Abstract
DNA and RNA oligomers that contain stretches of guanines can associate to form stable secondary structures including G-quadruplexes. Our study shows that the (UUAAAAGAAAAGGGGGGAU) RNA sequence, from the human immunodeficiency virus type 1 (HIV-1 polypurine tract or PPT sequence) forms in vitro a stable folded structure involving the G-run. We have investigated the ability of pyrimidine peptide nucleic acid (PNA) oligomers targeted to the PPT sequence to invade the folded RNA and exhibit biological activity at the translation level in vitro and in cells. We find that PNAs can form stable complexes even with the structured PPT RNA target at neutral pH. We show that T-rich PNAs, namely the tridecamer-I PNA (C4T4CT4) forms triplex structures whereas the C-rich tridecamer-II PNA (TC6T4CT) likely forms a duplex with the target RNA. Interestingly, we find that both C-rich and T-rich PNAs arrested in vitro translation elongation specifically at the PPT target site. Finally, we show that T-rich and C-rich tridecamer PNAs that have been identified as efficient and specific blockers of translation elongation in vitro, specifically inhibit translation in streptolysin-O permeabilized cells where the PPT target sequence has been introduced upstream the reporter luciferase gene.
INTRODUCTION
DNA and RNA oligomers that contain consecutive guanine (G) nucleotides are capable of folding into stable secondary structures such as G-quadruplexes, wherein four Gs are hydrogen bonded together into a roughly square planar array (1). G-quadruplexes are of remarkable stability and have been proposed to be involved in regulation of gene expression. For example, a DNA G-quadruplex structure formed in the c-myc promoter region functions as a transcriptional repressor element and an RNA G-quadruplex is believed to regulate alternative splicing of the pre-mRNA coding for hTERT, the reverse transcriptase component of the enzyme telomerase (2,3).
Human immunodeficiency virus type-1 (HIV-1) contains the 5′A4GA4G6A polypurine tract sequence (PPT) that is conserved in all HIV-1 strains and is present in the coding region of integrase (IN) and nef messenger RNAs. G-quadruplexes have been implicated in HIV-1 RNA dimerization (4) and recently were shown to occur in a reverse transcription intermediate, namely between the overlapping strands of the HIV-1 central DNA flap (5). It has been shown that HIV-1 nucleocapsid (NCp) and gp 120 envelope protein exhibit a high affinity for several tetramolecular quadruplexes (6,7). RNA quadruplexes are more stable than their DNA counterparts and in most cases no dissociation is experimentally observed for G tracts involving five guanine quartets (8).
In the present study, we have used peptide nucleic acid (PNA) targeted to the folded PPT sequence of HIV-1 messenger RNA. PNAs are DNA analogues in which the N-(2-aminoethyl) glycine units replace the deoxyribose phosphate backbone (9–11). PNAs are capable of sequence specific recognition of DNA and RNA, obeying the Watson–Crick hydrogen-bonding or/and Hoogsteen schemes (9–11). The neutral amide backbone of PNAs increases their binding affinity to DNA and RNA and the hybrid complexes exhibit high thermal stability (11–14). Short PNA probes were shown to be able to disturb and finally to bind folded RNA structures used as target sequences (15). Here, we show that pyrimidine PNAs overcome kinetic and thermodynamic obstacles and succeed to hybridize to folded PPT sequence and finally to unfold it. We have examined whether complexes formed with PNAs on the PPT sequence, likely triplex and duplex structures, affect RNA translation elongation in vitro. The cellular antisense activity of the best inhibitors in vitro was tested in streptolysin-O (SLO) permeabilized cells stably transformed with two reporter genes, the firefly luciferase (luc) and GFP that contain upstream of the reporter genes either the wild-type HIV-1 PPT target sequence, or a mutated HIV-2 PPT sequence, respectively.
MATERIAL AND METHODS
Oligonucleotides
The DNA, RNA and PNA oligonucleotides presented here were synthesized by Eurogentec (Seraing, Belgium). 13-mer-Acr PNA was synthesized as previously described (16). Sequences and names are given in Table 1. The oligoribonucleotides (RNA) were 5′ end-labelled with [γ32P] ATP (3000 Ci/mmol) and 10 units of T4 polynucleotide kinase.
Table 1.
Oligoribonucleotides and PNA used in this study
| A: Oligoribonucleotides used as PNA binding target for thermal denaturation experiments | |
| RNA-I-wt: | 5′AAAAGAAAAGGGG3′ |
| RNA-I-mut: | 5′AAAAGAAAAAGGA3′ |
| RNA-II-wt: | 5′AGAAAAGGGGGGA3′ |
| RNA-II-mut: | 5′AGAAAAAGAGAGA3′ |
| B: Oligoribonucleotides used as PNA binding target for gel shift assays | |
| RNA-III-wt: | 5′UUAAAAGAAAAGGGGGGAU3′ |
| RNA-III-mut: | 5′AUAAAAGAAAAAGGAGGAU3′ |
| C: PNAas used in this study | |
| 13mer-I PNA: | NH2-CCCCTTTTCTTTT-Lys |
| 13mer-II PNA: | NH2-TCCCCCCTTTTCT-Lys |
| 13mer-I-Acr PNA: | Acr-eg1-CCCCTTTTCTTTT-Lysb |
| 9mer PNA: | NH2-TTTTCTTTT-Lys |
| 9mer-Acr PNA: | Acr-AEEA-TTTTCTTTT-Lysc |
| 9mer-bis-PNA: | NH2-TTTTCTTTT-AEEA-TTTTCTTTT-Lysc |
| 11mer-bis-PNA: | NH2-CCTTTTCTTTT-AEEA-TTTTCTTTTCC-Lysc |
| 13mer-scr PNA: | NH2-TTTTCCTCTCCCT-Lys |
| D: ODNs used for RNAse H cleavage | |
| 15mer-PPT: | 5′ TCTTTTAAAATTGTG3′ |
| 15mer-PPT-I: | 5′ AATTTTCTTTAATTC3′ |
| 15mer-PPT-II: | 5′ TTATTCTTTAGTTTG3′ |
aPNA base sequences are written from N to C terminus.
beg1, 8-amino-3, 6 dioxaoctanoic acid.
cAEEA, 2-(2aminoethoxy) ethoxy acetic acid.
Spectroscopic experiments
Association and dissociation of the different PNA–RNA complexes were estimated by cooling/heating experiments, recording the UV absorbance as a function of temperature on Uvikon XL spectrophotometer (BioTek) with 1 cm optical pathlength quartz cuvettes. The temperature of cell holder was regulated by a Peltier thermosystem driven by LifePower tm software (DuSoTec GmbH) for the control and data acquisition. Samples were first heated to 95°C, then cooled down to 20°C and heated to 85°C at the rate of 0.5°C/min or 0.1°C/min with absorbance readings taken every 1°C at 260 and 400 nm. Samples were prepared in a buffer containing 100 mM KCl and 10 mM sodium cacodylate, pH 7. For cooling-melting temperature analysis, the baseline drift was corrected by subtracting absorbance at 400 nm from that at 260 nm. The cooling–melting curves were obtained by plotting the corrected absorbance at 260 nm versus temperature. The maximum of the first derivative of the cooling–melting curves (∂A/∂T) was taken as an estimation of Tass or Tdis values. The Tass and Tdis were estimated within, ±1°C accuracy.
Electrophoretic mobility shift assay
Unless otherwise indicated, the standard buffer (10 μl) for the PNA-binding assay contained 2 nM of 5′-end labelled RNAs in 50 mM Tris, pH 8. Samples were incubated with increasing concentrations of PNA at 25°C for 10 min. Gel electrophoresis was run at 25°C on a 15% polyacrylamide/bisacrylamide (19/1) non-denaturing gel or 12% polyacrylamide/7M urea denaturing gel. The gel and buffer contained TBE (100 mM Tris/90 mM Boric acid/1 mM EDTA) at pH 8.3. Following autoradiography a PhosphorImager was used for quantitation.
In vitro transcription and translation
The plasmid pRP159 (gift from Dr D. van Gent, Netherlands Cancer Institute, Amsterdam) was constructed by insertion of HIV-1 integrase gene (∼0.9 kb), into the pSP65 host vector (Promega) behind the SP6 promoter. In vitro transcription was performed on Nde-I linearized pRP159 using standard conditions (RiboScribe SP6 RNA Probe Synthesis Kit, Epicentre Technologies). The transcripts were translated in rabbit reticulocyte lysate purchased from Promega. Lysate (17 µl) was added to the reaction mixture (25 μl) supplemented with amino acid mix (1 μl, without methionine), 35S methionine (1 μl, 15 μCi/μl), PNA and transcript. The reaction mixture was incubated for 20 min at 37°C and 8 μl of the mixture was analysed on a 14% (w/v) Tris–Glycine, SDS–PAGE, NOVEX ™ pre-cast gel (Invitrogen). Translation kinetics experiments were carried out in the same reaction mixture. Aliquots (10 µl) were removed at the indicated times and quenched by addition of an equal volume of 2-fold concentrated loading buffer supplemented with β mercaptoethanol. Protein was quantitated using a Molecular Dynamics PhosphorImager.
Cell cultures
The CMV (+) PPT/HeLa cells stably contain two reporter genes, the firefly luciferase (Photinus pyralis) gene (luc) and the GFP gene, under the control of a bi-directional doxycycline-inducible CMV promoter. These cells contain the PPT sequence (5′AAAAGAAAAGGGGGGA) or a mutated sequence (5′ AAAAGAAGGGGAGGAA) upstream of the AUG translation start site of luciferase and GFP genes respectively (17). The CMV-luc/HeLa cells contain the reporter gene (luc) under the control of a doxycycline-inducible CMV promoter. There is no insertion of the PPT containing fragment in these cells. Both cell lines were grown in DMEM (Invitrogen) supplemented with 10% of fetal bovine serum, 2 mM glutamine, 50 U/ml of penicillin and 50 μg/ml of streptomycin. Cell culture medium was supplemented with G418 (500 μg/ml) and puromicin (2.5 μg/ml) to maintain the integrated target sequences.
Reversible permeabilization and luciferase assay
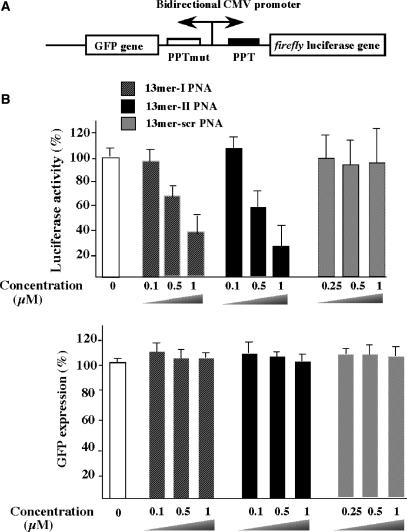
SLO (Institute of Medical Microbiology and Hygiene, Mainz, Germany) was used to reversibly permeabilize CMV/(+)PPT/HeLa or CMV-luc/HeLa cell lines toward PNAs according to a recently revised protocol (17–18). SLO was conserved in PBS buffer supplemented with 0.1% BSA. Cells were washed twice and re-suspended in HBSS (Hanks’ balanced salt solution with calcium and magnesium, 10 mM HEPES, and 1% fetal bovine serum). For each experiment, in a 48-well dishes, 1.3 × 105 cells in 100 µl were permeabilized by addition of an optimized amount of SLO (110 ng) and then incubated at 37°C for 15 min, in the presence of PNAs. Resealing was achieved by addition of 800 µl of DMEM supplemented with 10% fetal calf serum and further incubation at 37°C for 20 min. Cells were then transferred in 96-well dishes (4 × 104 cells/well) and cultured at 37°C for 4 h and then induced by addition of doxycycline (Sigma) and further cultured 40 h before quantifying firefly luciferase activity, GFP and total proteins. In the same time, cells were examined with respect to permeabilization efficiency and viability by flow cytometry (Facsort, Beckton Dickinson). Briefly, during permeabilization PNAs were replaced with FITC (20 µM) and after resealing cells were washed twice with PBS and passed through flow cytometer in PBS supplemented with propidium iodide (10 µg/ml) which permitted to determine the fraction of permeabilized and viable cells in the original culture. At the end of experiment, cells were harvested for lysate (passive lysis buffer, Promega) and both the firefly luciferase activity [expressed in relative light units (RLU)], GFP fluorescence and protein concentration [Bradford reagent from Bio-Rad, expressed in optical density (OD)] were measured using a spectrofluorimeter (Wallac Victor 2 Multi-label Counter, Perkin Elmer). The luciferase activity and GFP expression shown in Figure 6 were normalized to the absorbance data, that reflect the amount of proteins, and then expressed as a percentage compared with the luciferase activity and GFP fluorescence levels that were obtained in SLO-treated cells in the absence of PNA. Each data point was averaged over two replicates of three separate experiments.
Figure 6.
(A) CMV (+PPT)/HeLa cell lines contain bi-directional CMV promoter that allows expression of two reporter genes [firefly luciferase (luc) and GFP]. The PPT-wt sequence (5′AAAAGAAAAGGGGGGA3′) was inserted upstream of the luc gene and a mutated version (PPT-mut, 5′AAAAGAAGGGGAGGAA3′, the four mutations are underlined) was inserted upstream of the GFP gene. (B) PNA tridecamers are efficient inhibitors of translation. Cells were reversibly permeabilized by SLO in the presence of various PNAs (see ‘Materials and Methods’ section). PNA concentrations in final culturing conditions are indicated.
RESULTS AND DISCUSSION
RNA target characterization
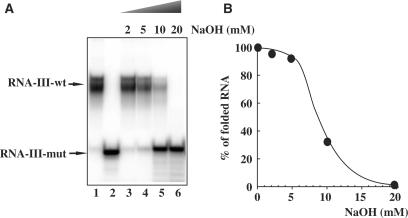
Figure 1 shows the electrophoretic mobility of the 19-mer, RNA-III-wt oligoribonucleotide containing the PPT sequence of the HIV-1 integrase coding sequence (Table 1). Several slow migrating species are detected with wild-type RNA (lane 1), while the derived 19-mer mutated RNA, RNA-III-mut, in which two adenines break the G-tract migrates faster and as a single species (lane 2). Such observations supports the involvement of the G-run in the structure of the RNA PPT sequence that likely adopts a quadruplex form. Indeed, stretches of guanines can associate through hydrogen bonding to form four-stranded structures. G-quadruplexes are of remarkable stability and have a preferential affinity for monovalent cations (typically K+ or Na+) that exhibit a suitable size for interaction with the electronegative carbonyl oxygen ring inside the G-quartet (19–21). Recently, Mergny et al. (20) have shown that the RNA sequence r-UG4U, forms extremely stable parallel RNA quadruplex that is much more stable than its DNA counterpart. The increased stability of RNA quadruplexes results from a faster association and a slower dissociation. Strand concentration and ionic environment plays an important role in the kinetics of quadruplex formation. In our case, the folded RNA structure was observed at very low 19-mer RNA-III-wt concentrations (2 nM) in the absence of added monovalent ions. One explanation is that this 19-mer RNA sequence is already folded after the synthesis and that during sample drying, setting in solution and dilution, the folded structure is maintained. This is not astonishing insofar as the lifetime of r-UG4U quadruplex at 37°C was estimated to be more than 100 years (20). The multiple bands of RNA-III-wt can be assessed to folded structures isomers. We have used in our studies different batches of synthesis and always observed the same profile of migration. NaOH addition to the RNA-III-wt showed a shift of the folded structures to an unfolded structure, which migrated at the same position as mutated RNA-III-mut (Figure 1A). Folded structures titration by NaOH allowed the determination of a mid-point transition from structured-RNA-III-wt to RNA-III-wt monomer at 9 mM NaOH (Figure 1B).
Figure 1.
RNA-III-wt is a 19-mer oligoribonucleotide that contains a G6 tract (lane 1). Two adenines break the G6 tract in RNA-III-mut oligomer (lane 2) (see Table 1 for sequences). (A) 32P-radiolabelled RNA-III-mut (1 μM) was incubated with the indicated amounts of NaOH for 15 min at 25°C and loaded on a 15% polyacrylamide gel. (B) Denaturation of RNA-III-wt by NaOH was quantified by PhosphorImager analysis.
Invasion of PPT RNA structure by complementary PNAs
Native gel-shift experiments
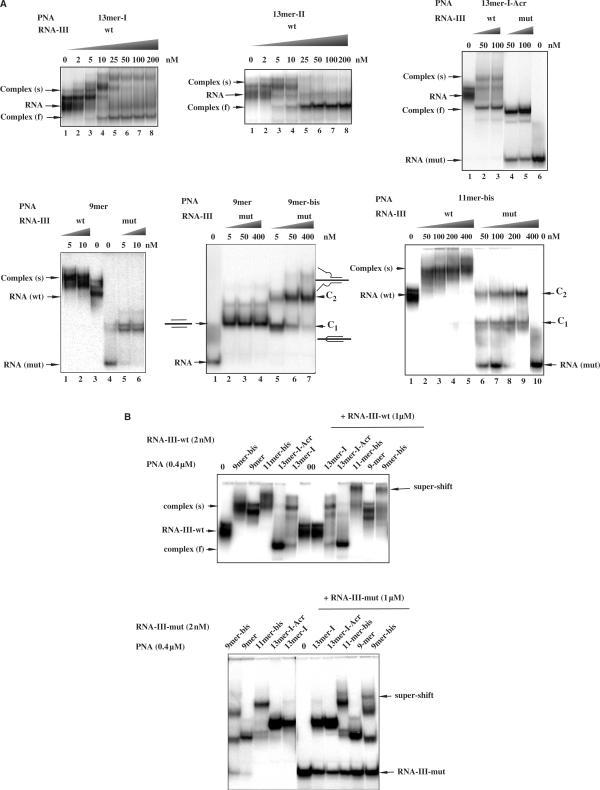
In order to determine whether short complementary PNAs can invade and bind to structured RNA targets, here RNA-III-wt, we have used electrophoretic mobility shift assays. Binding of PNAs to unfolded RNA-III-mut was also analysed. 32P-radiolabelled RNAs were incubated in the absence or in the presence of increasing concentrations of tridecamer PNAs, 13-mer-I and 13-mer-II. Figure 2A shows that at low concentrations (<25 nM) tridecamers form complexes that migrate slower than folded RNA [complex (s)] while at higher concentrations a discrete, faster migrating complexes [complex (f)] were predominant. We postulated that retarded low mobility species result from the fixation of PNAs to the A-rich region of the target sequence not engaged in the structure formed by the G-tract while high mobility complexes result from the invasion of RNA structures by 13-mer-I and 13-mer-II PNAs.13-mer-II PNA that can form six C.G base pairs invades slightly more efficiently folded RNA than 13-mer-I PNA that can form only four C.G base pairs with the PPT RNA target. The binding of the two tridecamer PNAs was also studied on the mutated target, RNA-III-mut. This RNA sequence is not stably structured and is complementary on 11 and 8 contiguous nucleotides with 13-mer-I and 13-mer-II PNAs, respectively. Then, binding of 13-mer-PNAs can be also observed on this RNA sequence, with complexes migrating at the same position as the ones observed on the wild-type sequence after PNA-induced unfolding (Figure 2A). Binding efficiencies of 13-mer-PNAs on the two RNA targets, wild type and mutated, was measured. For tridecamer-PNAs complexed to RNA-III-wt, we could not determine precisely the K50 values (that means the concentrations of PNAs required for the formation of 50% of complex) because of the multiplicity of the slow-mobility complexes, however these values are situated between 2 and 5 nM. On the contrary, K50 values can be determined on unstructured RNA-III mut; for unmodified and acridine-modified tridecamer-PNAs complexed to RNA-III-mut, K50 was determined around 4 nM.
Figure 2.
(A) Gel mobility-shift analysis of PNAs binding to their target RNA. A fixed concentration of the 32P-labeled RNA-III-wt and mismatches containing RNA-III-mut (2 × 10−9 M) were incubated in 50 mM Tris (pH 8) buffer with increasing concentration of PNA as indicated above the lanes. Slow migrating complexes (s) correspond to the complexes formed with folded RNA whereas higher electrophoretic mobility complexes (f) correspond to the invasion complexes that unfold the structured RNA target. C1 and C2 denote complexes involving one and two molecules of bis-PNAs respectively. (B) An excess of the RNA target sequence (2.5-fold excess relative to PNA concentration (400 nM)) was added to the complexes and incubated for 5 min. Bound and unbound complexes were separated in a 15% non-denaturing polyacrylamide gel at 20°C. RNA-III-wt (top) and RNA-III-mut (bottom) were used as targets.
To further characterize PNA binding to the folded PPT RNA sequence we used truncated PNA sequences, 9-mer and 11-mer. They are complementary both to the wild-type and the mutated sequence, to the A-rich region that is not involved in the structure of the PPT RNA. The 9-mer and 11-mer sequences were also used as bis-PNAs (see Table 1 for sequences), that were expected to bind efficiently to the PPT sequence. As expected, these short sequences bind to folded RNA-III-wt sequence and no unfolding was observed even at high PNA concentrations (Figure 2A), consistent with the fact that the target sequence is not involved in the structure. The 9-mer and 9-mer-bis PNAs, bind very efficiently to the wild-type and mutated RNA sequences. K50 values are situated in the range of 2–4 nM. On the mutated RNA target, the complexes are well resolved; with the monomeric PNA 9-mer an unique complex is observed whereas with both bis-PNAs, 9-mer and 11-mer-bis PNAs, two discrete shifted complexes (Figure 2A, C1 and C2) were observed. In the case of 11-mer-bis PNA, there are two bands in complex C1. Probably, 11-mer-bis PNA forms with RNA-III-wt two structurally isomeric C1 complexes. Similar structurally isomeric complexes formed between homopyrimidine bis-PNAs and single- and double-stranded DNA targets have been reported (22). At low bis-PNA concentrations, complex C1 was predominant while at high concentrations complex C2 was predominant. The complex C1 formed with 9-mer bis-PNA migrates as complex formed with 9-mer PNA and corresponds to the triplex formed with the A4GA4 purine sequence with two 9-mer PNAs. Complex C2 can be assigned to the triplex formation with two bis-PNAs as illustrated in Figure 2A. If it was the case, this complex would contain two free PNA strands that could form a PNA–RNA–PNA triplex with another RNA target. Indeed, upon addition of an excess of 19-mer RNA super-shifted complexes have been obtained with 9-mer bis-PNA and 11-mer bis-PNA (Figure 2B). Figure 2B shows that in the presence of 400 nM PNAs, almost the totality of the RNA-III-wt was converted in slow migrating (s) and fast migrating (f) complexes depending on PNA sequences. Upon addition of an excess RNA-III no change in the migration of complexes were observed except for complexes formed with 11-mer-bis and 9-mer-bis PNAs that were super-shifted (Figure 2B). These results support a model where each bis-PNA molecule participates in the triplex structure with only one arm; the second arm of each bis-PNA remains free (Figure 2A, complex C2). It is noteworthy that the lifetimes of complexes formed with RNA-III-wt are higher than those formed with RNA-III-mut as upon addition of an excess of target RNA followed by 5 min of incubation, partial dissociation of PNA–RNA-III-mut complexes was observed, whereas PNA–RNA-III-wt complexes were not dissociated (Figure 2B).
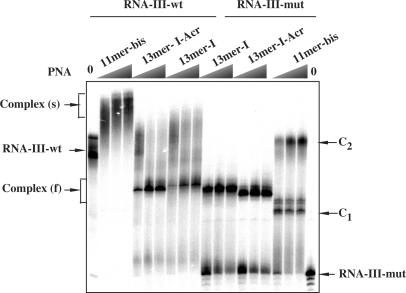
PNA2–RNA triplexes resist to denaturing gel conditions
In order to explore the stability of folded RNA and of its complexes with different PNAs we loaded the samples on denaturing acrylamide (7 M urea) gel. Figure 3 shows that wild-type RNA structures and complexes with PNA resist to the denaturing conditions. Complexes formed with mutated RNA are less stable in these conditions since complete shift of RNA was not achieved at the concentrations observed in non-denaturing gel.
Figure 3.
The 32P-labelled RNA-III-wt and RNA-III-mut were incubated in 50 mM Tris (pH 8) buffer with increasing concentration of PNA (50, 200 and 400 nM) and loaded on a 15% polyacrylamide, 7 M urea gel. Arrows indicate the migration position of wild-type and mutated RNAs and of complexes s, f, C1 and C2 as described in Figure 2 legend.
The cooling/melting temperatures of complexes formed on PPT RNA sequence with different PNAs
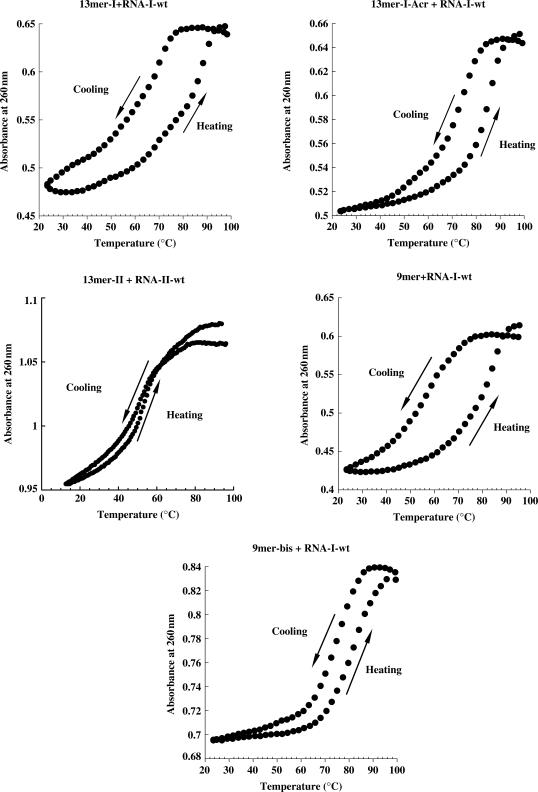
UV-melting curves were performed to further characterize the different complexes. For the two PNA tridecamers, 13-mer-I and 13-mer-II, the melting profiles are very different (Figure 4); the heating and cooling curves are superimposed for the 13-mer-II in contrast to that is observed for the 13-mer-I. In the latter case, temperatures of dissociation and association depend on the heating rate, supporting non-equilibrium conditions during experiment which gave rise to hysteresis phenomenon. This observation is consistent with duplex formation with 13-mer-II PNA and triplex formation with 13-mer-I PNA since triplex formation is known to be slower than duplex one (23). At acidic pH the melting curves of 13-mer-II PNA were similar to those obtained at neutral pH, suggesting that cytosine protonation is not involved in complex stability as expected for triplexes formed with C-rich third strand (data not shown). Altogether, these results suggest that tridecamer PNAs form at neutral pH stable complexes involving duplex (13-mer-II) or triplex (13-mer-I). To better characterize the complex formed with the 13-mer-I PNA on the PPT RNA, melting profiles were performed with the 9-mer PNA that forms a triplex on the A4GA4 sequence. In the models proposed for triplex formation with T-rich PNAs, Tass reflects the non-equilibrium formation of triplex while Tdis reflects in most cases the duplex dissociation (24). Our experiments are consistent with this model; Tdis values were almost the same (Tdiss (9-mer) = 84°C and Tdis (13-mer-I) = 88°C) but Tass are completely different (Tass (9-mer) = 58°C and Tass (13-mer-I) = 70°C).
Figure 4.
Melting profiles of the complexes formed with different PNAs (4 μM) as indicated above the melting curves and complementary RNAs (RNA-I-wt, RNA-II-wt) (2 μM). Samples were prepared in a buffer containing 100 mM KCl and 10 mM sodium cacodylate, pH 7. Cooling and heating profiles are not superimposed except for complexes formed with PNA 13-mer-II. Cooling and heating rates of 0.5°C/min were used in these experiments.
Finally, triplex and duplex-forming tridecamer PNAs discriminate wild-type target from mutated one. An important decrease on Tass (between 7°C and 12°C) of 13-mer-I, 13-mer-I-Acr and 13-mer-II PNAs to RNA-I-mut compared with RNA-I-wt was observed (Table 2).
Table 2.
UV-cooling-melting temperatures: temperatures of dissociation (Tdis) and association (Tass) in°C
| RNA-III* | wt | mut | |||||||
|---|---|---|---|---|---|---|---|---|---|
| PNA* | Tdis | Tass | Δa | Tdis | Tass | Δb | Tdis | Tass | Δa |
| 13mer-I | 88 | 70 | −18 | 78 | 65 | −13 | 83 | 58 | −25 |
| 13mer-I-Acr | 85 | 75 | −8 | 86 | 65 | −11 | |||
| 13mer-II | 52 | 52 | 0 | 45 | 45 | 0 | |||
| 11mer-bis | 87 | 80 | −7 | 74 | 69 | −5 | |||
| 9mer | 84 | 58 | −26 | 75 | 60 | −15 | |||
| 9mer-Acr | 86 | 59 | −27 | ||||||
| 9mer-bis | 82 | 78 | −4 | ||||||
*PNA and *RNA sequences in Table 1.
aΔ and bΔ = Tass − Tdis obtained in cooling–melting experiments with heating rates of 0.5°C/min and 0.1°C/min, respectively.
PNA–RNA complexes arrest translation elongation in vitro and induce synthesis of truncated proteins
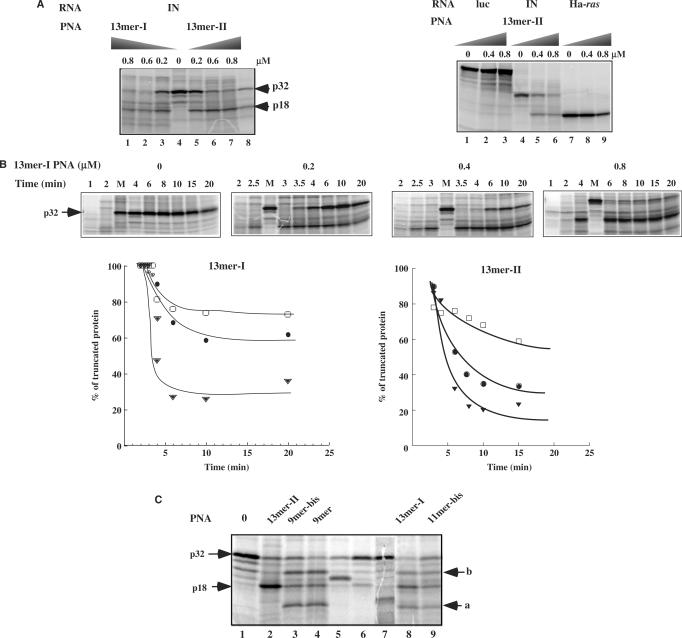
As expected, 13-mer-I PNA that forms a triplex with PPT sequence inhibits translation of HIV-1 integrase coding mRNA in vitro in a dose-dependent manner (Figure 5A). This inhibition is accompanied by the synthesis of a truncated protein. The truncated protein size (18 kDa) is similar to the size of the polypeptide chain obtained after RNase H cleavage of the RNA duplexed to a 15-mer phosphodiester oligonucleotide targeted to the PPT region (15-mer-PPT) (Figure 5A, lane 8). Interestingly, 13-mer-II PNA that forms a duplex with the PPT sequence is as efficient as 13-mer-I PNA to arrest translation elongation (Figure 5A). We have determined an IC50 value of 0.3 μM and 0.2 μM for PNA 13-mer-I and 13-mer-II, respectively. Both tridecamer PNAs did not affect the translation of Ha-ras and Pyralis Luciferase messenger RNAs (Figure 5A for 13-mer-II and data not shown for 13-mer-I).
Figure 5.
(A) (Left) Activity of different PNAs (Table 1) on translation of HIV-1-integrase mRNA in a rabbit reticulocyte lysate. In vitro transcribed HIV-1-integrase mRNA was translated in the absence of PNA (0) or at the concentrations indicated above the lanes, as described in ‘Materials and Methods’ section, and subsequently analysed by SDS–PAGE on a 14% polyacrylamide gel. To determine the site of translation arrest induced by 13-mer-I and 13-mer-II PNAs, translation was carried out in the presence of a 15-mer phosphodiester ODN complementary to PPT sequence (15-mer-PPT; see Table 1 for sequences) and RNase H (E.coli) (lane 8). Arrows indicate the position of HIV-integrase protein (p32, IN) and truncated proteins (18 kDa). (Right) Control experiments: Effect of 13-mer-II PNA on translation of Ha-ras and firefly luciferase (photinus pyralis) is shown. (B) (Top) Translation kinetics was determined in the absence (0) and in the presence of different concentrations of 13-mer-I PNA (0.2, 0.4 and 0.8 μM). Lane M: 20 min of translation in the absence of PNA. (Bottom) Percentages of truncated protein synthesized in the presence of 0.2 μM (filled triangle), 0.4 μM (filled circle), 0.8 μM (open box) of 13-mer-I and 13-mer-II PNAs as a function of translation time. (C) Translation arrest experiments carried out in the presence of 0.8 μM of 13-mer-I, 13-mer-II, 9-mer, 9-mer-bis and 11-mer-bis PNAs. Arrows (a) and (b) denote the position of secondary sites of translation arrest. Lanes 6, 7, 5: polypeptide chains synthesized in the presence of RNase H and of different 15-mer phosphodiester antisense ODNs that are complementary to the PPT sequence (15-mer-PPT) and to sequences upstream (15-mer-PPT-I) and downstream (15-mer-PPT-II) the PPT site, respectively; these ODNs are targeted to potential PNA binding sites (see Table 1 for15-mers sequences).
The time course for translation elongation arrest at a fixed PNA 13-mer-I concentration was also examined (Figure 5B). In the absence of PNA, translation full-length protein (p32, IN) synthesis was achieved in 4 min. In the presence of 13-mer-I PNA, the truncated p18 polypeptide was detected as early as 2 min after the initiation of translation. Truncated protein life-time is significantly higher when translation elongation was arrested with 13-mer-I PNA compared with 13-mer-II PNA: Figure 5B shows that depending on the PNA concentration used, the amount of truncated protein dropped to 80, 60 and 30% after 20 min of translation in the presence of 13-mer-I PNA whereas lower quantities (60, 40 and 20%) were obtained in the presence of 13-mer-II PNA. These results may reflect the longer lifetime of PNA2-RNA complexes versus the PNA–RNA complexes. These PNAs, 13-mer-I, 9-mer, 9-mer-bis, 11-mer-bis, share in common the T4CT4 sequence. HIV integrase coding mRNA contains a A3GA3 and A3GA2 sequences situated respectively upstream and downstream of the PPT sequence (25). Figure 5C shows that high concentrations of 13-mer-I PNA (>0.4 µM) arrest translation at the PPT site and also at these two other sites, upstream (arrow a, lanes 3, 4, 8, 9) and downstream (arrow b, lanes 3, 4, 8, 9) the PPT site. In order to evaluate the sizes of truncated proteins we have targeted PO-ODNs to the PPT and secondary sites in the presence of RNase H (Figure 5C, lanes 5–7). These two additional products were also observed with the shorter 9-mer, 9-mer-bis and 11-mer-bis PNAs (Figure 5C). It is noteworthy that the translation arrests at these secondary sites were more pronounced for the shorter PNAs, 9-mer and 9-mer-bis, compared with the 13-mer-I PNA that appeared more selective. PNA 13-mer-II that do not contain the T4CT4 sequence (see Table 1 for sequences) induces arrest of translation only at PPT site (Figure 5C, lane 2).
Antisense activity on SLO-permeabilized cells mediated by tridecamer PNAs at submicromolar concentrations
By using the CMV (+) PPT/HeLa cell line, containing two reporter genes, the firefly luciferase gene (luc) and the GFP gene, under the control of a bi-directional doxycycline-inducible CMV promoter, we carried experiments to study the intracellular antisense activity of tridecamer PNAs that were shown as specific and efficient translation inhibitors in vitro. Two inserts containing either the wild type HIV-1 PPT sequence or a mutated sequence (HIV-2 PPT) were cloned in the 5′ transcribed region of the luciferase and GFP genes respectively (Figure 6A). Therefore, it is an appropriate system to test sequence-specific cellular activity of anti-PPT molecules; PNAs targeted to the PPT sequence should induce inhibition of luciferase expression without affecting the GFP expression (Figure 6A). The PNAs were introduced into the cells by permeabilization of the cellular membrane following treatment with SLO. The SLO-based protocol is among the most efficient methods, to efficiently deliver oligonucleotides of various chemistries (17,18,26). A dose-dependent specific inhibition of luciferase expression was induced in the presence of PNA tridecamers in the SLO-permeabilized cells (Figure 6B). In these experiments, an important fraction of cells (∼95%) was permeabilized with an optimized amount of SLO without inducing important cell mortality (∼20%). These parameters were measured using flow cytometry analysis and propidium iodide labelling of dead cells and fluorescein labelling of permeabilized cells. Fluorescent microscopy studies without fixation carried out after SLO-treatment using fluorescein-labelled tridecamer PNA has shown a vesicular and nuclear localization of the PNA (data not shown). Similar PNA intracellular distribution after SLO permeabilization was observed using fixed cells (26). At 1 μM concentration, 13-mer-I and 13-mer-II PNA induced a strong inhibition of luciferase expression (∼70%) without affecting GFP expression (Figure 6B). Treatment with the control PNA containing a scrambled sequence did not affect luciferase activity (Figure 6B). Moreover, tridecamer PNAs did not inhibit luciferase activity of cells lacking the PPT insert (data not shown).
CONCLUSIONS
The interesting physicochemical properties and the in vitro biological activity of T-rich and C-rich tridecamer PNAs led us to test their intracellular antisense activity. Uncharged PNAs cannot straightforward be administrated into living cells using cationic polymers or lipids and the use of a DNA carrier is necessary to allow complexation with cationic lipids; delivery of PNAs can be also achieved by PNA attachment to cell-penetrating peptides (CPP). Therefore, it is difficult to make relevant comparisons of cellular activity of PNAs (27–29). In order to overcome this delivery problem we have used permeabilized cells; the SLO-based protocol is among the most efficient method existing at present, to efficiently deliver oligonucleotides of various chemistries in cells (30). Cellular experiments confirmed the in vitro translation experiments. The two tridecamer PNAs that form at neutral pH very stable complexes, likely involving duplex (13-mer-II) or triplex (13-mer-I) formation with target PPT RNA, specifically inhibit translation in SLO-permeabilized cells. In the present study, we show that tridecamer PNAs are as efficient as 18-mer phoshoramidate oligonucleotide, also administrated by a SLO procedure (18), to act as antisense molecules by binding to the complementary RNA target, present in the 5′-untranslated region of a firefly luciferase reporter gene. Moreover, while the phosphoramidate 18-mer, complementary to the PPT sequence did not exhibit, in vitro, any inhibitory effect when targeted to the coding portion of the mRNA, tridecamer PNAs very efficiently arrested translation elongation with concomitant synthesis of truncated peptide. Interestingly, this steric blockage of translation elongation was not only observed with T-rich triplex-forming PNAs as already described but also with C-rich duplex-forming PNAs. Our results open new perspectives for the development of such PNAs that can be targeted to any RNA sequence, to produce intracellularly truncated proteins of interest including dominant negative versions.
ACKNOWLEDGEMENTS
We thank Fabio Cannata for technical assistance. This work was supported by grant from CERC3 call for translational research proposals in chemistry ‘Nucleic Acids Chemistry’. F.B-H. is supported by the French Ministry for Research. Funding to pay the Open Access publication charges for this article was provided by INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 2.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gomez D, Lemarteleur T, Lacroix L, Mailliet P, Mergny J-L, Riou J-F. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004;32:371–379. doi: 10.1093/nar/gkh181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sundquist WI, Heaphy S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc. Natl Acad. Sci. USA. 1993;90:3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyonnais S, Hounsou C, Teulade-Fichou M-P, Jeusset J, Le Cam E, Mirambeau G. G-quartets direct assembly within a G-rich DNA flap. A possible event at the center of the HIV-1 genome. Nucleic Acids Res. 2002;30:5276–5283. doi: 10.1093/nar/gkf644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyonnais S, Gorelick RJ, Mergny J-L, Le Cam E, Mirambeau G. G-quartets direct assembly of HIV-1 nucleocapsid protein along single-stranded DNA. Nucleic Acids Res. 2003;31:5754–5763. doi: 10.1093/nar/gkg716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wyatt JR, Vickers TA, Roberson JL, Buckheit RW, Klimkait T, Debates E, Davis PW, Rayner B, Imbach JL, et al. Combinatorially selected guanosine-quartet structure is potent inhibitor of human immunodeficiency virus envelope mediated cell fusion. Proc. Natl Acad. Sci. USA. 1994;91:1356–1360. doi: 10.1073/pnas.91.4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mergny J-L, De Cian A, Ghelab A, Saccà B, Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine- substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 10.Larsen HJ, Bentin T, Nielsen PE. Antisense properties of peptide nucleic acid. Biochim. Biophys. Acta. 1999;1489:159–166. doi: 10.1016/s0167-4781(99)00145-1. [DOI] [PubMed] [Google Scholar]
- 11.Egholm M, Bucharrdt O, Christensen L, Buhrens C, Freir SM, Driver DA, Berg RH, Kim SK, Norden B, et al. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 12.Braasch DA, Corey DR. Novel antisense and peptide nucleic acid strategies for controlling gene expression. Biochemistry. 2002;41:4503–4510. doi: 10.1021/bi0122112. [DOI] [PubMed] [Google Scholar]
- 13.Karras JG, Maier MA, Lu T, Watt A, Manohoran M. Peptide nucleic acids are potent modulators of endogenous pre-mRNA splicing of the murine interleukin-5 receptor-alpha chain. Biochemistry. 2001;40:7853–7859. doi: 10.1021/bi010263l. [DOI] [PubMed] [Google Scholar]
- 14.Sazani P, Gemignani F, Kang SH, Maier MA, Manoharan M, Persmark M, Bortner D, Kole R. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat. Biotechnol. 2002;20:1228–1233. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]
- 15.Marin V, Armitage B. RNA guanine invasion by complementary and homologous PNA probes. J. Am. Chem. Soc. 2005;127:8032–8033. doi: 10.1021/ja051102y. [DOI] [PubMed] [Google Scholar]
- 16.Shiraishi T, Nielsen PE. Down-regulation of MDM2 and activation of p53 in human cancer cells by antisense 9-aminoacridine-PNA (peptide nucleic acid) conjugates. Nucleic Acids Res. 2004;32:4893–4902. doi: 10.1093/nar/gkh820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunet E, Corgnali M, Cannata F, Perrouault L, Giovannangeli C. Targeting chromosomal sites with locked nucleic acid-modified triplex-forming oligonucleotides: study of efficiency dependence on DNA nuclear environment. Nucleic Acids Res. 2006;34:4546–4553. doi: 10.1093/nar/gkl630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faria M, Spiller DG, Dubertret C, Nelson JS, White MR, Scherman D, Hélène C, Giovannangeli C. Phosphoramidate oligonucleotides as potent antisense molecules in cells and in vivo. Nat. Biotechnol. 2001;19:40–44. doi: 10.1038/83489. [DOI] [PubMed] [Google Scholar]
- 19.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 20.Mergny J-L, De Cian A, Ghelab A, Saccà B, Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyonnais S, Hounsou C, Teulade-Fichou M-P, Jeusset J, Le Cam E, Mirambeau G. G-quartets assembly within a G-rich DNA flap. A possible event at the center of the HIV-I genome. Nucleic Acids Res. 2002;30:5276–5283. doi: 10.1093/nar/gkf644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen GI, Bentin T, Larsen HJ, Nielsen PE. Structural isomers of bis-PNA bound to a target in duplex DNA. J. Mol. Biol. 2001;307:67–74. doi: 10.1006/jmbi.2000.4487. [DOI] [PubMed] [Google Scholar]
- 23.Mergny J-L, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 24.Lesnik EA, Risen LM, Driver DA, Griffith CG, Sprankle K, Freir SM. Evaluation of pyrimidine PNA binding to ssDNA targets from nonequilibrium melting experiments. Nucleic Acids Res. 1997;25:568–574. doi: 10.1093/nar/25.3.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sénamaud-Beaufort C, Leforestier E, Saison-Behmoaras TE. Short pyrimidine stretches containing mixed base PNAs are versatile tools to induce translation elongation arrest and truncated protein synthesis. Oligonucleotides. 2003;13:465–478. doi: 10.1089/154545703322860780. [DOI] [PubMed] [Google Scholar]
- 26.Faruqi AF, Egholm M, Glazer PM. Peptide nucleic acid-targeted mutagenesis of a chromosomal gene in mouse cells. Proc. Natl Acad. Sci. USA. 1998;95:1398–1403. doi: 10.1073/pnas.95.4.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamilton SE, Simmons CG, Kathiriya IS, Corey DR. Cellular delivery of peptide nucleic acids and inhibition of human telomerase. Chem. Biol. 1999;6:343–351. doi: 10.1016/S1074-5521(99)80046-5. [DOI] [PubMed] [Google Scholar]
- 28.Wolf Y, Pritz S, Abes S, Bienert M, Lebleu B, Oehlke J. Structural requirements for cellular uptake and antisense activity of peptide nucleic acids conjugated with various paptides. Biochemistry. 2006;45:14944–14954. doi: 10.1021/bi0606896. [DOI] [PubMed] [Google Scholar]
- 29.Bendifallah N, Rasmussen FW, Zachar V, Ebbesen P, Nielsen PE, Koppelhus U. Evaluation of cell-penetrating peptides (CPPs) as vehicles for intracellular delivery of antisense peptide nucleic acid (PNA) Bioconjugate Chem. 2006;17:750–758. doi: 10.1021/bc050283q. [DOI] [PubMed] [Google Scholar]
- 30.Giles RV, Ruddell CJ, Spiller DG, Green JA, Tidd DM. Single base discrimination for ribonuclease H-dependent antisense effects within intact human leukemia cells. Nucleic Acids Res. 1995;23:954–961. doi: 10.1093/nar/23.6.954. [DOI] [PMC free article] [PubMed] [Google Scholar]