucleic acids, are very important not only for nucleic acid metabolism (1,2) but also for a variety of biotechnologies (3–5). For this reason, many studies have examined their structure, stability and function (6–10). Most of these studies, however, have been carried out under relatively dilute conditions. These are quite different to those inside a cell, where up to 40% of the total volume is taken up by macromolecules (11,12).
As numerous studies have shown, molecular crowding dramatically increases the association between biomolecules and substantially affects biomolecular reaction rates (13–27). For example, Zimmerman and Pheiffer (14) showed that DNA ligase activity is enhanced by molecular crowding conditions. Wenner and Bloomfield (16), in contrast, found that crowding agents have negligible effects on EcoRV activity. Thus, molecular crowding has different effects on different types of nucleic acid metabolizing enzymes. For this reason, systematic studies of the effects of molecular crowding on enzyme activity, structure and stability for a series of nucleases are needed to predict the nuclease behavior in cellular conditions and to understand how molecular crowding affects protein functions in the presence of nucleic acids.
In the current study, we systematically investigated the effects of molecular crowding on DNA hydrolysis by various nucleases. We found that the cleavage of both a large DNA (plasmid) and a short DNA oligonucleotide by DNase I was increased in the presence of polyethylene glycol (PEG). An increase in activity by molecular crowding was found for endonucleases (DNase I and S1 nuclease) but not for exonucleases (exonucleases I and III). Kinetic analyses showed that molecular crowding affected the maximal velocity (Vmax) but not the Michaelis constant (Km) for the nucleases. Overall, our results indicate that molecular crowding has different effects on the catalytic activities of exonucleases and endonucleases.
MATERIALS AND METHODS
Materials
PEG 4000, PEG 8000 and PEG 20000 [average molecular weight = 3000, 8000 and 20000, respectively (28)] were purchased from Wako Pure Chemicals (Osaka, Japan) and used without further purification. DNase I from bovine pancreas was purchased from Invitrogen (Carlsbad, CA, USA). Exonuclease I from Escherichia coli, S1 nuclease from Asergillus oryzae and exonuclease III from E. coli were purchased from Takara Bio (Tokyo, Japan). Purified DNase I for the circular dichroism (CD) studies was purchased from Roche (Mannheim, Germany). All other reagents were of reagent grade.
Substrate DNAs
Plasmid [pcDNA 3.1(+)] was purchased from Invitrogen. E. coli strains were grown aerobically in Luria–Bertani medium in the presence of 100 μg/ml ampicillin at 37°C with shaking at 250 r.p.m. E. coli Top 10 cells (Invitrogen, CA, USA) were used for plasmid propagation. HPLC-grade oligonucleotide substrates of 29-mer sequences, 5′-ACGATATCTCCCTATAGTGAGTCGTATTA-3′ and 5′-TAATACGACTCACTATAGGGAGATATCGT-3′, were purchased from Hokkaido System Science (Sapporo, Japan). These sequences were designed using mfold to avoid folding into undesired structures (29). For quantitative analysis of the hydrolysis reaction of nuclease, the sequence 5′-ACGATATCTCCCTATAGTGAGTCGTATTA-3′ was labeled with 6-carboxyfluorescein at its 5′-end, although the 6-carboxyfluorescein could affect the rate of cleavage by a nuclease. A 29-mer double-stranded DNA (dsDNA) was prepared by annealing (incubation at 90°C for 5 min, followed by cooling at 1°C/min to the reaction temperature) of 5′-ACGATATCTCCCTATAGTGAGTCGTATTA-3′ with 5′-TAATACGACTCACTATAGGGAGATATCGT-3′. The 29-mer dsDNA was used as substrate in assays of DNase I and exonuclease III. The sequence 5′-ACGATATCTCCCTATAGTGAGTCGTATTA-3′ was used as a single-stranded DNA (ssDNA) substrate in the assays of S1 nuclease and exonuclease I.
Assay of DNA hydrolysis by nucleases
Because the optimal condition (cations, pH and temperature) for DNA hydrolysis by each nuclease varies, it is difficult to study all the enzymes using a single-standard buffer. DNase I was assayed in a buffer of 2.5 mM MgCl2, 0.5 mM CaCl2 and 10 mM Tris–HCl (pH 7.5) at 25°C. The reactions were performed with 0.01–0.1 U of DNase I and 0.5 μg of plasmid [pcDNA 3.1(+)] or 20 μM dsDNA (29-mer) as a substrate. S1 nuclease was assayed in 280 mM NaCl, 1 mM ZnSO4 and 30 mM sodium acetate buffer (pH 4.6) at 37°C. The reactions were performed with 0.15 U of S1 nuclease and 10 μM ssDNA. Exonuclease III was assayed in a buffer of 5 mM MgCl2, 10 mM 2-mercaptoethanol and 50 mM Tris–HCl (pH 8.0) at 37°C. The reactions were performed with 1–10 U of exonuclease III and 20 μM dsDNA. Exonuclease I was assayed in a buffer of 6.7 mM MgCl2, 10 mM 2-mercaptoethanol and 67 mM glycine-KOH (pH 9.5) at 37°C. The reactions were performed with 0.5–5 U of exonuclease I and 10 μM ssDNA.
The reactions were terminated by adding gel loading buffer containing 100 mM EDTA, 45% (w/v) sucrose and 0.03% (w/v) bromophenol blue, followed by cooling on ice. The mixtures were resolved by 20% native polyacrylamide gel electrophoresis (PAGE) for short DNA oligonucleotides (29-mer dsDNA and ssDNA) or 0.8% agarose gel electrophoresis for the plasmid DNA reactions. The voltage for the native PAGE and for agarose gel electrophoresis was 200 and 100 V, respectively. The short DNAs were visualized using a Fuji Film FLA-5100 phosphorimager (Fuji Film Co., Tokyo, Japan). The plasmid DNA was visualized by staining with ethidium bromide. The fluorescence intensities of bands (LAU/mm2) were calculated using Multi Gauge ver. 2.2 software (Fuji Film Co.). DNA hydrolysis was evaluated according to the amount of residual substrate (%) because the product bands were too diffuse to analyze quantitatively. The amount of the residual substrate was estimated as follows (30): Residual substrate (%) = [fluorescence intensity (LAU/mm2) of the substrate band after reaction] × 100/[fluorescent intensity (LAU/mm2) of the substrate band before reaction]. All experiments were repeated at least three times. The experimental error was <2%.
For kinetic studies, the dsDNA and ssDNA concentrations were 0.1–20 μM for DNase I and 0.1–10 μM for exonuclease I. The initial rates (v) were estimated from the data for the first 10% of the reaction. The values of v were plotted versus the DNA (substrate) concentration to allow calculations of the kinetic parameters (Km and Vmax). The kinetic parameters were calculated from the non-linear best fit of the data to the Michaelis–Menten equation v = Vmax [S]/(Km + [S]), where [S] indicates substrate concentration, using Origin software (Microcal Software Inc., Northampton, MA, USA).
CD measurements
The structure and stability of DNase I in the absence and presence of PEG was studied by CD analysis. CD spectra were recorded using a Jasco J-820 spectrometer equipped with a Peltier temperature control system (Jasco, Tokyo, Japan). The cuvette-holding chamber was flushed with a constant steam of dry N2 gas to avoid water condensation on the cuvette exterior. Data were collected from 195 to 255 nm with a 1-s response time and a 1-nm bandwidth using a 0.1-cm quartz cuvette. The CD measurements for 0.5 μM DNase I were carried out in 10 mM Tris–HCl (pH 7.5) in the absence and presence of 20% (w/v) PEG 4000 at 25°C. Each spectrum shown is the average of five individual scans and is corrected for the spectrum of the buffer. For thermal denaturation, the CD signal at 222 nm was monitored for a 1°C/min temperature rise from 5 to 95°C.
Thermodynamic stability of DNase I and exonuclease I
The thermodynamic stability of the nucleases was studied by performing stability assays in the absence and presence of PEG. The thermal stability of DNase I was assayed in 10 mM Tris–HCl (pH 7.5) in the absence and presence of PEG 4000, and that for exonuclease I was assayed in 10 mM 2-mercaptoethanol and 67 mM glycine-KOH (pH 9.5) in the absence and presence of PEG 8000. The proteins were incubated at 60°C, and hydrolysis was assayed as described above (‘Assay of DNA hydrolysis by nucleases’). After 10 min of hydrolysis, the reaction mixture was resolved and visualized as described above. The thermal stability of DNase I and exonuclease I at 60°C was assessed according to the residual activity, which was calculated as follows (31): Residual activity (%) = [Fluorescence intensity (LAU/mm2) of the substrate band before the reaction with heat treatment − Fluorescence intensity (LAU/mm2) of the substrate band after the reaction with heat treatment] × 100/[Fluorescence intensity (LAU/mm2) of the substrate band before the reaction without heat treatment − Fluorescence intensity (LAU/mm2) of the substrate band after the reaction without heat treatment]. All experiments were repeated at least three times. The experimental error was <8%.
RESULTS
Effect of molecular crowding on DNase I activity
DNase I, an endonuclease, can cleave single- and double-stranded DNAs in a non-sequence specific manner (5). Plasmid DNA is widely used as a substrate in studies of DNase I. In the current studies, we examined the effect of molecular crowding on nucleases. To mimic the aqueous cellular environment, we used PEG because it is inert and because it is commercially available in a wide range of molecular weights (11).
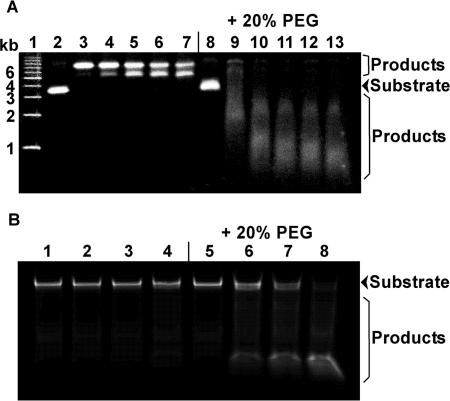
We first examined the effect of 20% (w/v) PEG on the hydrolysis of supercoiled plasmid DNA [pcDNA 3.1(+)] by DNase I at 25°C. Figure 1A shows the DNA fragments after electrophoresis on a 0.8% (w/v) agarose gel. The two bands in lanes 2 and 3 correspond to supercoiled and open circular plasmid DNA (32). During the reaction, two distinct bands were observed in the absence of PEG 20000 (lanes 3–7). According to previous observations (32), the slower and faster migrating bands correspond to open circular and linear plasmid DNA, respectively. Although open circular and linear plasmid DNAs were observed, diffuse bands corresponding to products degraded by DNase I were not observed. These results show that, in the absence of PEG, 0.01 U of DNase I was not sufficient for hydrolyzing the plasmid DNA within 5 min. In contrast, in the presence of PEG 20000, the plasmid DNA was cleaved rapidly after 1 min (lane 9), indicating that it was already degraded. These results demonstrate for the first time that molecular crowding significantly enhances the hydrolysis of supercoiled plasmid DNA by DNase I.
Figure 1.
(A) Hydrolysis of plasmid [pcDNA 3.1(+)] DNA by DNase I (0.01 U) in the absence or presence of 20% (w/v) PEG 20000 at 25°C. Lane 1 shows the DNA size marker. Lanes 2, 3, 4, 5, 6 and 7 show 0, 1, 2, 3, 4 and 5 min, respectively, of hydrolysis by DNase I in the absence of PEG 20000. Lanes 8, 9, 10, 11, 12 and 13 show 0, 1, 2, 3, 4 and 5 min, respectively, of hydrolysis by DNase I in the presence of 20% (w/v) PEG 20000. (B) Hydrolysis of a short oligonucleotide DNA (29-mer dsDNA) by DNase I (0.1 U) in the absence or presence of 20% (w/v) PEG 20000 at 25°C. Lanes 1, 2, 3 and 4 show 0, 1, 4 and 10 min, respectively, of hydrolysis by DNase I in the absence of PEG 20000. Lanes 5, 6, 7 and 8 show 0, 1, 4 and 10 min, respectively, of hydrolysis by DNase I in the presence of 20% (w/v) PEG 20000.
In addition, the migration of the substrate bands under dilute conditions (lane 2) was slightly different from their migration under crowded conditions (lane 8). We found that the plasmid DNA mobility decreased slightly with increasing concentrations of PEG 20000 (Supplementary Figure S1). This may reflect a slight difference in the conformation of the plasmid DNA induced by PEG 20000 or a slower entry into the gel of DNA molecules from a PEG solution.
To quantitatively evaluate the effect of molecular crowding on DNA hydrolysis by DNase I, we used a short (29-mer) dsDNA oligonucleotide as a substrate. Figure 1B shows native PAGE of the 29-mer dsDNA after reaction with DNase I in the absence and presence of 20% (w/v) PEG 20000 at 25°C. Before the reaction, the migration of the substrate DNA was the same in dilute (lane 1) and crowded conditions (lane 5). This result supports previous studies showing that PEG does not significantly affect the thermodynamic stability of short DNA duplexes (27). We did not observe a substantial change in the migration of the dsDNA before or after the DNase I reaction under dilute conditions (lane 1–4), showing that, in the absence of PEG, 0.01 U of DNase I is not sufficient to hydrolyze the 29-mer dsDNA within 10 min. In contrast, bands migrating faster than that of the substrate DNA are clearly visible in the presence of PEG 20000 (lane 5–8). The result shows that dsDNA hydrolysis by DNase I was greatly enhanced by the addition of 20% (w/v) PEG 20000. Therefore, it appears that molecular crowding increases the cleavage yield of DNase I not only for the large DNA (plasmid) but also for the short (29-mer) DNA oligonucleotide.
In addition, we observed the accumulation of product DNA (lanes 6–8). We found that the size of the product DNA was ∼10 bp (Supplementary Figure S2). Suck and Oefner (33) reported that both sides of the DNA double helix at the cleavage point contact DNase I over a span of 10 bp. Thus, 10-bp dsDNA regions on both sides of the cleavage point are required for higher activity. This can explain the accumulation of ∼10-bp dsDNAs in our experiments.
Effect of molecular crowding on the hydrolytic activity of various nucleases
We next examined the influence of molecular crowding on the two main types of nucleases (exo and endo), including S1 nuclease, an endonuclease for ssDNA (34); exonuclease III, an exonuclease for dsDNA (35,36); exonuclease I, an exonuclease for ssDNA (37) and DNase I, an endonuclease for both ssDNA and dsDNA (5).
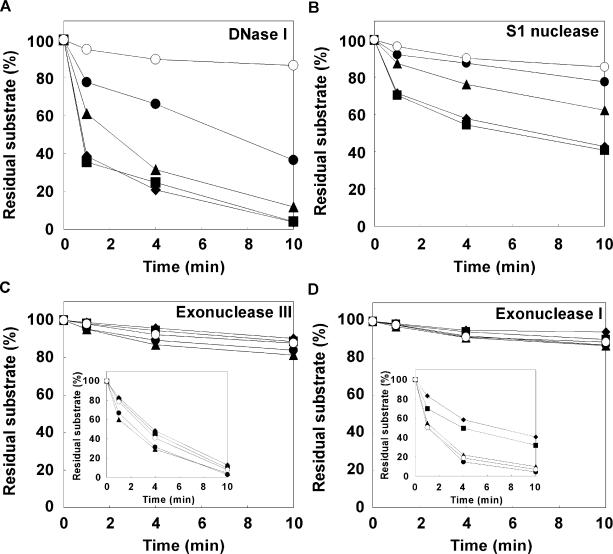
In the case of DNase I, the amounts of residual dsDNA after 10 min in the presence of 20% (w/v) PEG 200, 4000, 8000 and 20000 was estimated to be 12, 4, 10 and 10%, respectively. Thus, PEG 4000 was the most effective of the tested PEGs at enhancing the hydrolysis reaction (Supplementary Figure S3A). For this reason, we further investigated the amount of residual 29-mer dsDNA in the presence of 0–20% (w/v) PEG 4000 versus the time using DNase I at 25°C (Figure 2A). In the absence of PEG 4000, the amount of residual dsDNA after 10 min was estimated to be 87%. In contrast, the amount of residual dsDNA after 10 min in the presence of 5, 10, 15 and 20% (w/v) PEG 4000 was estimated to be 37, 12, 4 and 4%, respectively. These results show that hydrolysis of dsDNA by DNase I is greatly enhanced by PEG.
Figure 2.
Amount of residual DNAs in the presence of 0% (w/v) (open circles), 5% (w/v) (closed circles), 10% (w/v) (closed triangles), 15% (w/v) (closed squares) and 20% (w/v) (closed diamonds) of PEG and (A) 0.1 U DNase I, (B) 0.15 U S1 nuclease, (C) 1 U exonuclease III (inset shows 10 U), (D) 0.5 U exonuclease I (inset shows 5 U). A ssDNA was used as a substrate for S1 nuclease and exonuclease I and a dsDNA was used as a substrate for DNase I and exonuclease III. PEG 4000 was used as the crowding agent for DNase I and S1 nuclease reactions, and PEG 8000 was used for exonucleases III and I. Error bars (smaller than ±2%) were omitted for clarity.
In the case of S1 nuclease, amongst PEG 200, 4000, 8000 and 20000, PEG 4000 was the most effective at enhancing the rate of hydrolysis (Supplementary Figure S3B). Thus, we further investigated the amount of residual 29-mer ssDNA in the presence of 0–20% (w/v) PEG 4000 versus time using S1 nuclease (Figure 2B). Similar to the results for DNase I, PEG 4000 enhanced the hydrolysis of ssDNA by S1 nuclease. Although the substrates for these two nucleases are different, molecular crowding had similar effects, indicating that the effect is not due to changes in substrate structure. Instead, the results indicate that molecular crowding generally enhances DNA hydrolysis by endonucleases regardless of the substrate structure.
Next, we investigated the effects of molecular crowding on DNA hydrolysis by exonucleases. Figure 2C and D show the amount of residual DNAs versus time for exonucleases III and I, respectively, in the presence of 0–20% (w/v) PEG 8000 (Supplementary Figure 3C and D shows the effect of molecular weight of PEG on the activities of exonucleases III and I). There was little hydrolysis of the substrate DNAs by exonucleases III and I at low enzyme concentrations (0.5–1 U) in both the dilute and molecular crowding conditions. This indicates that the effects of molecular crowding on the hydrolysis reactions of exonucleases were relatively small compared with the effects on the endonuclease reactions. Therefore, to further investigate the effects of molecular crowding on hydrolysis by exonucleases, we increased the amount of enzyme to 10 U for exonuclease III and to 5 U for exonuclease I. Surprisingly, PEG did not enhance the activity of exonuclease III, even at the high enzyme concentration (inset in Figure 2C): after 10 min, the amounts of residual 29-mer dsDNA using 10 U of exonuclease III at 0, 5, 10, 15 and 20% (w/v) PEG 8000 were estimated to be 8, 3, 5, 10 and 13%, respectively. The results show that DNA hydrolysis by exonuclease III is not enhanced by molecular crowding.
Interestingly, DNA hydrolysis by exonuclease I was strongly inhibited by molecular crowding with PEG 8000 (inset in Figure 2D): after 10 min, the amount of residual 29-mer ssDNA at 0, 5, 10, 15 and 20% (w/v) PEG 8000 after 10 min was estimated to be 7, 4, 10, 32 and 41%, respectively. Thus, it appears that molecular crowding generally enhances endonuclease activities but does not affect or inhibit exonuclease activities.
Thermodynamic stabilities of the endonuclease and exonuclease under dilute and molecular crowding conditions
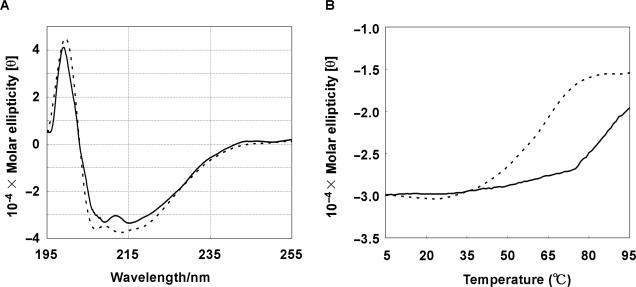
Although we found that molecular crowding enhances endonuclease activity rather than exonuclease activity, the origin of the difference in the effects remains unclear. Because we already demonstrated that the structure and stability of the substrate DNAs are not critical factors in the molecular crowding effect, we examined whether molecular crowding affects the thermodynamic stability of the nucleases. We first compared the structure and stability of an endonuclease (DNase I) in dilute and molecular crowding conditions by CD. Figure 3A shows CD spectra of DNase I in the absence and presence of 20% (w/v) PEG 4000 at 25°C. Both CD spectra have negative peaks around 215 and 208 nm and a positive peak around 198 nm, showing that α-helices are the dominant structure in DNase I in dilute and molecular crowding conditions. These results are identical with those reported by Ajitai and Venyaminov (38). In addition, PEG 4000 did not cause a peak shift, indicating that molecular crowding did not affect the secondary structure of DNase I. On the other hand, molecular crowding strongly affected the stability of the DNase I structure. Figure 3B shows melting curves for the α-helical DNase I structure in the absence and presence of 20% (w/v) PEG 4000. The melting temperature (Tm) of the DNase I structure in the absence of PEG 4000 was estimated to be 60°C. Surprisingly, the Tm of the DNase I structure was >80°C in the presence of 20% (w/v) PEG 4000. This demonstrates that molecular crowding stabilizes the structure of DNase I.
Figure 3.
(A) CD spectra for 0.02 mg/ml DNase I in the absence (dotted line) and presence (solid line) of 20% (w/v) PEG 4000 at 25°C. (B) Thermal denaturation curves for DNase I in the absence (dotted line) and presence (solid line) of 20% (w/v) PEG 4000. The changes in the ellipticity were monitored at 222 nm. All measurements were performed in 50 mM HEPES (pH 7.2).
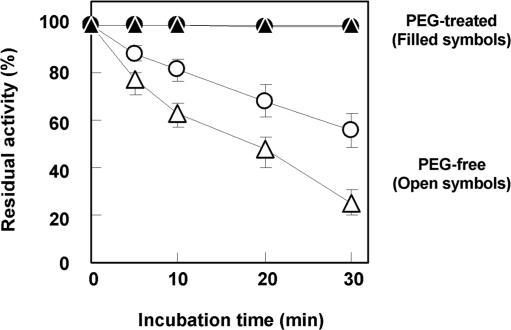
We did not measure the CD spectra of exonuclease I because purified exonuclease I is not commercially available. Thus, to investigate how molecular crowding affects the stability of exonuclease I, we carried out a heat stability test for both exonuclease I and DNase I in dilute and molecular crowding conditions. In these experiments, DNase I and exonuclease I were incubated for 0–30 min at 60°C in the absence and presence of PEG (PEG 4000 or 8000). After the incubation, we carried out a 10-min hydrolysis reaction. Figure 4 shows the residual DNase I and exonuclease I activities versus time of incubation at 60°C. After a 30-min incubation, the residual activities of DNase I and exonuclease I in the absence of PEG were estimated to be 56 and 26%, respectively. The inactivation of the nucleases by incubation for 30 min at 60°C agrees with the observations of Chow and Resnick (31). On the other hand, the residual activities of DNase I and Exonuclease I in the presence of PEGs (PEG 4000 or 8000, respectively) remained around 100%. These results show that PEG preserves the active structures of DNase I and exonuclease I. The stabilization of DNase I by molecular crowding observed in the heat stability assay agrees with the CD results showing that molecular crowding with PEG increased the Tm of DNase I from 60°C to >80°C. Therefore, these results indicate that the structure of both exonuclease I and DNase I are stabilized by molecular crowding. Although elucidation of the mechanism of the stabilization by molecular crowding is interesting and demands further study, it appears that the difference in the effects of molecular crowding on the endonucleases and exonucleases is not due to effects on their thermodynamic stability.
Figure 4.
Residual activities of DNase I and exonuclease I after incubation for 0–30 min at 60°C. Hydrolysis by DNase I in the absence (open circles) or presence (closed circles) of 20% (w/v) PEG 4000 was carried out for 10 min at 25°C. Hydrolysis by exonuclease I in the absence (open triangles) or presence (closed triangles) of 20% (w/v) PEG 8000 was carried out for 10 min at 37°C.
Kinetic parameters for DNA hydrolysis by DNase I and exonuclease I in dilute and molecular crowding conditions
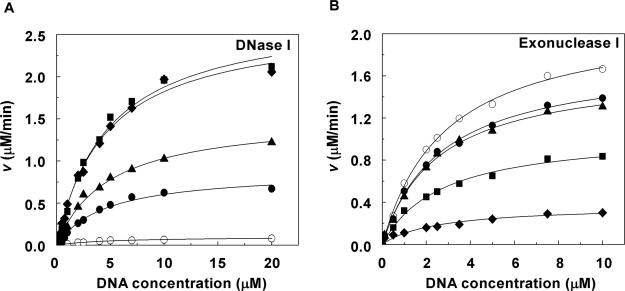
We further investigated the origin of the different effects of molecular crowding on the endonucleases and the exonucleases by determining the kinetic parameters for the hydrolysis reactions in dilute and molecular crowding conditions. We measured the initial velocity (v) of the hydrolysis reaction at various concentrations of DNA substrate in the absence of PEG and in presence of PEG 4000 at 25°C for DNase I or in the presence of PEG 8000 at 37°C for exonuclease I. The value of v for DNase I increased as the concentration of PEG 4000 increased (Figure 5A). In contrast, the value of v for exonuclease I decreased as the concentration of PEG 8000 was increased (Figure 5B). Table 1 lists the Km and Vmax of DNA hydrolysis by DNase I and exonuclease I at the various concentrations of PEG. The Km values of DNase I and exonuclease I for the substrate DNA in the absence of PEG were estimated to be 5.0 and 2.9 μM, respectively, and in the presence of 20% (w/v) PEGs, they were estimated to be 4.5 and 2.3 μM, respectively. The Km values of DNase I and exonuclease I were not significantly affected by the addition of PEG. On the contrary, the Vmax of DNase I increased from 0.1 to 2.7 μM/min as the concentration of PEG 4000 was raised from 0 to 20% (w/v), whereas that of exonuclease I decreased from 2.2 to 0.4 μM/min as the concentration of PEG 8000 was raised from 0 to 20% (w/v). These effects of molecular crowding on the Vmax are consistent with the results of equilibrium analyses of DNase I and exonuclease I (Figure 2A and D). These kinetic parameters reveal that molecular crowding influences the hydrolytic activity of the nucleases by directly affecting catalysis as indicated by the changes in the Vmax.
Figure 5.
Initial velocities (v) for the DNase I (A) and exonuclease I (B) hydrolysis reactions in the presence of 0% (w/v) (open circles), 5% (w/v) (closed circles), 10% (w/v) (closed triangles), 15% (w/v) (closed squares) and 20% (w/v) PEG (closed diamonds). The DNase I reaction was carried out in a buffer containing 100 mM NaCl, 5 mM MgCl2 and 50 mM HEPES (pH 7.2) at 25°C in the absence or presence of PEG 4000. The exonuclease I reaction was carried out in a buffer containing 100 mM NaCl, 6.7 mM MgCl2, 10 mM 2-mercaptoethanol and 67 mM glycine-KOH (pH 9.5) at 37°C in the absence or presence of PEG 8000. The dsDNA and ssDNA concentrations in kinetic assays were 0.1–20 μM for DNase I and 0.1–10 μM for exonuclease I. The value of v was plotted versus the concentration of substrate DNA. Error bars (smaller than ±1%) were omitted for clarity.
Table 1.
Kinetic parameters of DNase I and exonuclease I in the absence and presence of PEGsa
| PEG concentration | DNase I | Exonuclease I | ||
|---|---|---|---|---|
| % (w/v) | Km (μM) | Vmax (μM/min) | Km (μM) | Vmax (μM/min) |
| 0 | 5.0 ± 0.8 | 0.1 ± 0.0 | 2.9 ± 0.2 | 2.2 ± 0.1 |
| 5 | 4.6 ± 0.6 | 0.9 ± 0.0 | 2.7 ± 0.2 | 1.8 ± 0.1 |
| 10 | 4.8 ± 0.5 | 1.5 ± 0.1 | 2.7 ± 0.2 | 1.7 ± 0.1 |
| 15 | 4.8 ± 0.6 | 2.8 ± 0.1 | 2.7 ± 0.1 | 1.1 ± 0.1 |
| 20 | 4.5 ± 0.4 | 2.7 ± 0.1 | 2.3 ± 0.2 | 0.4 ± 0.1 |
aPEG 4000 was used as a crowding agent for DNase I, and PEG 8000 was the crowding agent for exonuclease I. The DNase I and exonuclease I reactions were carried out at 25 and 37°C, respectively.
DISCUSSION
Our equilibrium studies of DNA hydrolysis indicate that molecular crowding enhances the activity of endonucleases but has little effect on exonuclease activity. Moreover, we found that molecular crowding affected neither the thermodynamic stabilities of the nuclease structures nor their binding to substrate DNAs. Kinetic analyses demonstrated that molecular crowding affected catalysis. Therefore, the difference in the reaction mechanism and/or type of reaction (endo or exo) may account for the different effects of molecular crowding on endonucleases and exonucleases.
Because of the importance of nucleases in vivo and in vitro, many studies have examined their reaction mechanisms. For DNase I, Glu75 in the active site accepts a proton from His131, which in turn accepts a proton from a water molecule positioned close to the substrate DNA. Then, the nucleophile attacks the phosphorous of the substrate DNA, cleaving the P-O-3′ bond (33). This proton acceptor–donor chain (Glu75–His131–water), which is critical for the catalytic activity, is also found in the active site of exonuclease I as Glu17–His181–water (39). Since the reaction mechanisms for DNase I and exonuclease I are almost identical, it is surprising that molecular crowding had opposite effects on endonucleases and exonucleases. Thus, it is difficult to determine the reason for the opposite effects on the basis of the cleavage mechanisms.
On the other hand, the reaction types mediated by DNase I (endo) and exonuclease I (exo) are distinct. Endonucleases can randomly hydrolyze internal sites in DNA substrates, whereas exonucleases remove terminal nucleotides. We therefore suspect that the origin of the different effects of molecular crowding is the different reaction type (endo versus exo) rather a difference in the reaction mechanism. Wenner and Bloomfield (16) reported that molecular crowding increases the Vmax of EcoRV (endo-type). Conversely, the Vmax of T7 DNA polymerase activity and the nick-translation reaction of DNA polymerase I (both exo-type) are reduced by molecular crowding (27,40) (Supplementary Table 1). Although the effects of other factors, such as the dielectric constant, viscosity of the solution and flexibility of the protein structure on enzyme activities should be examined, our current findings and these previous results suggest that the different effects of molecular crowding on endonucleases and exonucleases are related to the reaction type. These results and considerations lead us to conclude that molecular crowding generally increases the catalytic activity of endonucleases and decreases or does not affect the catalytic activity of exonucleases. These findings should help clarify in general how molecular crowding affects protein functions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported in part by Grants-in-Aid for Scientific Research and the ‘Academic Frontier’ Project (2004–09) from MEXT, Japan. Funding to pay the Open Access publication charges for this article was provided by Fine Co. Ltd.
Conflict of interest statement. None declared.
REFERENCES
- 1.Napirei M, Karsunky H, Zevnik B, Stephan H, Mannherz HG, Moroy T. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat. Genet. 2000;25:177–181. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- 2.Napirei M, Wulf S, Mannherz HG. Chromatin breakdown during necrosis by serum Dnase I and the plasminogen system. Arthritis Rheum. 2004;50:1873–1883. doi: 10.1002/art.20267. [DOI] [PubMed] [Google Scholar]
- 3.Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974;11:1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klug A, Lutter LC. The helical periodicity of DNA on the nucleosome. Nucleic Acids Res. 1981;9:4267–4283. doi: 10.1093/nar/9.17.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laskowski M. The Enzymes. 1981. Deoxyribonuclease I. Vol. 4, 3rd edn, Academic Press, NY, pp. 289–311. [Google Scholar]
- 6.Serpersu EH, Shortle D, Mildvan AS. Kinetic and magnetic resonance studies of active-site mutants of staphylococcal nuclease: factors contributing to catalysis. Biochemistry. 1987;26:1289–1300. doi: 10.1021/bi00379a014. [DOI] [PubMed] [Google Scholar]
- 7.Judice JK, Gamble TR, Murphy EC, de Vos AM, Schultz PG. Probing the mechanism of staphylococcal nuclease with unnatural amino acids: kinetic and structural studies. Science. 1993;261:1578–1581. doi: 10.1126/science.8103944. [DOI] [PubMed] [Google Scholar]
- 8.Hibino Y, Kusashio E, Terakawa T, Sugano N. Enhancement of an Mg(2+)-dependent nuclease activity in rat liver cells exposed to cisplatin. Biochem. Biophys. Res. Commun. 1994;202:749–756. doi: 10.1006/bbrc.1994.1994. [DOI] [PubMed] [Google Scholar]
- 9.Mannherz HG, Peitsch MC, Zanotti S, Paddenberg R, Polzar B. A new function for an old enzyme: the role of DNase I in apoptosis. Curr. Top. Microbiol. Immunol. 1995;198:161–174. doi: 10.1007/978-3-642-79414-8_10. [DOI] [PubMed] [Google Scholar]
- 10.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 11.Zimmerman SB, Minton AP. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 12.Minton AP. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J. Biol. Chem. 2001;276:10577–10580. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- 13.Minton KW, Karmin P, Minton AP. Nonspecific stabilization of stress-susceptible proteins by stress-resistant proteins: a model for the biological role of heat shock proteins. Proc. Natl Acad. Sci., USA. 1982;79:7107–7111. doi: 10.1073/pnas.79.23.7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmerman SB, Pheiffer BH. Macromolecular crowding allows blunt-end ligation by DNA ligases from rat liver or Escherichia coli. Proc. Natl Acad. Sci. USA. 1983;80:5852–5856. doi: 10.1073/pnas.80.19.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison B, Zimmerman SB. Stabilization of T4 polynucleotide kinase by macromolecular crowding. Nucleic Acids Res. 1986;14:1863–1870. doi: 10.1093/nar/14.4.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wenner JR, Bloomfield VA. Crowding effects on EcoRV kinetics and binding. Biopyhs. J. 1999;77:3234–3241. doi: 10.1016/S0006-3495(99)77154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minton AP. Implications of macromolecular crowding for protein assembly. Curr. Opin. Struct. Biol. 2000;10:34–39. doi: 10.1016/s0959-440x(99)00045-7. [DOI] [PubMed] [Google Scholar]
- 18.Ellis RJ. Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol. 2001;11:114–119. doi: 10.1016/s0959-440x(00)00172-x. [DOI] [PubMed] [Google Scholar]
- 19.Morar AS, Wang X, Pielak GJ. Effects of crowding by mono-, di-, and tetrasaccharides on cytochrome c-cytochrome c peroxidase binding: comparing experiment to theory. Biochemistry. 2001;40:281–285. doi: 10.1021/bi002296r. [DOI] [PubMed] [Google Scholar]
- 20.Hatters DM, Minton AP, Howlett GJ. Macromolecular crowding accelerates amyloid formation by human apolipoprotein C-II. J. Biol. Chem. 2002;277:7824–7830. doi: 10.1074/jbc.M110429200. [DOI] [PubMed] [Google Scholar]
- 21.Miyoshi D, Nakao A, Sugimoto N. Molecular crowding regulates the structural switch of the DNA G-quadruplex. Biochemistry. 2002;41:15017–15024. doi: 10.1021/bi020412f. [DOI] [PubMed] [Google Scholar]
- 22.Shtilerman MD, Ding TT, Lansbury PT., Jr Molecular crowding accelerates fibrillization of alpha-synuclein: could an increase in the cytoplasmic protein concentration induce Parkinson's disease? Biochemistry. 2002;41:3855–3860. doi: 10.1021/bi0120906. [DOI] [PubMed] [Google Scholar]
- 23.Sasahara K, McPhie P, Minton AP. Effect of dextran on protein stability and conformation attributed to macromolecular crowding. J. Mol. Biol. 2003;326:1227–1237. doi: 10.1016/s0022-2836(02)01443-2. [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi D, Matsumura S, Nakano S, Sugimoto N. Duplex dissociation of telomere DNAs induced by molecular crowding. J. Am. Chem. Soc. 2004;126:165–169. doi: 10.1021/ja036721q. [DOI] [PubMed] [Google Scholar]
- 25.Nakano S, Karimata H, Ohmichi T, Kawakami J, Sugimoto N. The effect of molecular crowding with nucleotide length and cosolute structure on DNA duplex stability. J. Am. Chem. Soc. 2004;126:14330–14331. doi: 10.1021/ja0463029. [DOI] [PubMed] [Google Scholar]
- 26.Miyoshi D, Karimata H, Sugimoto N. Drastic effect of a single base difference between human and tetrahymena telomere sequences on their structures under molecular crowding conditions. Angew. Chem. Int. Ed. 2005;24:3740–3744. doi: 10.1002/anie.200462667. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki Y, Miyoshi D, Sugimoto N. Effect of molecular crowding on DNA polymerase activity. Biotechnol. J. 2006;1:440–446. doi: 10.1002/biot.200500032. [DOI] [PubMed] [Google Scholar]
- 28. Note that there is sometimes a discrepancy between the PEG number and the average molecular weight. The average molecular weight was provided by the manufacturer and is based on their experimental measurements.
- 29.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love JD, Hewitt RR. The relationship between human serum and human pancreatic DNase I. J. Biol. Chem. 1979;254:12588–12594. [PubMed] [Google Scholar]
- 31.Chow TYK, Resnick MA. Purification and characterization of an endo-exonuclease from Saccharomyces cerevisiae that is influenced by the RAD52 gene. J. Biol. Chem. 1987;262:17659–17667. [PubMed] [Google Scholar]
- 32.Johnson PH, Grossman LI. Electrophoresis of DNA in agarose gels. Optimizeing separations of conformational isomers of double- and single-stranded DNAs. Biochemistry. 1977;16:4217–4225. doi: 10.1021/bi00638a014. [DOI] [PubMed] [Google Scholar]
- 33.Suck D, Oefner C. Structure of DNase I at 2.0 Å resolution suggests a mechanism for binding to and cutting DNA. Nature. 1986;321:620–625. doi: 10.1038/321620a0. [DOI] [PubMed] [Google Scholar]
- 34.Gite S, Reddy G, Shankar V. Active-site characterization of S1 nuclease II. Biochem. J. 1992;288:571–575. doi: 10.1042/bj2880571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richardson CC, Schildkraut CL, Aposhian HV, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid polymerase of Escherichia coli. J. Biol. Chem. 1964;239:222–232. [PubMed] [Google Scholar]
- 36.Richardson CC, Kornberg A. A deoxyribonucleic acid phosphatase-exonuclease from Escherichia coli. I. purification of the enzyme and characterization of the phosphatase activity. J. Biol. Chem. 1964;239:242–250. [PubMed] [Google Scholar]
- 37.Lehman IR. The deoxyribonucleases of Escherichia coli. I. Purification and properties of a phosphodiesterase. J. Biol. Chem. 1960;235:1479–1487. [PubMed] [Google Scholar]
- 38.Ajtai K, Venyaminov SU. CD study of the actin DNase I complex. FEBS Lett. 1983;151:94–96. doi: 10.1016/0014-5793(83)80350-0. [DOI] [PubMed] [Google Scholar]
- 39.Breyer WA, Matthews BW. Structure of Escherichia coli exonuclease I suggests how processivity is achieved. Nat. Struct. Biol. 2000;7:1125–1128. doi: 10.1038/81978. [DOI] [PubMed] [Google Scholar]
- 40.Zimmerman SB, Harrison B. Macromolecular crowding increases binding of DNA polymerase to DNA: An adaptive effect. Proc. Natl Acad. Sci. USA. 1987;84:1871–1875. doi: 10.1073/pnas.84.7.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.