Abstract
The transcription elongation factor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) sensitivity-inducing factor (DSIF) regulates RNA polymerase II (RNAPII) processivity by promoting, in concert with negative elongation factor (NELF), promoter-proximal pausing of RNAPII. DSIF is also reportedly involved in transcriptional activation. However, the role of DSIF in transcriptional activation by DNA-binding activators is unclear. Here we show that DSIF acts cooperatively with a DNA-binding activator, Gal4-VP16, to promote transcriptional activation. In the absence of DSIF, Gal4-VP16-activated transcription resulted in frequent pausing of RNAPII during elongation in vitro. The presence of DSIF reduced pausing, thereby supporting Gal4-VP16-mediated activation. We found that DSIF exerts its positive effects within a short time-frame from initiation to elongation, and that NELF does not affect the positive regulatory function of DSIF. Knockdown of the gene encoding the large subunit of DSIF, human Spt5 (hSpt5), in HeLa cells reduced Gal4-VP16-mediated activation of a reporter gene, but had no effect on expression in the absence of activator. Together, these results provide evidence that higher-level transcription has a stronger requirement for DSIF, and that DSIF contributes to efficient transcriptional activation by preventing RNAPII pausing during transcription elongation.
INTRODUCTION
Transcription elongation plays an important role in gene expression in both prokaryotes and eukaryotes. In recent years, several lines of evidence have indicated that transcription elongation is not only a rate-limiting step in gene expression, but also a dynamic and highly regulated process that also impacts downstream events, such as mRNA processing, and RNA surveillance and export (1,2). The importance of such regulation is illustrated by the discovery that a variety of elongation factors contribute to development, differentiation and disease progression (3). Intensive study of transcription elongation factors has identified a number of regulatory mechanisms by which RNAPII efficiently elongates RNA, regardless of impediments (2,3).
DSIF is a heterodimer protein complex composed of Spt4 and Spt5, and is conserved among eukaryotes. DSIF exerts both negative and positive effects on elongation by directly binding to RNAPII through the KOW domain of Spt5 (4–6). DSIF negatively regulates transcription by acting in concert with NELF to mediate promoter proximal pausing of RNAPII (7,8). The positive regulatory activity of DSIF has been shown to stimulate transcription processivity through an as-yet undefined mechanism. DSIF progresses along with RNAPII to downstream regions of transcribed genes, and phosphorylation of the C-terminal repeat (CTR) region of Spt5 plays a key role in converting DSIF from a repressor to a positive regulator (9–13), suggesting possible mechanisms of regulation of the activator functions of DSIF.
Analysis of Drosophila embryo polytene chromosomes using immunostaining demonstrated that Spt5 localizes to active sites of transcription, and co-localizes with the phosphorylated form of the large subunit of RNAPII during elongation (11,14). Saunders et al. (15) reported that Drosophila Spt5 tracks with the RNAPII elongation complex along chromatin in vivo. In addition to its interaction with RNAPII, genetic and biochemical evidence indicates that DSIF also associates with other components of the transcriptional machinery, such as TFIIF, TFIIS, CSB, Spt6, FACT, Chd1 and the Paf complex, and with factors involved in mRNA maturation and surveillance, such as the mRNA capping enzyme, cap methyltransferase, and the nuclear exosome (2,16–19). Although the biochemical characteristics of DSIF are consistent with its function as a general transcription elongation factor, recent studies suggest that it also has a role in the regulation of development, and in gene-specific regulation. For example, zebrafish carrying a point mutation or deletion of spt5 display a pleiotropic but highly neuron-specific pattern of defects, suggesting the involvement of DSIF in development (20). In Drosophila embryos, an Spt5 missense mutation has locus-specific effects on transcription, suggesting that Spt5 affects gene expression selectively (21). Moreover, microarray analysis of both zebrafish and human Spt5 knockdown cells showed changes in expression of only a small subset of genes (unpublished data).
The above discrepancies may be explained by assuming that there is a stronger requirement for DSIF during high-levels of transcriptional activity (22). This idea is supported by studies of hsp, c-fos and HIV genome activation. Induction of heat shock gene transcription causes massive recruitment of Spt5 to hsp loci (11). Drosophila and zebrafish carrying Spt5 null alleles show defects in their heat shock response (21,23). Knockdown of DSIF in human cells causes a significant defect in c-fos transcriptional activation in response to epidermal growth factor, while having a negligible effect on c-fos expression under basal conditions (9). DSIF has also been implicated in Tat-mediated transactivation of HIV genome transcription. Tat is a viral activator that binds cis-acting TAR elements in nascent RNAs, and stimulates elongation of HIV genes. Knockdown of Spt5 in human cells decreases Tat-mediated transactivation and HIV-1 replication, but does not significantly affect cell viability (24). DSIF cooperates with Tat by preventing premature RNA release at terminator sequences, suggesting a possible mechanism of action of DSIF in regulating HIV transcription (25). The transcription of most cellular genes, however, is thought to be activated by DNA-binding activators. It is not clear whether DSIF exerts similar effects when working with DNA-binding activators.
In this report, we used in vitro transcription assays of Gal4-VP16, a DNA-binding transcriptional activator, to investigate the requirement for DSIF in transcriptional activation. Gal4-VP16 interacts with general transcription factors and the Mediator complex to stimulate initiation (26–29). It has also been implicated in the stimulation of elongation, probably through its interaction with TFIIH (30). We demonstrated that in the absence of DSIF, Gal4-VP16-mediated transcriptional activation causes more pausing during elongation than that which occurs during basal transcription. DSIF supported full transcriptional activation by reducing pausing of RNAPII during elongation. We also showed that transcriptional activity requires DSIF in vivo. In cultured HeLa cells, Gal4-VP16-induced expression of a reporter gene was significantly decreased upon Spt5 knockdown. In the absence of the VP16 activation domain, reporter gene expression was at basal levels, and was not affected substantially by Spt5 knockdown. Co-expression of the DNA-binding competitor of Gal4-VP16, Gal4DBD, which blocked transcriptional activation of the reporter gene, diminished the requirement for DSIF. These results suggest that DSIF regulates transcription elongation in response to transcriptional activation by DNA-binding activators. In addition, we showed that DSIF exerts its positive effect within a short time-frame from initiation to elongation, and that NELF is not involved in the positive regulatory effect of DSIF.
MATERIALS AND METHODS
Preparation of recombinant proteins
An expression plasmid encoding recombinant Histidine (His)-tagged DSIF (His-DSIF) was constructed by combining sequences for His-tagged human Spt4 (hSpt4) and hSpt5 in a single expression plasmid. The co-expression construct was generated using pET-hSpt4 and pET-hSpt5 (4). pET-hSpt5 was digested by AatII and NdeI to generate the hSpt5 coding sequence fragment. pET-14b was digested using NcoI and AatII to eliminate the His-tag sequence, and then ligated to the hSpt5 fragment to generate pT7hSpt5. pET-hSpt4 was digested using SphI and BamHI and inserted into pT7hSpt5 that had been digested with SphI and BglII.
Recombinant His-DSIF (His-hSpt4/hSpt5) was expressed in E. coli BL21-CodonPlus (DE3)-RIL (Stratagene). After induction with 1 mM IPTG for 4 h at 30°C, cells were harvested and lysed, and then lysates were loaded onto a Ni-NTA column (Qiagen). Recombinant His-DSIF was purified under native conditions according to the protocols in the QIAexpressionist handbook (Qiagen). Proteins eluted from the Ni-NTA column were loaded onto a 1 ml Mono Q column and eluted with a linear gradient of 100 to 1000 mM HGKEDP [20 mM HEPES (pH 7.9), 20% glycerol, 100–1000 mM KCl, 0.2 mM EDTA, 1 mM DTT, 1 mM PMSF]. The fractions were analyzed by SDS–polyacrylamide gel electrophoresis (PAGE), and fractions containing recombinant His-DSIF were dialyzed against 100 mM HGKEDP, and stored at −80°C until use. Coexpression of hSpt4 and hSpt5 was done to address the formation of insoluble aggregates, and avoid the denaturation/renaturation process used in a previous purification protocol (4).
His-GAL4 (1–94)-VP16 (413–490) was expressed in E. coli and purified as described by Reece et al. (31). Flag-NELF was purified as previously described (8). Cdk9 and Cyclin T1 subunits of P-TEFb were coexpressed in Sf9 cells with baculoviral vectors and purified as previously described (9).
Construction of the plasmid pG5MLPDG
The plasmid pG5MLPDG was generated by replacing the G-free cassette of pG5MLP (32) with the double G-free cassettes fragment from the plasmid pSLG402 (9).
In vitro transcription assays
Concentrated P1.0 fractions were prepared as described previously (33,34). In vitro transcription reactions using the concentrated P1.0 fraction and plasmid DNA templates were carried out as described previously (9,34). Briefly, in reactions using pG5MLP as a template, 12.5 μl reaction mixtures containing 125 ng DNA (32) and the concentrated P1.0 fraction were prepared in the presence or absence of recombinant DSIF and Gal4VP16 in TRX buffer [25 mM Tris–HCl (pH 7.9), 10% (v/v) glycerol, 50 mM KCl, 0.5 mM DTT and 0.5 mM EDTA]. Reactions were incubated for 40 min at 30°C. NTPs and 80 μM 3′-OMe-GTP in TRX buffer were then added, and the mixture was incubated for the indicated times. Where indicated, 1.5 mM each of ATP, UTP and CTP were added and reactions were incubated for an additional period of time. In Figure 3D, pG5MLPDG was used as a template. Transcription reaction was allowed to proceed for 20 min in the presence of 60 μM ATP, 600 μM GTP, 600 μM CTP, 5 μM UTP and 5 μCi of [α-32P]UTP (800 Ci/mmol). G-free RNA fragments derived from transcripts were isolated after RNase T1 treatment, deproteinized, precipitated with ethanol and analyzed using 8% acrylamide denaturing gels, as previously described (4). In Figures 1F and 3E, transcripts were quantified by a phosphorimager (Molecular Dynamics, Storm 860).
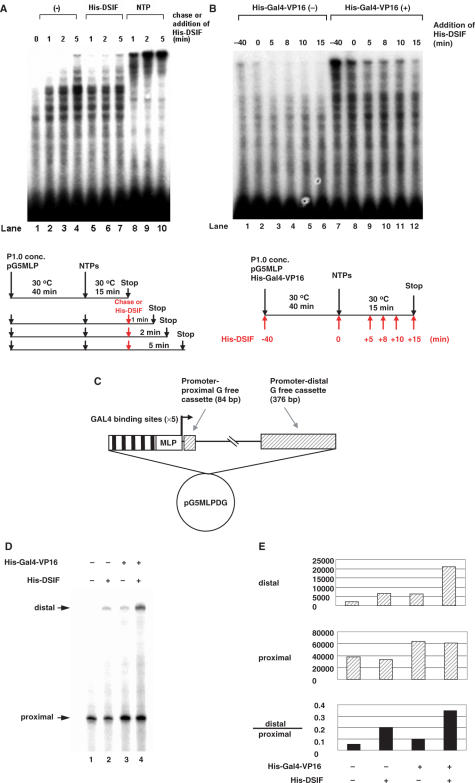
Figure 3.
DSIF does not exert its positive activity on late transcription complexes that are paused downstream. (A) In vitro transcription assays were carried out using P1.0 and pG5MLP as the template, as shown in the lower diagram. After a 40-min pre-incubation step, 60 μM ATP, 10 μM CTP, 1 μM UTP and 5 μCi [α-32P]UTP (800 Ci/mmol) were added to initiate transcription, and reactions were allowed to proceed for 15 to 20 min. In lanes 5 to 7, 7.5 ng of His-DSIF was added after 15 min of initiation/elongation, and reactions were further incubated for 1, 2 and 5 min, respectively. In lanes 8 to 10, a chase experiment was carried out as described for Figure 1C. The incubation time after the first 15-min initiation/elongation is labeled at the top. (B) In vitro transcription assays were carried out as described for panel A, except that 7.5 ng of His-DSIF was added at the indicated time points, as shown in the lower diagram. His-Gal4-VP16 was added together with P1.0 to the reactions in lanes 7 to 12. (C) Schematic representation of the plasmid pG5MLPDG. The template produces transcripts containing two G-free cassettes under the control of the same promoter and GAL4-binding sites as in pG5MLP. The promoter-proximal and -distal G-free cassettes of 84 and 376 bp in length are located 40 and 1522 bp downstream of the transcription start site, respectively. (D) In vitro transcription reaction was carried out using pG5MLPDG as a DNA template with concentrated P1.0. His-DSIF (7.5 ng) and His-Gal4-VP16 (150 ng) were added as indicated. Arrows indicate promoter-proximal (40–124) and -distal (1512–1888) fragments of transcripts. (E) The promoter-proximal and -distal fragments of transcripts in D were quantified using a phosphorimager, and the amounts (in arbitrary units) were illustrated in bars. Quantitative presentation of the ratio between the distal and proximal G-free cassettes was illustrated in the bottom panel.
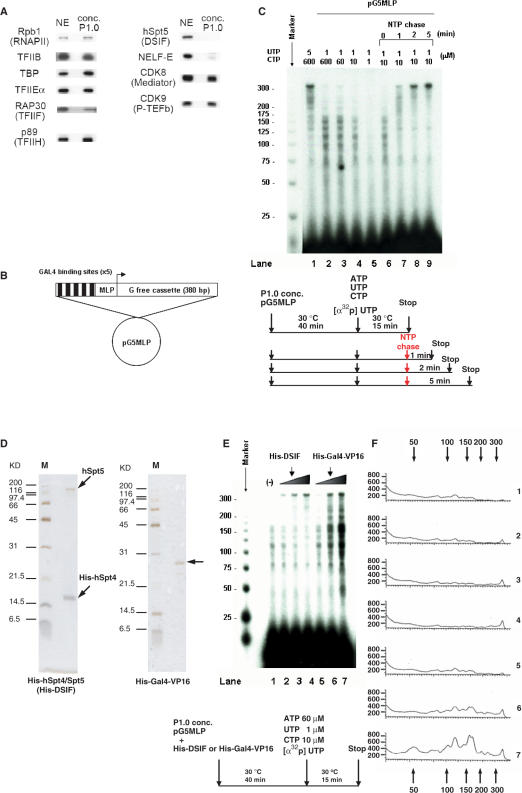
Figure 1.
Transcription elongation pauses in low concentrations of nucleotide precursors. (A) Western blot analysis showed that the concentrated P1.0 fraction contains RNAPII, general transcription factors, Mediator, and P-TEFb, but lacks DSIF and NELF. (B) Schematic representation of pG5MLP, showing the five Gal4-binding sites followed by the adenovirus major late promoter (MLP), and a 380 bp G-free cassette. (C) Transcription assays were carried out using P1.0 and pG5MLP as the template, with 60 μM ATP, 5 μCi of [α-32P]UTP (800 Ci/mmol) and the indicated concentrations of UTP and CTP. Chase reactions were carried out after 15 min of initiation/elongation, by further incubation for the indicated times with 1.5 mM ATP, CTP and UTP, as shown schematically in the lower diagram. (D) Silver staining of purified recombinant His-DSIF and His-Gal4-VP16 proteins. (E) In vitro transcription assays were carried out using P1.0 and pG5MLP as the template, in the absence or presence of His-DSIF (7.5 ng in lane 2; 30 ng in lane 3; 120 ng in lane 4) and His-Gal4-VP16 (50 ng in lane 5; 100 ng in lane 6; 150 ng in lane 7), as shown schematically in the lower diagram. Numbers to the left indicate the positions of markers (nucleotides). (F) Products in panel E was quantified by a phosphorimager, and the amounts (in arbitrary units) were plotted against the length. The numbers at the top and the bottom indicate the positions of markers (nucleotides). The numbers to the right correspond to the lane numbers in panel E.
Immunodepletion of P-TEFb from concentrated P1.0 fraction
Immunodepletion was performed by incubating 100 μl concentrated P1.0 fraction containing 0.2% NP-40 and 350 mM KCl with 3.5 μg of anti-CDK9 antibodies at 4°C for 30 min, followed by incubation with 30 μl protein-G Sepharose beads (GE Healthcare). After removal of the beads, the fraction was incubated with 3.5 μg fresh anti-CDK9 antibodies again for 30 min at 4°C, followed by three rounds of incubation with 30 μl protein-G Sepharose beads. The depleted fraction was dialyzed against 100 mM HGKEDP prior to analysis in Western blotting and in vitro transcription assays.
Luciferase assay
The following sequences were inserted into pBluescript SK+ (Stratagene) carrying the mouse U6-promoter (9) to generate the shRNA expression plasmids pBS-U6-hSpt5 (No. 1) and pBS-U6-hSpt5 (No. 2), respectively: U6-P160-1 (No. 1), 5′-GAACTGGGCGAGTATTACAttcaagagaTGTAATACTCGCCCAGTTCtt-3′ [sequences in uppercase correspond to nucleotides (nts) 406–424 of hSpt5 mRNA]; U6-P160-2 (No. 2), 5′-GGCTATATCGGTG TGGTGAttcaagagaTCACCACACCGATGTAGCCtt-3′ (sequences in uppercase correspond to nts 2155–2173 of hSpt5 mRNA.
The reporter gene plasmid carried a HindIII–HindIII DNA fragment (420 bp) derived from the yeast GAL1–GAL10 promoters, the adenovirus early region 4 (E4) promoter sequence (−33 to +9), and the luciferase gene derived from the PicaGene PGV-B plasmid (Toyo-ink).
HeLa S3 cells were maintained in DMEM (Invitrogen) supplemented with 10% fetal calf serum and l-glutamate. Cells (8 × 104 cells) were plated in 24-well plates and transfected with a total of 552 ng of DNA [50 ng of reporter plasmid, 2 ng of pCG-GAL4DBD (1–94) or pCG-GAL4 (1–94)-VP16 (413–490), kindly provided by Dr W. Herr (35) and 500 ng of pBluescript carrying the U6-promoter] using Lipofectamine 2000 (Invitrogen). In Figure 5B, the cells were harvested at the indicated times post-transfection. In Figure 5C, the indicated amounts of Gal4DBD expression plasmid were used. HeLa cells were transferred to fresh medium in a 12-well plate 24 h post-transfection and harvested 72 h post-transfection and luciferase activity was measured. Luciferase activity from triplicate experiments was normalized to protein amounts; data represents normalized values.
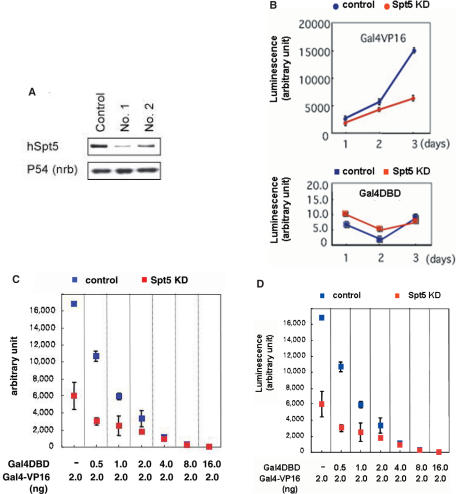
Figure 5.
Gal4-VP16-activated transcription requires Spt5. (A) HeLa cells were transfected with one of the three constructs: a control plasmid, pBS-U6-hSpt5 (No. 1) or pBS-U6-hSpt5 (No. 2). Proteins were isolated from cells 72 h post-transfection, and subjected to Western blot analysis using an antibody against hSpt5 (upper panel). p54/nrb was examined as loading control (lower panel). (B) HeLa cells were co-transfected with a reporter plasmid carrying Gal4-binding sites and the luciferase gene, pCG-GAL4-VP16 (upper panel) or pCG-GAL4 (lower panel), and an shRNA-expressing plasmid for control (blue) or hSpt5 knockdown (KD) cells (red). Transfected cells were collected at the indicated times after transfection, and luciferase activity was measured. Data represents the means ± standard deviation (SD) of three independent experiments. (C) Reporter gene assays were performed as described for (B), except that the indicated amounts of a Gal4 (1–94) expression plasmid were co-transfected with the Gal4VP16 expression plasmid into HeLa cells. Luciferase activity 72 h after transfection is shown. Data represents the means ± SD of three independent experiments. (D) Based on the results in panel C, the effects of hSpt5 knockdown on the reporter gene expression were calculated.
RESULTS
DSIF and Gal4-VP16 have distinct roles in stimulating transcription
We employed an in vitro transcription assay developed previously in our lab to analyze the stimulatory activity of DSIF and Gal4-VP16. The system was based on previous work showing that DSIF stimulated elongation under conditions of limited nucleotide triphosphates (NTPs) (4). A phosphocellulose eluate (P1.0) derived from HeLa cell nuclear extracts (NEs) was generated, as previously described, containing RNAPII, general transcription factors and P-TEFb, but lacking DSIF and NELF (Figure 1A). A supercoiled plasmid DNA template containing five Gal4-binding sites upstream of an adenovirus major late promoter (MLP), and a 380-bp G-free cassette downstream of the promoter, was used as the template (Figure 1B).
In the absence of DSIF, synthesis of full length transcripts was efficiently carried out in P1.0 in the presence of 60 μM ATP, 600 μM CTP, 5 μM UTP and 5 μCi [α32P] UTP (Figure 1C, lane 1). Reducing the concentrations of UTP and CTP resulted in inefficient synthesis of the full length transcripts, and the appearance of short transcripts ranging in size from 100 to 200 nt (Figure 1C, lanes 2 to 4). Further reducing the concentrations of UTP and CTP to 1 μM significantly decreased the amount of shorter transcripts (Figure 1C, lane 5). This was consistent with the previous results obtained using a different template (4). In order to determine whether these results reflected transcriptional pausing or termination, the reaction was carried with the addition of a 1.5 mM ATP, CTP and UTP chase (Figure 1C, lanes 6 to 10). Under these conditions, short transcripts were extended and gradually disappeared, and full-length transcripts appeared after 2 min incubation with the high concentrations of NTPs. These results suggested that the short transcripts generated in this assay resulted from pausing of RNAPII, rather than transcription termination, and that under conditions of limited concentrations of NTPs, RNAPII is prone to pausing in vitro.
Upon the addition of increasing amounts of recombinant histidine-tagged DSIF (His-DSIF) (Figure 1D) to the reaction, there was a reduction in short transcripts, and full-length transcripts were efficiently produced (Figure 1E, lanes 1 to 4). This result was also consistent with the previous results (4), and suggested that DSIF stimulates elongation by reducing pausing. In contrast to DSIF, His-Gal4-VP16 (Figure 1D) increased the synthesis of full-length transcripts as well as short transcripts of various lengths (Figure 1E, lanes 5 to 7). Quantification of the transcripts using a phosphorimager (Figure 1F) revealed that the addition of His-DSIF resulted in a 3- to 6-fold increase in the amount of full-length transcripts and a concomitant decrease in the amount of shorter transcripts. Transcripts of 120 nt, for example, decreased by approximately 2.5-fold (Figure 1F, lanes 1 and 4). The addition of His-Gal4-VP16 resulted in a 2- to 8-fold increase in the amount of full-length transcripts, as well as an increase in the amount of shorter transcripts (i.e. 120 nt transcripts increased 3-fold, Figure 1F, lanes 1 and 7). These results suggested that DSIF and Gal4-VP16 regulate different steps of transcription.
DSIF and Gal4-VP16 act in a cooperative manner
To determine whether the short transcripts generated in the presence of Gal4-VP16 were the products of paused RNAPII, we performed a chase experiment in the presence of Gal4-VP16 (Figure 2A). Short transcripts generated in the presence of His-Gal4-VP16 disappeared after the 2-min chase period (Figure 2A, lanes 3 and 4), demonstrating that they were due to paused RNAPII.
Figure 2.
DSIF and Gal4-VP16 stimulate transcription cooperatively. (A) Transcription assays were carried out as described for Figure 1E. Chase reactions were carried out as described for Figure 1C for 2 min, as shown in the lower diagram. (B) Transcription assays were carried out as described for Figure 1E, except that the initiation/elongation time was changed, as shown in the lower diagram. Initiation/elongation time is indicated at the top. Lanes 2, 4, 6, 8, 10 and 12 contained 7.5 ng of recombinant His-DSIF. Lanes 3, 4, 7, 8, 11 and 12 contained 150 ng of His-Gal4-VP16.
To study the effect of DSIF and Gal4-VP16 in more detail, we varied the incubation time after NTP addition (Figure 2B). When neither DSIF nor Gal4-VP16 was present, there was a negligible difference between the products of an 11.5- and 15-min reaction, wherein RNAPII paused between nts +100 and +200. After a 30-min reaction, more transcripts of various sizes, including full-length transcripts, were generated (Figure 2B, lanes 1, 5 and 9). These results suggested that longer incubation times permit more initiation events. In the absence of DSIF, synthesis of full-length transcripts took over 15 min, whereas in the presence of DSIF, less than 11.5 min were needed (Figure 2B, lane 2). More full-length products appeared after 30 min incubation in the presence of DSIF compared to the absence of DSIF, which was likely due to a higher level of processivity (Figure 2B, lanes 2, 6 and 10). When His-Gal4-VP16 was added to the reaction instead of His-DSIF, in an 11.5-min reaction, transcripts were generated in the size range of 100–200 nt; full-length transcripts first appeared after 15 min, which was later than their appearance in the DSIF-containing reactions (Figure 2B, lanes 3, 7 and 11). In addition, in contrast to DSIF-containing reactions, small-sized transcripts increased significantly over time, suggesting that in the presence of His-Gal4-VP16, initiation occurred efficiently, and the elongation step was rate-limiting. Concomitant addition of His-DSIF and His-Gal4-VP16 increased the amount of full-length products compared to addition of either His-DSIF or His-Gal4-VP16 alone at all time points (Figure 2B, lanes 4, 8 and 12), and the size distribution of the transcripts was more like that seen in reactions containing His-DSIF, rather than His-Gal4-VP16. These results showed that DSIF and Gal4-VP16 act cooperatively to stimulate a high-level of transcriptional activation. When a DNA template lacking Gal4-binding sites was used in the reaction, there was no stimulation of transcription (data not shown). Together, these results indicated that the cooperative function Gal4-VP16 and DSIF requires that both molecules act on the same DNA molecule.
Prior incubation of DSIF with transcription complex is critical to its positive activity
DSIF associates with its target genes upon transcriptional induction, such as during heat-shock induction in Drosophila (11), possibly through recruitment by activators to elongation complexes. On the other hand, we have shown here that DSIF reduces transcriptional pausing equally well in the presence or absence of an activator, suggesting that activators may be dispensable for the recruitment and function of DSIF. We next examined the mechanism of recruitment of DSIF to elongation complexes, and whether activators have any effect on its activity.
As shown earlier (Figure 1C), when high concentrations of cold NTP were added 15 min after transcription was allowed to initiate, paused transcripts were efficiently extended (Figure 3A, lanes 8 to 10). When DSIF was instead added at the same time point, no stimulatory effect was observed (Figure 3A, lanes 5 to 7), suggesting that DSIF is unable to exert its stimulatory activity on the elongation complex at this stage. We therefore changed the time points at which DSIF was added to the reaction, to determine the stage at which DSIF exerted its positive effect (Figure 3B). Compared to reactions in which DSIF was added 40 min before the addition of NTPs (Figure 3B, lane 1), the positive activity of DSIF was attenuated when it was added concomitantly with NTPs, and was abolished when added afterward (Figure 3B, lanes 2 to 6). This indicated that DSIF exerts its positive effect within a short window of time from initiation to elongation. The presence of Gal4-VP16 increased the amount of transcripts, but did not change the effect of DSIF, indicating that the activator does not alter the time-frame in which DSIF functions (Figure 3B, lanes 7 to 12).
Although DSIF is thought to regulate transcription elongation, the above result suggests that DSIF may in fact affect a step before elongation, such as promoter clearance. To examine this possibility, we carried out transcription reactions by using a DNA template that produces long transcripts containing double G-free cassettes (Figure 3C). RNase T1 treatment of the transcripts allows simultaneous quantification of promoter-proximal and -distal regions. The level of promoter-proximal transcripts can generally be equated with the level of transcription initiation, while the ratio of promoter-distal to promoter-proximal transcripts reflects the efficiency of elongation. As reported previously (9,20), DSIF led an increase of efficiency of transcription elongation by 3.8-fold, while it had negligible effect on the synthesis of promoter-proximal transcripts (Figure 3D, lanes 1 and 2), suggesting that DSIF does not influence transcription initiation. On the other hand, Gal4-VP16 activated the synthesis of both promoter-proximal and -distal transcripts with only a marginal effect on the distal-to-proximal ratio, suggesting that Gal4-VP16 mainly affects transcription initiation (Figure 3D, lane 3). Gal4-VP16 and DSIF showed selective effects on initiation and elongation, respectively, even at the highest concentrations examined (data not shown). Collectively, these results are consistent with the previous view and suggest that Gal4-VP16 and DSIF together enhance overall transcription by accelerating different steps in transcription.
NELF does not affect the positive activity of DSIF
It has been established that DSIF negatively regulates transcription elongation by acting in concert with NELF (7,8). In contrast, how DSIF stimulates transcription elongation is largely unknown except that P-TEFb-mediated phosphorylation of the Spt5 subunit of DSIF is responsible for converting DSIF from a repressor to a positive regulator (9). One possible model is that promoter-proximal pausing leads to processive elongation thereafter, possibly serving as a checkpoint for this subsequent process. We therefore examined the roles of NELF and P-TEFb in the positive activity of DSIF. We purified Flag-epitope-tagged NELF (FLAG-NELF) (Figure 4A) from a Flag-NELF-E-expressing HeLa cell line derivative, and added it to the transcription assay (Figure 4B). There was no appreciable effect of addition of NELF on DSIF-activated transcription, or basal transcription levels in the absence of DSIF (Figure 4B, lanes 3 to 6). In the presence of DRB, an inhibitor of the P-TEFb kinase, the positive activity of DSIF was significantly inhibited (Figure 4B, lanes 2 and 8), suggesting that P-TEFb is critical for the positive regulatory effect of DSIF. Addition of NELF resulted in a much stronger inhibitory effect and the generation of transcripts of less than 150 nt (Figure 4B, lanes 9 and 10). Based on the results of previous studies, this effect is likely due to promoter-proximal pausing induced by NELF and DSIF upon inhibition of P-TEFb activity by DRB (4,36). Consistent with the previously published results (36), NELF was unable to induce such pausing in the absence of DSIF, even in the presence of DRB (Figure 4B, lanes 5, 6, 11 and 12). These results indicated that the positive activity of DSIF is dependent on P-TEFb and independent of NELF.
Figure 4.
NELF does not affect the positive activity of DSIF. (A) Silver staining of purified Flag-NELF. (B) In vitro transcription assays were carried out as described for Figure 1E, except that His-DSIF was added to reactions of lanes 2 to 4 and 8 to 10; Flag-NELF was added to the reactions of lanes 3 to 6 and 9 to 12 and DRB was added to the reactions of lanes 7 to 12. Numbers on the left indicate the positions of markers (nucleotides). (C) Western blot analysis of CDK9, RNAPII and CDK8 in the concentrated P1.0 fraction either mock depleted or depleted with anti-CDK9 antibody. (D) In vitro transcription assays were carried out as described in Figure 1E, except that P-TEFb- or mock-immunodepleted concentrated P1.0 fraction was used. P-TEFb was added back prior to incubation as indicated. (E) In vitro transcription assays were carried out using P-TEFb-depleted concentrated P1.0 fraction, His-DSIF and pG5MLP as a template. His-DSIF and Flag-NELF were combined with the mixture during the pre-incubation, while P-TEFb was added 2 min after addition of nucleotides, as shown in the diagram.
To further confirm the above results, we depleted P-TEFb from P1.0 fraction (Figure 4C) and added it back prior to preincubation (Figure 4D) or during elongation (Figure 4E), The latter experiment was carried out in order to exclude the possibility that the presence of P-TEFb during preincubation and initiation steps may cause phosphorylation of RNAPII or DSIF and thus prevent the effect of NELF on elongation. As expected, P-TEFb was critical for the stimulatory activity of DSIF (Figure 4D, lanes 4 to 6). When P-TEFb was absent, NELF cooperated with DSIF to repress elongation (Figure 4E, lanes 3 to 6), and the addition of P-TEFb after the start of transcription alleviated this repression (Figure 4E, lanes 7 to 10). Importantly, with P-TEFb added back, the full-length transcripts were synthesized to similar extent regardless of the presence of NELF, suggesting that the occurrence of NELF- and DSIF-induced promoter-proximal pausing has no influence on the subsequent DSIF- and P-TEFb-induced processive elongation.
DSIF is critical to Gal4-VP16-mediated activation in vivo
To investigate the role of DSIF in transcriptional activation in vivo, we used a reporter gene assay in HeLa cells, in which the expression of the reporter gene is controlled by Gal4-VP16. In this system, we predicted that if Gal4-VP16-mediated transcriptional activation involves the positive activity of DSIF, depletion of the large subunit of DSIF, hSpt5, would reduce expression of the reporter gene. We transfected HeLa cells with a plasmid expressing a short hairpin RNA (shRNA) targeting hSpt5, together with Gal4-VP16 or Gal4DBD expression plasmids, and a luciferase reporter plasmid with Gal4-binding sites upstream of the promoter. Western blot analysis showed an 80% decrease in the amount of hSpt5 in cells transfected with shRNA No. 1, compared to cells transfected with a control plasmid, 72 h after transfection (Figure 5A). A second shRNA targeting a different sequence in hSpt5 similarly knocked down hSpt5 protein levels in cells, indicating that the effect of shRNAs was gene-specific. We used shRNA No. 1 in the following experiments. Knockdown of hSpt5 caused a significant reduction in Gal4-VP16-activated expression of the reporter gene 72 h after transfection (Figure 5B, upper panel), consistent with in vitro results that DSIF cooperates with Gal4-VP16 to achieve high-level activation. In contrast, when cells were transfected with a plasmid encoding Gal4DBD instead of Gal4-VP16, knockdown of hSpt5 had little effect on the low-level expression of the reporter gene (Figure 5B, lower panel).
The above results indicated that Spt5 is essential under conditions where transcriptional activity is induced, but not for transcription under basal conditions. To examine the requirement for DSIF more precisely, we adjusted the expression levels of the luciferase reporter gene by co-expressing varying amounts of Gal4DBD with Gal4-VP16. The Gal4DBD(1–94) is able to bind DNA templates, and is believed to interfere with Gal4-VP16-mediated activation in a competitive manner. As shown in Figure 5C, expression of increasing amounts of Gal4DBD diminished expression of the reporter gene in both hSpt5 knockdown and control cells, suggesting that Gal4-VP16-mediated activation was competitively suppressed by Gal4DBD. Note that the expression level of the luciferase reporter gene in hSpt5 knockdown cells approached that of control cells with increasing amounts of Gal4DBD. Figure 5D shows the ratio of luciferase activity in hSpt5 knockdown cells relative to control cells, illustrating quantitatively the results in Figure 5C. These results indicated that different levels of transcription have different requirements for DSIF.
Together, the results of both in vivo and in vitro assays showed that transcription at a higher level imposes a stronger requirement for DSIF.
DISCUSSION
In the current study, we demonstrated that DSIF acts cooperatively with a DNA-binding activator, Gal4-VP16, to activate transcription in vitro, by reducing pausing. Using a reporter gene assay in cultured HaLa cells, we showed that Spt5 knockdown reduced Gal4-VP16-induced expression of the reporter gene, but had a negligible effect on expression in the absence of the activation domain. The requirement for DSIF was diminished by suppressing transcriptional activation through expression of a DNA-binding competitor of Gal4-VP16, Gal4DBD, suggesting that DSIF is an elongation factor targeting sites of active transcription, but has minimal involvement in basal transcription. In addition, we showed that DSIF exerts its positive effect within a short time-frame from initiation to elongation but does not affect initiation, and that NELF is not involved in the positive activity of DSIF. These results elucidate the role of DSIF in transcriptional activation, giving additional weight to the correlation between DSIF and sites of active transcription, and support the idea that transcription at high-levels has a stronger requirement for DSIF. Thus, expression of genes that are undergoing active transcription is likely to be affected more by the loss of function of DSIF compared to genes undergoing basal-level transcription.
Regulation of transcription elongation and kinetics of activation
Our data suggests that Gal4-VP16 stimulates transcription initiation but is unable to overcome pausing during elongation, and that DSIF is required to reduce pausing, thereby cooperating with the activator to promote transcription.
As to why DSIF would be required only for high-level transcription, it is possible that when transcription occurs at a very low level, preinitiation or initiation may be the rate-limiting step, and therefore, the efficiency of the subsequent elongation step (i.e. the presence or absence of DSIF) may not contribute substantially to the overall rate of RNA synthesis. Upon transcriptional induction, however, the rates of preinitiation and initiation may be elevated by the action of activators, and the efficiency of the elongation step may become critical to the overall rate of RNA synthesis. Although it is not clear to what extent this model is applicable to cellular genes, it has been shown that expression of c-fos has similar requirements for DSIF as the reporter gene studied here (9). The general applicability of this model may be tested by carrying out microarray analysis of DSIF knockdown samples prepared under various induction conditions.
How then does DSIF play important roles in activated transcription? The following explanations may account for the significant requirement for DSIF: (a) activators may directly recruit DSIF to target genes, making the effect of DSIF significant; (b) activators may indirectly recruit DSIF through other proteins; (c) activators may cause a modification of DSIF which enhances its positive activity.
As to the first and second possibilities, several groups have reported the recruitment of DSIF to various genes, including heat shock genes, c-fos, junB and MAP kinase phosphotase-1 (MKP-1), upon extracellular stimuli (9–11,37). However, no evidence so far has been found to show direct physical interactions between DSIF and activators. We also failed to detect any interaction between Gal4-VP16 and DSIF using a GST pull-down assay or a cross-linked immunoprecipitation experiment (data not shown). Moreover, Gal4-VP16 neither increased the stimulatory effect of DSIF (Figure 3D) nor changed the time-frame of DSIF recruitment (Figure 3B), suggesting that Gal4-VP16 may not directly recruit DSIF. Since DSIF binds to RNAPII (4), DSIF may be recruited secondarily through its interaction with RNAPII upon transcriptional induction. This idea is supported by the observation that the DSIF density changes dynamically corresponding to similar change of RNAPII around promoter regions of hsp70, c-fos, junB and MKP-1 during activation (9–11,37).
Methylation of the Spt5 subunit of DSIF may negatively regulate the association with RNAPII (38). In contrast, P-TEFb-mediated phosphorylation of Spt5 is critical for the positive activity of DSIF (9,13). DSIF may undergo these modifications during activation of transcription. As to the third possibility, the protein arginine methyltransferases PRMT1 and PRMT5, which are capable of methylating Spt5, associate with cytokine-inducible promoters under basal conditions and disassociate from the promoters after activation (38). In contrast, P-TEFb has been reported to be recruited to promoter regions during the activation of transcription (11,37). Several transcription factors, such as CIITA, NF-κB, Myc and STAT3, have been shown to interact with and recruit P-TEFb to target gene promoters (39–42). During Tat-transactivated HIV transcription, P-TEFb is recruited by Tat to stimulate elongation (43). However, we did not find any interaction between P-TEFb and Gal4-VP16 in a GST pull-down assay and a cross-linked immunoprecipitation experiment (data not shown). Yang et al. (44) have reported that P-TEFb interacts with Mediator through Brd4, which may be implicated in the recruitment of P-TEFb. The Mediator is a multiprotein complex recruited to promoters via activators, including Gal4-VP16, and serves as a molecular bridge between activators and RNAPII (45–47). Activators may stimulate the recruitment of P-TEFb through Mediator to modulate phosphorylation of the CTD of RNAPII as well as of DSIF during a post-initiation stage.
DSIF exerts its positive effect within a short time window from initiation to elongation
We showed that DSIF was unable to exert its positive effect in vitro when added to transcription reactions at later time points. This finding can be interpreted in several ways. DSIF may only associate with early elongation complexes, and may not be able to exert its positive effect on mature elongation complexes. This may occur in one of several ways. First, a factor that helps DSIF to enter transcription complexes, such as Mediator, may not exist in late elongation complexes. Mediator does not appear to walk along with the elongation complex to downstream regions of genes. Rather, it may act as a bridging molecule between activators and DSIF, and thus may help recruit more DSIF to activated genes. Functional links between DSIF and Mediator have been reported recently, although their physical interaction has not yet known (48). Second, RNAPII may undergo a conformational change that inhibits its interaction with DSIF after a specific time-frame of DSIF action. Thirdly, other factors recruited at a later stage of transcription may mask the binding site on RNAPII that is involved in its interaction with DSIF.
Alternatively, prior incubation of DSIF with transcription complexes may be necessary for its action. This model would involve a slow mechanism of action, such as modification of DSIF. Since P-TEFb-mediated phosphorylation of the CTR of hSpt5 activates the positive activity of DSIF (9), the time required for this phosphorylation event may delay the kinetics of DSIF action. Furthermore, since P-TEFb is prone to be released from transcription complexes during elongation (49,50), DSIF may not be phosphorylated efficiently in mature elongation complexes because of the lack of P-TEFb.
ACKNOWLEDGEMENTS
We are indebted to Drs R.G. Roeder, S. Hirose and T. Kokubo for many helpful discussions, Dr Q. Zhou for the antibody to CDK9 and the protocol for immunodepletion. We are grateful to Dr T. Yung for many helpful suggestions to our manuscript preparation. We also thank S. Kamijo and T. Yamada for shRNA expression vectors, Y. Chen for recombinant DSIF protein preparation and members of the Handa Lab for helpful suggestions. We thank Drs W. Herr and D. Reinberg for providing expression vectors of Gal4-fusion proteins in human cells and the Gal4VP16 protein in E. coli, respectively. This study was supported by a Grant-in Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology (to T.W. and H.H.) and a Tokyo Tech. Award for Challenging Research to T. W. In addition, this study was supported in part by a Grant from the 21st Century COE Program from the Ministry of Education, Culture, Sports, Science and Technology, Special Coordination Funds for Promoting Science and Technology from the Japan Science and Technology Agency and a grant for Research and Development Projects in Cooperation with Academic Institutions from the New Energy and Industrial Technology Development Organization (to H.H).
Funding to pay the Open Access publication charges for this article was provided by XXX.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sims RJ, 3rd, Mandal SS, Reinberg D. Recent highlights of RNA-polymerase-II-mediated transcription. Curr. Opin. Cell Biol. 2004;16:263–271. doi: 10.1016/j.ceb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 2.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 3.Shilatifard A, Conaway RC, Conaway JW. The RNA polymerase II elongation complex. Annu. Rev. Biochem. 2003;72:693–715. doi: 10.1146/annurev.biochem.72.121801.161551. [DOI] [PubMed] [Google Scholar]
- 4.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. J. Biol. Chem. 1999a;274:8085–8092. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- 6.Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999b;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 8.Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim DK, Hasegawa J, Omori M, et al. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell. Biol. 2003;23:1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 10.Aida M, Chen Y, Nakajima K, Yamaguchi Y, Wada T, Handa H. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol. Cell. Biol. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 13.Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplan CD, Morris JR, Wu C, Winston F. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 2000;14:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saunders A, Werner J, Andrulis ED, Nakayama T, Hirose S, Reinberg D, Lis JT. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301:1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- 16.Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc. Natl Acad. Sci. USA. 2004;101:7572–7577. doi: 10.1073/pnas.0401493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR, 3rd, Hartzog GA. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- 19.Pei Y, Shuman S. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J. Biol. Chem. 2002;277:19639–19648. doi: 10.1074/jbc.M200015200. [DOI] [PubMed] [Google Scholar]
- 20.Guo S, Yamaguchi Y, Schilbach S, Wada T, Lee J, Goddard A, French D, Handa H, Rosenthal A. A regulator of transcriptional elongation controls vertebrate neuronal development. Nature. 2000;408:366–369. doi: 10.1038/35042590. [DOI] [PubMed] [Google Scholar]
- 21.Jennings BH, Shah S, Yamaguchi Y, Seki M, Phillips RG, Handa H, Ish-Horowicz D. Locus-specific requirements for Spt5 in transcriptional activation and repression in Drosophila. Curr. Biol. 2004;14:1680–1684. doi: 10.1016/j.cub.2004.08.066. [DOI] [PubMed] [Google Scholar]
- 22.Winston F. Control of eukaryotic transcription elongation. Genome Biol. 2001;2:REVIEWS1006. doi: 10.1186/gb-2001-2-2-reviews1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keegan BR, Feldman JL, Lee DH, Koos DS, Ho RK, Stainier DY, Yelon D. The elongation factors Pandora/Spt6 and Foggy/Spt5 promote transcription in the zebrafish embryo. Development. 2002;129:1623–1632. doi: 10.1242/dev.129.7.1623. [DOI] [PubMed] [Google Scholar]
- 24.Ping YH, Chu CY, Cao H, Jacque JM, Stevenson M, Rana TM. Modulating HIV-1 replication by RNA interference directed against human transcription elongation factor SPT5. Retrovirology. 2004;1:46. doi: 10.1186/1742-4690-1-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bourgeois CF, Kim YK, Churcher MJ, West MJ, Karn J. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol. Cell. Biol. 2002;22:1079–1093. doi: 10.1128/MCB.22.4.1079-1093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikeda K, Stuehler T, Meisterernst M. The H1 and H2 regions of the activation domain of herpes simplex virion protein 16 stimulate transcription through distinct molecular mechanisms. Genes Cells. 2002;7:49–58. doi: 10.1046/j.1356-9597.2001.00492.x. [DOI] [PubMed] [Google Scholar]
- 27.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier JL, Triezenberg SJ, Reinberg D, Flores O, et al. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol. Cell. Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodrich JA, Hoey T, Thut CJ, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 29.Mittler G, Stuhler T, Santolin L, Uhlmann T, Kremmer E, Lottspeich F, Berti L, Meisterernst M. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. Embo J. 2003;22:6494–6504. doi: 10.1093/emboj/cdg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blau J, Xiao H, McCracken S, O’Hare P, Greenblatt J, Bentley D. Three functional classes of transcriptional activation domain. Mol. Cell. Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reece RJ, Rickles RJ, Ptashne M. Overproduction and single-step purification of GAL4 fusion proteins from Escherichia coli. Gene. 1993;126:105–107. doi: 10.1016/0378-1119(93)90596-u. [DOI] [PubMed] [Google Scholar]
- 32.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 33.Kim DK, Inukai N, Yamada T, Furuya A, Sato H, Yamaguchi Y, Wada T, Handa H. Structure-function analysis of human Spt4: evidence that hSpt4 and hSpt5 exert their roles in transcriptional elongation as parts of the DSIF complex. Genes Cells. 2003;8:371–378. doi: 10.1046/j.1365-2443.2003.00638.x. [DOI] [PubMed] [Google Scholar]
- 34.Endoh M, Zhu W, Hasegawa J, Watanabe H, Kim DK, Aida M, Inukai N, Narita T, Yamada T, et al. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol. Cell. Biol. 2004;24:3324–3336. doi: 10.1128/MCB.24.8.3324-3336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das G, Hinkley CS, Herr W. Basal promoter elements as a selective determinant of transcriptional activator function. Nature. 1995;374:657–660. doi: 10.1038/374657a0. [DOI] [PubMed] [Google Scholar]
- 36.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 37.Fujita T, Ryser S, Tortola S, Piuz I, Schlegel W. Gene-specific recruitment of positive and negative elongation factors during stimulated transcription of the MKP-1 gene in neuroendocrine cells. Nucleic Acids Res. 2007;35:1007–1017. doi: 10.1093/nar/gkl1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwak YT, Guo J, Prajapati S, Park KJ, Surabhi RM, Miller B, Gehrig P, Gaynor RB. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol. Cell. 2003;11:1055–1066. doi: 10.1016/s1097-2765(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 39.Kanazawa S, Okamoto T, Peterlin BM. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity. 2000;12:61–70. doi: 10.1016/s1074-7613(00)80159-4. [DOI] [PubMed] [Google Scholar]
- 40.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 41.Eberhardy SR, Farnham PJ. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 2001;276:48562–48571. doi: 10.1074/jbc.M109014200. [DOI] [PubMed] [Google Scholar]
- 42.Giraud S, Hurlstone A, Avril S, Coqueret O. Implication of BRG1 and cdk9 in the STAT3-mediated activation of the p21waf1 gene. Oncogene. 2004;23:7391–7398. doi: 10.1038/sj.onc.1207972. [DOI] [PubMed] [Google Scholar]
- 43.Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 45.Uhlmann T, Boeing S, Lehmbacher M, Meisterernst M. The VP16 activation domain establishes an active mediator lacking CDK8 in vivo. J. Biol. Chem. 2007;282:2163–2173. doi: 10.1074/jbc.M608451200. [DOI] [PubMed] [Google Scholar]
- 46.Malik S, Roeder RG. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- 47.Gold MO, Tassan JP, Nigg EA, Rice AP, Herrmann CH. Viral transactivators E1A and VP16 interact with a large complex that is associated with CTD kinase activity and contains CDK8. Nucleic Acids Res. 1996;24:3771–3777. doi: 10.1093/nar/24.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malik S, Barrero MJ, Jones T. Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator. Proc. Natl Acad. Sci. USA. 2007;104:6182–6187. doi: 10.1073/pnas.0608717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 50.Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]