Abstract
Formation of the stable, strand separated, ‘open’ complex between RNA polymerase and a promoter involves DNA melting of approximately 14 base pairs. The likely nucleation site is the highly conserved −11A base in the non-template strand of the −10 promoter region. Amino acid residues Y430 and W433 on the σ70 subunit of the RNA polymerase participate in the strand separation. The roles of −11A and of the Y430 and W433 were addressed by employing synthetic consensus promoters containing base analog and other substitutions at −11 in the non-template strand, and σ70 variants bearing amino acid substitutions at positions 430 and 433. Substitutions for −11A and for Y430 and W433 in σ70 have small or no effects on formation of the initial RNA polymerase-promoter complex, but exert their effects on subsequent steps on the way to formation of the open complex. As substitutions for Y430 and W433 also affect open complex formation on promoter DNA lacking the −11A base, it is concluded that these amino acid residues have other (or additional) roles, not involving the −11A. The effects of the substitutions at −11A of the promoter and Y430 and W433 of σ70 are cumulative.
INTRODUCTION
Transcription in bacteria is catalyzed by DNA-dependent RNA polymerase (RNAP). In order to form an initiation-competent promoter complex, the multi-subunit bacterial RNAPs require the participation of sigma factors. This class of initiation factors binds to the RNAP in solution prior to any DNA interaction. For historic reasons, the bacterial RNAP is referred to as the ‘core’ enzyme, while the complex of core and sigma is called the ‘holo’ enzyme. Studies with Escherichia coli RNAP have indicated that formation of the initiation-competent, stable ‘open’ complex (RPo), in which a 14 base pair (bp) region of the promoter DNA spanning positions −11 to +3 (1) has been melted, is a multi-step process (1,2). The initial, unstable, ‘closed’ complex (RPc) between holo RNAP and promoter DNA isomerizes to the open complex in a reaction that includes two kinetically significant intermediates (I1, I2) and involves conformational changes in both the promoter DNA and the RNAP (2–5):
| Scheme 1 |
Here both RPc and I1 have short half lives, of the order of seconds in transcription compatible buffers, while I2 and RPo are much longer lived (half lives of minutes or hours) (2,3).
Sigma factors are involved in both the recognition of specific promoter elements and the initiation of the promoter DNA strand separation, which enables the template strand to base pair with nucleoside triphosphates, the precursors of the RNA. Highly conserved aromatic amino acids, positioned on one side of an alpha helix in region 2.3 of the sigma factor (6–9) have been implicated in the melting process (10–13). Of these, especially the Y430 and W433 (E. coli σ70 numbering) have been extensively studied. Both the Y430 and the W433 have generally been assumed to interact with the highly conserved −11A residue in the −10 region. Substitutions at both positions have been found to be deleterious for promoter DNA melting (10,11,13,14), but remarkably, the Y430A substitution was also found to facilitate formation of complexes of RNAP with short model templates at low temperatures (15), suggesting an inhibitory role for the Y430 residue under some circumstances. The effects of substitutions for Y430 and W433 have been found to be cumulative (11,12,16). A substantial amount of experimental evidence supports a crucial role for the −11A base on the non-template strand in initiating the process of promoter DNA strand separation (17–23), but not in formation of the closed complex (17). It is thought that flipping the −11A out of the DNA helix and into a hydrophobic pocket on the sigma factor, nucleates DNA melting (13,18,24–26). Consistent with such a role for the −11A, mismatches at −11 were found to partially compensate for the deleterious effects on open complex formation of non-template strand substitutions at −11 (22,27). From studies of the effects of base analog substitutions at −11, it was concluded that the N1 of the −11A is important in the nucleation of promoter DNA melting (21).
In much of the previous work different assay conditions and promoter DNAs were used, making some of the results difficult to compare. Thus the roles of Y430 and W433 have remained unclear. Here we revisit and extend the prior work using uniform assay conditions and template DNAs. Our results have led to the following novel insights: (i) The Y430 and W433 residues have important functions beyond any direct interaction they may have with the −11A. (ii) Substitutions for the −11A and the Y430 and W433 have similar effects in directly inhibiting the rate limiting step in the formation of an open RNAP-promoter complex. (iii) The effects of substitutions at −11A and at Y430 and W433 are cumulative.
MATERIALS AND METHODS
Materials
Oligodeoxynucleotides were synthesized by Invitrogen, Integrated DNA Technologies [2AP (2 amino purine) and Oligonucleotides abasic at positions −11 or −8], or TriLink (Nebularine oligos). (γ-33P) ATP was purchased from Perkin Elmer, DNA modifying enzymes from either New England Biolabs or Roche and E. coli RNAP core from EpiCenter. All chemicals were from Sigma, Fisher or Amresco.
Protein purification and characterization
Mutagenesis of the rpoD gene (16) and sigma expression vectors (11) are as described. Escherichia coli σ factors were purified using the protocol of Zhi and Jin (28) that does not involve protein denaturation, with minor modifications: a sonicator was used to perform the cell lysis instead of using a French press; GE Healthcare HiTrap chelating HP columns were used for the Ni2+ column chromatography. As in our hands, there was no significant difference between purified sigma factors before and after ion-exchange chromatography, we routinely omitted this step. However, buffer and salt content of the storage buffer for all purified sigma factors is similar regardless of whether this step was included. The final concentrations of the dialysis buffer components are 10% glycerol, 50 mM Tris-HCl, pH 8, 0.1 mM EDTA, 0.1 mM DTT, 0.01% Triton X-100 and 350 mM NaCl. After concentrating the protein in this buffer, an equal volume of pure glycerol was added. With the above procedure there is a detectable but insignificant contamination of the σ70 with core RNAP.
Reconstitution reactions
The optimal ratio of the purified sigma factors to core enzyme for reconstitution of holo RNAP was determined by titrating varying amounts of sigma with core RNAP and monitoring formation of a stable, heparin-resistant complex between the RNAP and a strong promoter. The fold-excess of sigma over core required for maximal extents of stable complex formation differed for the various preparations of sigma factors used. Purified RNAP core (400 nM) and σ factors were incubated on ice for 1 h with an excess of sigma as determined in the assays performed above. All concentrations were adjusted using storage buffer.
DNA labeling and annealing
DNA oligonucleotides, purified as described (29), were 5′ end-labeled with 33P by polynucleotide kinase in a reaction containing γ-33P-ATP (29). Unincorporated (γ-33P) ATP was removed using BioRad Micro Bio-Spin 6 Chromatography Columns. Annealing of complementary DNA strands was performed in a reaction containing 25 mM Tris-HCl, pH 7.9, 50 mM NaCl, 100 nM 33P-labeled DNA, and 150 nM unlabeled complementary strand. Reactions were incubated at 90°C to 95°C in a heat block for 5 min, followed by slow cooling to room temperature. The concentration of the annealed DNA is expressed as the concentration of the limiting, radiolabeled strand.
Electrophoretic mobility shift assay (EMSA)
Each reaction (10 μl) contained, in Fork binding buffer (FBB: 30 mM Hepes, pH 7.5, 1 mM DTT, 0.1 mg/ml BSA, 100 mM NaCl, 0.1 mM EDTA, pH 8, 1% glycerol), 10 nM annealed DNA, and 50 nM of RNAP holoenzyme. Reactions were started by addition of RNAP and incubated at room temperature for 10 min. To assay for formation of stable complexes, reactions were challenged with 200 μg/ml of heparin by adding 1 μl from a 2 mg/ml stock and incubated for an additional 10 min. For reactions without a heparin challenge, 1 μl of ddH2O was added and incubation was allowed to proceed for 10 additional minutes. For loading, 2 μl of non-denaturing dye solution (45% glycerol, 50 mM sucrose, 0.1% BPB and 0.1% XCFF) was added to each reaction and 9 μl was applied to a running 4% non-denaturing gel poured and run in TAE buffer (40 mM Tris-acetate and 1 mM EDTA). Gels were run at low voltage (90–100 V) for 1–2 h at room temperature. After drying, the gel was analyzed by PhosphorImaging (Molecular Dynamics) using ImageQuant 5.2 software to quantify the radiolabeled DNA that is free and RNAP-bound, in order to determine the fraction of DNA bound by RNAP. Error was determined from half of the spread of the values (two experiments), or the standard deviation (three or more experiments).
Obtaining binding and rate constants by EMSA
All of the following experiments were performed at room temperature in the FBB described in the section earlier.
Kd: the equilibrium dissociation constant, was determined by titrating a constant annealed DNA concentration of 2 nM with RNAP (a range of 4 nM to 200 nM final concentration). Reactions were incubated, heparin challenged (10 min, 200 μg/ml), and loaded onto non-denaturing gels as described before. After PhosphorImager analysis of the radiolabeled bound and free DNA, fractions bound (y) were plotted against RNAP concentration using Kaleidagraph version 3.52 and fit to the following hyperbolic equation: y = ymax/(1+(Kd/[RNAP])) + yo.
koff: The first-order rate constant, koff, for dissociation of stable RNAP-promoter complexes was determined by mixing final DNA and RNAP concentrations of 10 nM and 50 nM, respectively, in a 65 μl volume. After a 10 min incubation at room temperature, 2.6 μl of 5 mg/ml heparin (final concentration of 200 μg/ml) was added and aliquots were removed after 0.5, 1, 2, 5, 10 and 30 min and added to 2 μl of non-denaturing dye before loading 10 μl onto a native gel as described before. After PhosphorImager analysis of the radiolabeled bound and free DNA, fractions bound in heparin-stable complexes (determined as indicated before) were plotted against time using Kaleidagraph version 3.52 and fit to the following sum of exponentials: y = A1*(exp(−koff1*t)) + A2*(exp(−koff2*t)), where A1 and A2 are the amplitudes for the first and second decay process, with rate constants koff1 and koff2, respectively. The first event is likely the fast dissociation of RNAP bound in non-specific and closed complexes and the second, with rate constant koff2, (reported here as koff) the dissociation of stable complexes. All errors were calculated as described for the EMSA assays. Half lives were calculated as t1/2 = 0.69/koff.
kon: the second-order rate constant for formation of a stable complex, kon, was calculated from the experimentally determined pseudo first-order rate constant (kobs) for association of RNAP and promoter DNA to form a stable complex, and koff, the first-order rate constant for dissociation of the stable complex. To determine kobs, promoter DNA and RNAP at final concentrations of 10 nM and 50 nM, respectively, were mixed in a 10 μl volume and the reaction was incubated for 0.5, 1, 2, 5, 10 or 30 min before adding 1 μl of 2 mg/ml heparin (final concentration of 200 μg/ml) for 30 s (to remove all closed complexes). Addition of dye and loading of reactions is as described before. Fractions of DNA bound (determined as indicated before) were plotted against time using Kaleidagraph version 3.52 and fit to the following pseudo first order equation: y = ymax*(1 − exp(−kobs*t)). From the experimental values of kobs, kon was then calculated using the following equation: kobs = kon[RNAP] + koff.
KMnO4 probing
Reactions contained 10 nM annealed DNA in Tris Binding Buffer (TBB: 40 mM Tris-HCl, pH 8, 40 mM KCl, 1 mM MgCl2; we found that the KMnO4 reactions were more efficient in this buffer (30) despite slightly reduced extents of heparin-resistant binding) and were started by the addition of RNAP to 50 nM (final volume 20 μl). Incubation was carried out for 10 min at room temperature. Heparin challenge was then performed for an additional 10 min by adding 1 μl of a 2 mg/ml stock to obtain a final concentration of 100 μg/ml. For experiments without heparin, 1 μl of ddH2O was added instead. Then 1 μl of a freshly made 21 mM stock of KMnO4 was added for a final concentration of 1 mM. After 30 s, 5 μl of stop solution containing 1.5 M NaOAc, pH 8, 4 mg/ml glycogen, and 300 mM β-mercaptoethanol was added and reactions were placed on ice. After ethanol precipitation, the dried pellets were re-disolved in 70 μl of 1M piperidine, and the solutions incubated for 20 min at 90°C. Reactions were stopped by placing on dry ice. Ten microliter of 5 M LiCl was added to the thawing reactions, and the DNA was ethanol precipitated again. The dried pellets were taken up in ddH2O, dried again, dissolved in 8 μl of formamide dye (10 mM NaOH, 1 mM EDTA, 80% formamide, 0.1% XCFF, 0.1% BPB) and loaded onto a 10% sequencing gel, poured and run in TBE (89 mM Tris, 89 mM boric acid, 2 mM EDTA). After electrophoresis, the gel was dried and the bands were revealed by PhosphorImaging (Molecular Dynamics). Analysis of the images was performed using ImageQuant 5.2 software. To compare extents of RNAP-induced melting of a promoter duplex, the −1T band was used to quantify strand opening, and the radioactivity of this band was divided by the radioactivity of full length Duplex DNA. A normalized background value, established by using the same quantification procedure with a lane containing labeled DNA without added RNAP, was subtracted. Finally, all results were re-normalized to the value obtained for WT RNAP with Duplex DNA. Error was determined by taking the standard deviation (three or more determinations), or half of the spread (two determinations).
RESULTS
DNA templates and RNAP used in this study
Our goal was to arrive at a better understanding of the roles of amino acid residues Y430 and W433 of σ70, and of the −11A in promoter DNA melting. Our approach has been to combine σ70 containing substitutions of A, L, F, W or H for Y430 and of A, L, F, Y or H for W433 with promoter DNAs bearing substitutions for the −11A. The DNA substrates used in this work are shown in Figure 1A. We have carried out most experiments using a promoter (‘Duplex’) that is truncated in the downstream direction at position +1, the start site of transcription. In the non-template strand these DNAs had the −11A or substitutions of 2AP, G, purine, or an abasic nucleotide (Figure 1B). Another template (‘Mismatch’) had an A at −11 in the template strand, leading to a mismatch in the templates that contained A, 2AP, or purine at −11 in the non-template strand. Duplex templates containing a G at −11 of the non-template strand had a −11C in the template strand, and a G at this position for the Mismatch DNA. The promoter used has consensus −35 and −10 regions [the latter includes the upstream TG element, and is thus an extended −10 (31,32)], to ensure tight RNAP binding in the closed complex, so that effects of substitutions would mostly be on DNA melting rather than closed complex formation. Our previous work (33) showed that substitutions at the −10 and −35 that decreased the similarity of these regions to their consensus sequences, greatly reduced binding affinity for RNAP in the closed complex. The rationale for the use of truncated templates is that the effects of substitutions in both promoter DNA and σ70 are more pronounced, as compared to a longer DNA template (to position + 20) of the same sequence (data not shown).
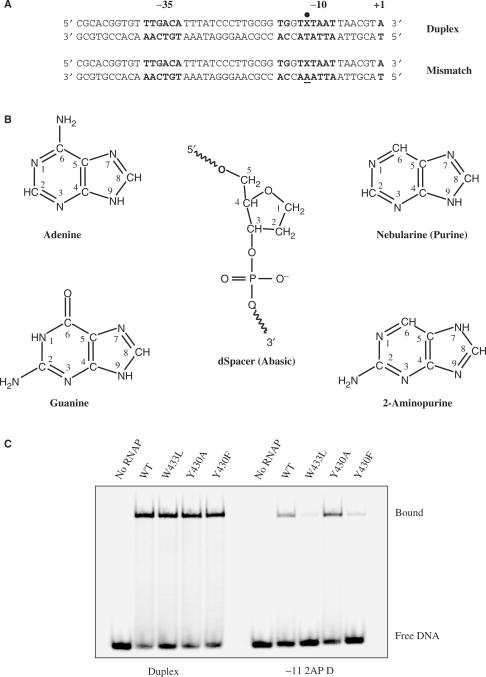
Figure 1.
DNA templates used and sample EMSA. (A) Duplex is a truncated version of a longer promoter sequence used in earlier work (16). It lacks any DNA downstream of position +1. The −35 and extended −10 elements (i.e. TGXTATAAT) as well as the +1 transcription start site are shown in bold, and the −11 position is marked by a dot. Mismatch DNA contains an A, instead of a T, at −11 in the template strand, resulting in a single mismatch at this position when the base on the non-template strand is A or 2AP. The same terminology was also applied if the non-template strand contained a purine (Nebularine) or was abasic at −11. For the −11G Duplex the template strand has a −11C, and for the Mismatch −11G, a −11G. (B) The structure of Adenine and other −11 substituents used in this work. dSpacer (Integrated DNA Technologies) models an abasic site. (C) Example of an EMSA gel for reaction mixes containing radiolabeled Duplex or −11 2AP Duplex and RNAP containing WT or substituted σ70. Subsequent to incubation of RNAP and radiolabeled DNA at room temperature, the mixtures were subjected to a heparin challenge (200 μg/ml for 10 min) prior to loading onto a non-denaturing polyacrylamide gel.
The effects of substitutions at −11A of promoter DNA, and σ70 Y430 and W433 on stable complex formation
Stable complex formation at room temperature for some combinations of promoter and σ70 substitutions are shown in Figure 1C. In this experiment, prior to loading binding reactions consisting of RNAP and labeled DNA on the gel, they were challenged for 10 min by added heparin. Heparin binds tightly to free RNAP and is a competitor with DNA for the binding to the enzyme. Due to the fact that the heparin is added in great excess, it will sequester any free RNAP that forms because of dissociation of RNAP promoter-complexes. Thus only stable complexes survive the heparin challenge. Stable complex formation between RNAP and Duplex DNA is seen to be greatly affected by the identity of the base at −11 of the non-template strand. The gel image in Figure 1C shows that stable complex formation is decreased for WT RNAP-DNA complexes containing the 2AP substitution at −11 compared to the Duplex DNA (A at −11). Figures 2 and 3 summarize the results collected for all experiments carried out with various combinations of DNA templates and RNAP bearing substitutions in σ70. The groups of bars represent different DNA templates and the bars within each group, the various amino acid substitutions. We tested purine for its lack of substituents, 2AP as an A-analog but with the amino group at the 2—instead of at the 6 position, G, which in addition to the 2-amino group also has a 6 carbonyl group, and dSpacer which has di-deoxyribose, without an attached base (Figure 1B). 2AP has found much use for its fluorescent properties (34–37), but has been a very useful probe for RNAP promoter interactions as well (19–21).
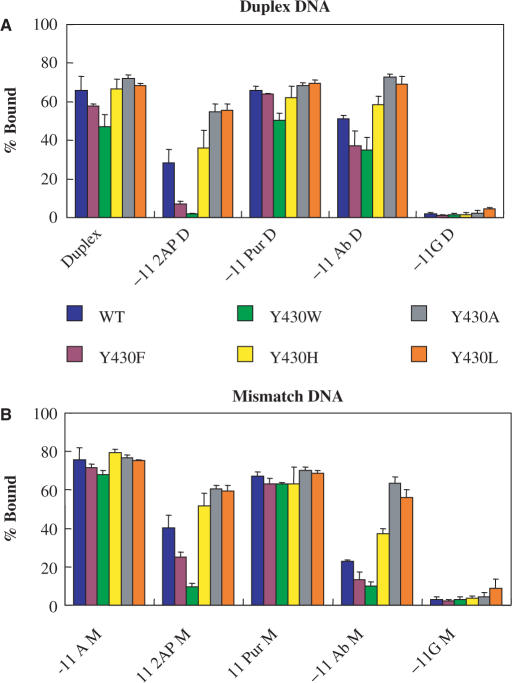
Figure 2.
Effects of substitutions at Y430 of σ70 on stable complex formation. Duplex and Mismatch DNA bearing different substitutions at −11 (−11 2AP D = 2-Aminopurine Duplex; −11 Pur D = Purine Duplex; −11 Ab D = Abasic Duplex; −11G D = −11 Guanine Duplex; M is Mismatch) were incubated at room temperature with RNAP containing WT or mutant σ70 with substitutions at position 430. Amounts of label in the complex and free DNA bands were determined by PhosphorImaging. The bars indicate the percentage of radioactivity in the complex bands (values are the averages of two or more independent experiments; errors were computed as half the differences between the duplicates for two experiments and standard deviations for three or more experiments). For details of the assay conditions see the Materials and Methods section and the legend to Figure 1C. Each group of bars represents the collection of σ70 variants tested. (A) Duplex templates. (B) Mismatch templates.
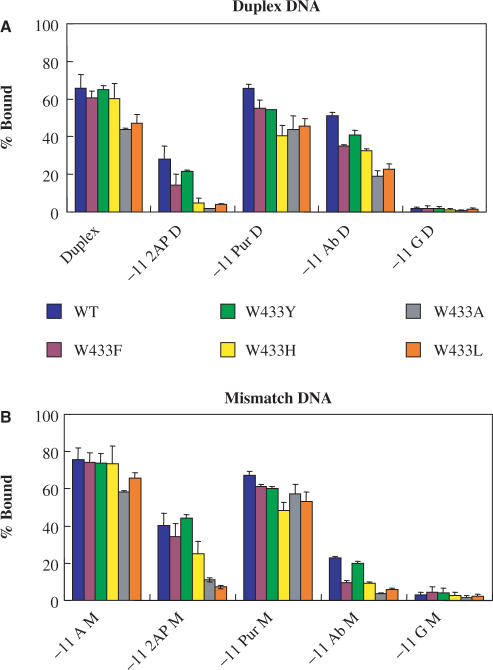
Figure 3.
Effects of substitutions at W433 of σ70 on stable complex formation. Determination of stable complex formation of Duplex and Mismatch DNA bearing different substitutions at −11 with RNAP containing WT or mutant σ70 with substitutions at position 433. The assay conditions are as indicated in the legend to Figure 2, and in the Materials and Methods Section. Each group of bars represents the collection of σ70 variants tested. (A) Duplex templates. (B) Mismatch templates.
For Duplex DNA binding to WT RNAP, among the substitutions tested, −11 purine was similar to −11A and −11 abasic was slightly worse (Figures 2A and 3A). Here the destabilization of the −11 base pairing would (partially) compensate for any loss of contacts due to the removal of the −11A (22,38). Formation of a stable complex was inhibited considerably by both the −11 2AP, and the −11 G. The latter two had the greatest effect of any single change in either promoter DNA or σ70 tested in our experiments (Figures 2A and 3A: compare bars within the Duplex group with the first bar of the −11 2AP and −11G groups). This is likely due to steric hindrance by substituents not present at the same positions in the canonical −11A (the NH2 at the 2 position of 2AP and G, and the additional carbonyl O at the 6 position of G). The absence of the 6 amino group may not be as important, as despite its absence, the promoter with −11 purine behaved similarly to that with the −11A. The −11G substitution may have been particularly detrimental due to the presence of an H at the N1, absent at the N1 of both A and 2AP (Figure 1B), which had previously been shown to be important for open complex formation (21). Consistent with prior observations (22,27), a mismatch at position −11 for some of the σ70 variants slightly improves stable complex formation with the −11A, −11 2AP and, to a lesser extent, −11G Duplexes (Figure 2B). This effect can be interpreted to reflect the greater ease of rotation of a −11A out of the helix if it is not base paired to a T. Interestingly, the Abasic Duplex DNA with an A at −11 of the template strand (called ‘Abasic Mismatch’) was more impaired in stable complex formation than the Abasic Duplex with a T at the same position. One possible interpretation of this result is that the template strand −11T of the −11 AT base pair is recognized by the RNAP, another, that the abasic substitution has sequence-dependent structural and thermodynamic consequences (22,39).
When RNAP mutants are used containing substitutions for Y430 of σ70, small defects are seen with the Duplex or Purine Duplex DNA (Figure 2A), particularly for Y430F and Y430W. These defects become much more pronounced when paired with DNA containing other substitutions at −11A, particularly with the −11 2AP. The effects of the substitutions at 430 of σ70 on stable complex formation by RNAP with the −11 2AP Duplex can be summarized as follows: (i) the σ70 with the Y430A (see also Figure 1C) and Y430L substitutions tolerate the −11 2AP to a greater extent than does WT σ70, (ii) the Y430H substitution has a small effect and (iii) the Y430F (see also Figure 1C) and Y430W substitutions are deleterious. Unexpectedly, a very similar pattern is seen for the DNA containing the −11 abasic substitution for Duplex DNA. This indicates that the Y430 residue could not have as its sole function a role dependent on the −11 A base. Results with the same −11 substitutions for Mismatch DNA (Figure 2B) are similar to the results obtained with WT RNAP, in that a slight improvement in stable complex formation is seen for most templates. All Y430 RNAP mutants, as well as WT RNAP, show large defects in stable binding to the −11G Duplex and Mismatch DNA and are therefore difficult to compare.
In a similar series of experiments to those shown in Figure 2A and B, the effects of substitutions at residue W433 were studied with the −11A substituted DNA templates (Figure 3A and B). For stable binding to Duplex DNA, W433A and W433L are slightly defective, while W433F, Y, and H behave similarly to WT RNAP under our conditions (Figure 3A). Like the Y430 mutant RNAP, these defects became more pronounced when paired with the −11 substituted DNA templates. The amino acid preferences for the 433 position are different from those at the 430 position in that an aromatic amino acid is preferred in the former, while a small to medium sized aliphatic amino acid for the latter. As with the Y430 mutant RNAP, a range of effects for substitutions at W433 was observed even when the DNA used was abasic for −11 in the non-template strand. Thus, like Y430, W433 must have a role other than that involving the −11A. The results for stable binding to the −11 Purine Duplex, the −11G Duplex (Figure 3A), and the Mismatch DNA templates (Figure 3B) by the W433 mutant RNAP are similar to those obtained for the Y430 RNAP (Figure 2A and B).
Our results clearly demonstrate RNAP's preference for certain base analogs over others at the −11 position of non-template DNA. To verify the unique properties of the −11 base, experiments similar to those shown in Figures 2 and 3 for substitutions at −11 of Duplex were also carried out with Duplex containing substitutions for the −8A (data not shown). The −8 position is conserved to a lesser extent in E. coli σ70 promoters (40) than the −11 (56% versus 76%), and would therefore be expected to be more tolerant to base substitutions. Based on modeling, the −8A is also positioned in the DNA helix far enough away from the Y430 and W433 amino acids such that we would not expect an interaction to occur between the two (5). In contrast to the data shown in Figures 2 and 3, it was found that for every substitution except −8G, the results were very similar to those seen with the −8A Duplex and Mismatch. The −8G substitution had deleterious consequences, but not as severe as the −11G. Thus, stable complex formation was largely insensitive to substitution at the −8 position on the non-template strand. These results confirm the important role of the −11 base on this strand. All of the above substitutions at the −11 and −8 position were also introduced in fork DNA (non-template strand to +1 and template strand to −12), but no significant differences between DNA templates or RNAP mutants were found at room temperature. The one exception was for substitution of −11G, which showed similar RNAP binding patterns as described before but just a 1.5 to 2-fold decrease in overall binding compared to −11A fork DNA.
For comparison, we have also carried out experiments with different promoter DNAs. With longer DNA sequences identical to the Duplex (Figure 1A) through position +1, but with an extension to +20 (16), under the same conditions, no effects of substitutions at 430 and 433 of σ70 on stable complex formation at room temperature were observed. When a 2AP was introduced at the −11 position, the effect of the substitution was also slight, and substitutions in σ70 had small effects. This is in sharp contrast to the results presented in Figures 1C, 2 and 3. With a similar, long template containing a total of three non-consensus substitutions in regions −10 and −35, some differential effects of the substitutions were seen, although the overall binding was much reduced even for the WT RNAP (data not shown). To determine whether the presence of the extended −10 TG sequence affected our results, a version of the Duplex DNA without the TG sequence was also employed. It displayed diminished binding of wt RNAP under the standard conditions used in Figures 2 and 3, but the patterns of the substitutions tested (W433L, Y430A and Y430F), with Duplex, and −11Ab Duplex (both with CA instead of TG) was not altered, including Y430A binding better than WT to the −11Ab Duplex (data not shown).
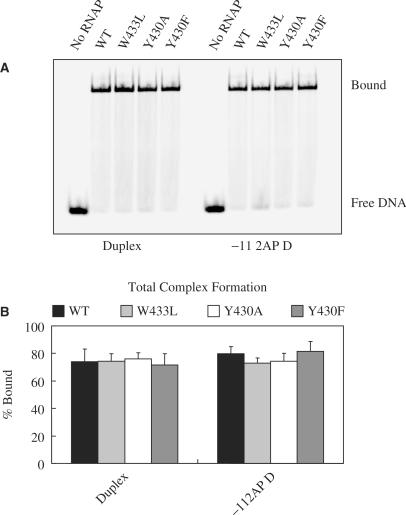
Little or no effect of substitutions in promoter DNA or σ70 on complex formation in the absence of a heparin challenge
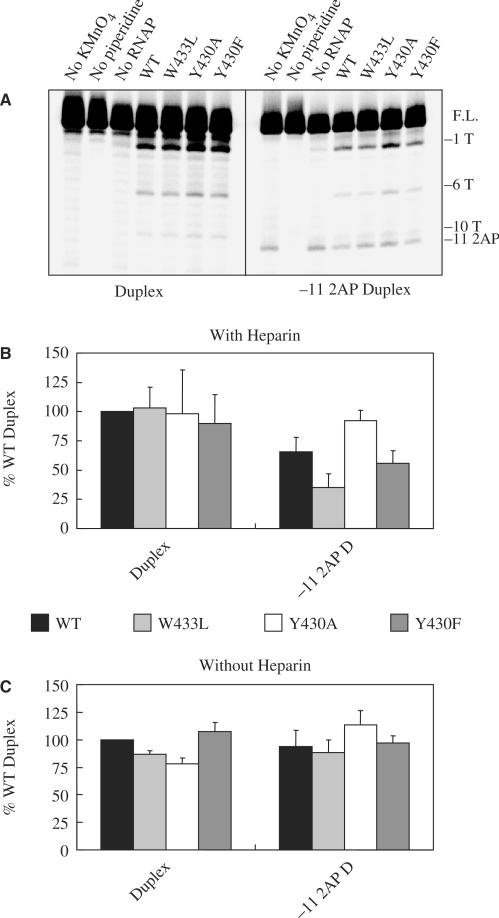
In order to try and pinpoint the step(s) in Scheme 1 at which the substitutions in promoter DNA or σ70 exert their effects, we performed binding experiments in the absence of a heparin challenge to determine whether complex formation in general (i.e. both closed and open complexes) was affected. The experiments shown in Figure 4 as well as those shown in Figures 5, 6 and 7 were carried out with a subset of promoter DNAs and σ70. The results in Figure 4A (gel image) and 4B (quantification by Phosphor Imaging) demonstrate that for both DNAs (Duplex and −11 2AP Duplex), total complex formation is similar for RNAP with WT and the three mutant σ70. If the RNAP and DNA are incubated for 30 s instead of 10 min, similar amounts of total complex formation are detected (data not shown). Thus defects in stable complex formation (Figures 1C, 2 and 3) cannot be explained by invoking effects of the promoter—or σ70 substitutions on DNA binding. Total complex formation was also not affected for RNAP binding to −11 Abasic Duplex DNA (data not shown). For the −11 G Duplex, small (less than 2-fold) differences among the various RNAP in total complex formation became apparent (data not shown), although any correlation between binding and stable complex formation is difficult to assess in view of the very low extents of stable complex formation seen with this template.
Figure 4.
Complex formation in the absence of a heparin challenge. (A) Gel image. RNAP with different amino acids residues at positions 430 and 433 in σ70, and either Duplex or −11 2AP Duplex DNA were incubated at room temperature. After complex formation the samples were loaded onto a non-denaturing polyacrylamide gel without prior challenge with heparin. (B) Quantification of the results. Work-up of samples was as indicated in Materials and Methods and the legend to Figure 2.
Figure 5.
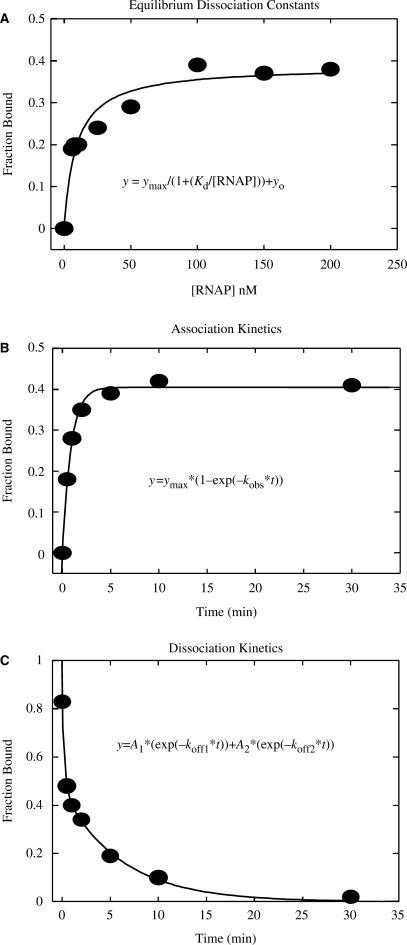
Sample data for the determination of Kd, kobs and koff for the WT −11 2AP Duplex. (A) Kd determination by titrating 2 nM of 33P radiolabled DNA with different concentrations of RNAP. Determination of complex formation was by EMSA, following a challenge with heparin. Fitting to the equation shown in the figure yielded Kd = 9.4 nM. (B) Determination of kobs used 50 nM RNAP and 10 nM DNA. Samples were analyzed by EMSA after a 30 sec. challenge by heparin. Fitting to the equation shown in the figure yielded a kobs = 1.1 min −1. (C) Determination of koff by challenging a mixture of 10 nM Duplex DNA and 50 nM RNAP that had been incubated for 10 min with heparin (added at time = 0), and subsequent analysis of aliquots taken at various time intervals, for the presence of heparin-resistant complexes. Fitting to the equation shown in the figure yielded a koff = 0.163 min−1. Experimental conditions are as indicated in Materials and Methods.
Figure 6.
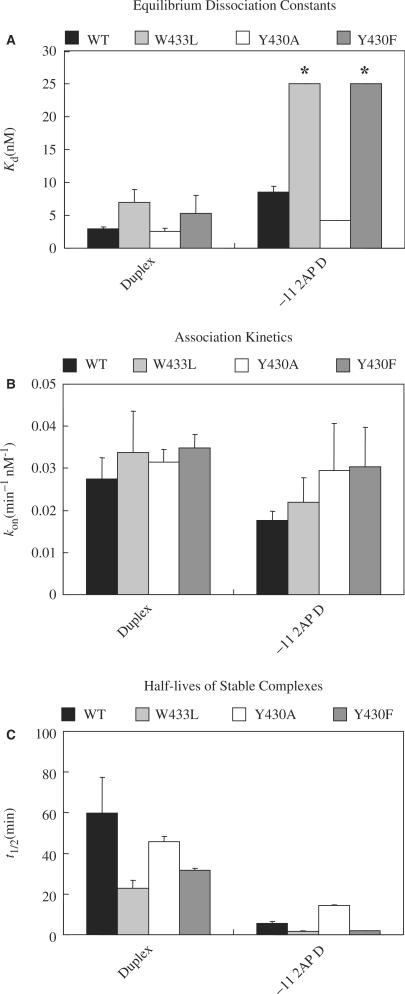
Effects of DNA and σ70 substitutions on equilibrium and kinetics of stable complex formation. (A) Kd. The asterisks above two of the bars are to indicate that these are lower estimates for the values of Kd. (B) kon (calculated from values of kobs and koff). (C) Half lives, t1/2, of the complexes, as calculated from the koff values: koff = 0.010, 0.031, 0.015 and 0.022 min−1 respectively for complexes of Duplex with WT, W433L, Y430A and Y430F RNAP, and 0.134, 0.469, 0.049 and 0.326 min−1 respectively for complexes of −11 2AP Duplex with WT, W433L, Y430A and Y430F RNAP. See the legend to Figure 5 and the Materials and Methods for experimental detail and calculation of kon and t1/2.
Figure 7.
DNA strand separation by wild-type or mutant RNAP at Duplex or −11 2AP Duplex DNA. Before KMnO4 treatment, all DNAs were incubated with RNAP for 10 min at room temperature prior to a challenge with heparin for an additional 10 min. Two to three independent experiments were performed, and the error bars indicate the spread between two experiments or the standard deviations of three experiments. (A) Typical gel with samples loaded subsequent to a heparin challenge. The bands marked −11 2AP are due to cleavage at the 2AP by the piperidine in the absence of exposure to KMnO4. F.L. stands for full length DNA. (B) Quantification of experiments such as that shown in A (See ‘Materials and Methods’). (C) Quantification of the experiments not involving a heparin challenge.
Substitutions at positions −11 of promoter DNA and 430 and 433 of σ70 affect the life times of stable RNAP-promoter complexes
The above experiments established that substitutions at −11, and at Y430 and W433 did not affect RNAP binding to promoter DNA in the absence of a heparin challenge, but rather had an effect on the fraction of RNAP promoter complexes that was heparin-resistant. To quantify the effects of the substitutions, we determined the equilibrium binding constants for formation of stable complexes (i.e. after incubation of RNAP and promoter DNA, the complexes were subjected to a 10 min challenge with heparin prior to loading of the reaction mixture on a non-denaturing gel). An example experiment is shown in Figure 5A and the quantification of the data is shown in Figure 6A. It is seen that the −11 2AP substitution increases the Kd for stable complex formation to WT RNAP by almost 3-fold, indicating a significant weakening of the complex. As compared to RNAP containing WT σ70, the W433L and Y430F substitutions in σ70 result in smaller (approximately 2-fold) increases in Kd for the −11A Duplex, in agreement with the data shown in Figures 2 and 3. More pronounced effects of the substitutions became evident with the −11 2AP Duplex: the binding affinity of the W433L and Y430F RNAP was so weak that only a lower limit of about 25 nM could be established for Kd. With the −11 2AP Duplex, the complex was more stable (smaller Kd) for the Y430A RNAP, consistent with the data shown in Figures 2 and 3. The Kd determinations also demonstrate that the effects of the substitutions at −11 of the promoter and 430 and 433 of σ70 are cumulative, i.e. the mutant RNAP exacerbates the effects of the −11 2AP substitution.
To gain further insight into the steps in Scheme 1 affected by the substitutions, and in view of the possibility that the Kd data may have been obtained near the tight-binding limit of our experiments, where discrimination between different Kd values may not have been optimal, we measured the kinetics of stable complex formation. In these experiments we determined the RNAP concentration-dependent rate parameter, kobs, that was then converted to values of kon. Studies on the rate of dissociation of the complexes were carried out to get the rate constant, koff and the complex half lives. The koff/kon ratio provides another estimate of the dissociation constant. Sample experiments are shown in Figures 5B and C, respectively. The data are collected in Figures 6B and C (values for koff are given in the legend to Figure 6C). Within the error of our experiments, neither the 2AP substitutions in promoter DNA nor the σ70 substitutions in RNAP significantly affect kon. However, they lead to an increase in koff by as much as a factor of 15. The experimental conditions for the equilibrium and kinetic experiments differed: the glycerol concentration was greater in the former. Thus the Kd values determined cannot be compared to the koff/kon ratio (and the similarities of the extents of complex formation in Figure 6 are fortuitous). Even so the effects of the substitutions would be expected to be comparable. However, from the kinetic results, some substitutions would be anticipated to cause an approximately 15-fold increase in Kd, but direct measurements only show a 3-fold increase, in agreement with the supposition that we were operating near the lower limit of detectable values for Kd. For the truncated promoter DNAs used here, the effects of the σ70 substitutions were most notable from the koff values for complexes formed with the −11 2AP Duplex: compared with the WT RNAP, the RNAP with the W433L and Y430F had a greater koff and shorter half life by factors of 3.5 and 2.4, respectively, while the Y430A substitution resulted in complexes with a smaller koff and greater half life, by a factor of 2.8. We conclude that the substitutions at Y430 and W433 of σ70 affect the stability of RNAP-promoter complexes.
Effects of the presence of a G or lack of a base at −11
We additionally performed kinetic experiments using WT RNAP and both −11 Abasic Mismatch and −11G Duplex promoter DNA, as well as WT and W433L RNAP with −11 Abasic Duplex. We were unable to detect any significant differences in on—or off rates for the interaction of WT and W433L RNAP with −11 Abasic Duplex DNA, as compared to Duplex DNA (data not shown), despite slightly reduced levels of overall heparin resistant binding (Figures 2A and 3A). Complexes of WT RNAP and −11 Abasic Mismatch DNA were slightly more stable than those of −11 2AP Duplex DNA (Half-life of 11.3 min for −11 Abasic Mismatch compared to 5.4 min for −11 2AP Duplex) (data not shown), while both are less stable than complexes containing the Duplex or −11 Abasic Duplex DNAs. Thus the reduced extent of stable binding seen with the −11 Abasic Mismatch DNA compared to the −11 Abasic Duplex (Figures 2 and 3) is due to the decreased stability of the former complex. Consistent with the slow on-rate observed in prior work (41), we were not able to determine a meaningful kon for the interaction of WT RNAP and the −11G Duplex due to the low amounts of heparin resistant binding and for the same reason, no koff either.
Substitutions in the promoter or σ70 do not affect the extent of strand separation in stable complexes
In Scheme 1 there are two types of stable complexes: the intermediate I2 with, at most, a few melted base pairs at the upstream edge of the DNA region that becomes single stranded in the open complex, and the fully strand-separated open complex RPo. Under most conditions the equilibrium between these complexes heavily favors the open complex. In order to determine whether the substitutions in the Duplex DNA or σ70 might have shifted the complex towards the I2 form, we determined their effects on the relative extents of promoter strand separation by subjecting RNAP-promoter complexes to KMnO4 probing (42) both with and without a prior challenge with heparin. A sample gel for the determination of the extent of strand separation of complexes surviving a 10 min heparin challenge is shown in Figure 7A. As a measure of RNAP-induced promoter DNA melting, the intensity of the band generated by piperidine-induced cleavage at the oxidized −1T in single stranded regions was quantified, and expressed as percent of the amount of radioactivity in uncleaved DNA. The values are shown in Figure 7B for samples that were heparin-challenged, and in 7C for samples that were not. The presence of the −11 2AP does not affect strand opening in the absence of a heparin challenge, but the extent of strand opening is reduced after incubation with heparin prior to loading the samples on the gel. The presence of the −11 2AP only affects the extent of strand separation when the equilibrium is skewed to the left by the addition of heparin, which binds to free RNAP and inhibits promoter binding. These results are similar to those obtained by EMSA with and without heparin challenge (compare left bars of the Duplex and −11 2AP Duplex in Figures 2A and 3A, with left bars of the same DNAs in Figure 4B).
Just as was observed by EMSA (Figures 2 and 3), with the −11A Duplex the differential abilities of the RNAP with substitutions in their σ70 to orchestrate heparin-stable strand separation of promoter DNA are masked under our conditions (see left-hand clusters of bands and bars in Figures 7A and B, respectively). Again the use of the −11 2AP Duplex allows better differentiation among the σ70 with substitutions at 430 and 433: For the complexes formed with the W433L mutant, a smaller fraction of the radioactivity is in cleaved DNA bands than for complexes formed with WT σ70, while with the Y430A a larger fraction of the radioactivity is found in the KMnO4 generated bands. The Y430F is similar to the WT σ70 in this regard. This substitution's lack of deleterious effects here, as compared to the EMSA assays, may be partially due to the lower concentrations of heparin used (100 µg/ml in the KMnO4 experiments versus 200 µg/ml for the EMSA). For W433L and Y430A, the results closely parallel those obtained with the EMSA assay, which monitored formation of stable RNAP-promoter complexes. In the absence of a heparin challenge, no significant differences were seen in the KMnO4 patterns between W433L, Y430A and Y430F for either the −11A or the −11 2AP Duplexes. This is in agreement with the results from the EMSA experiments on the extent of formation of complexes in the absence of a heparin challenge (Figure 4B). We have no data that, for any combination of substitutions in σ70 and Duplex DNA tested, would support the accumulation of complexes that survive a heparin challenge, but yet are not fully strand separated (e.g. I2). Indeed we also have observed that stable complexes, even if containing both mutant σ70 and a 2AP substitution at −11, can initiate RNA synthesis. When tested on a template that is extended in the downstream direction compared to that shown in Figure 1, the complexes were able to make the abortive product UpApU from UpA and UTP in proportion to their ability to form heparin resistant complexes (data not shown).
DISCUSSION
Amino acid sequence preferences at positions 430 and 433, and the roles of the Y430 and W433 residues
The observation that the Y430F substitution is deleterious to formation of a stable complex between RNAP and promoter DNA containing −11 2AP, would suggest that the tyrosine OH group plays an unknown but important role in formation of a stable RNAP-promoter complex. With the consensus promoter that extends to +20, at low temperatures, the RNAP containing the Y430A substitution (but not Y430L) is deficient in open complex formation (data not shown): the same is the case for a similarly long promoter with non-consensus −10 and −35 regions at room temperature. Such behavior had previously been observed with the long non-consensus promoter for Y430A RNAP (11). Similarly, Juang and Helmann (10) found that for B. subtilis σA, both the Y189A and Y189L (Y 189 is the ortholog of σ70 Y430) substitutions had detrimental effects on open complex formation with a full sized promoter. With the short Duplex promoter sequence, the RNAP containing σ70 substitutions Y430A and Y430L are better than the WT sequences of σ70 at forming a stable complex. In this respect, our results on stable complex formation at room temperature, are consistent with those from Gralla's group, who investigated total complex formation (no heparin challenge) near 0°C; under these conditions the vast majority of the complexes are expected to be unstable, closed, complexes (15). The latter group concluded that for their short DNA templates, the Y430 had an inhibitory effect, which would be relieved when the Y430 residue was substituted by A (15). This would also be the case for the Y430L substitution. We speculate that Y430's role is to be inserted between the bases of the DNA to initiate promoter bending. Then, for the Duplex promoter which would be difficult to bend as it lacks any sequence downstream of +1 to serve as a handle for the bending, it might be less detrimental to form a stable complex with RNAP containing σ70 with an A or L at position 430. As they are smaller and more flexible they would be less likely to be forced into the DNA helix, than the Y430 of WT σ70. The OH of Y430 conceivably could aid in this process by engaging in H-bond formation. It has been suggested (21) that RNAP may compete with, and weaken, the hydrogen bonding between the −11A-T pair prior to base flipping. Because the Y430 substitutions to A or L facilitate open complex formation with Duplex DNA, it is not likely that the hydroxyl of Y430 is engaging in such a competing hydrogen bond interaction with the −11A.
It had been assumed that the Y430 and W433 residues played a direct role in flipping the −11 base out of the DNA helix and into a pocket on the RNAP (12,13). The fact that we see a range of activities for the various substitutions at positions 430 and 433, even with a DNA that is abasic at the −11 position of the non-template strand, sets limits on models for the roles of each of the residues W433 and Y430. A priori, there are multiple possibilities, including the following: (i) No interactions with the −11A; (ii) interaction(s) only to −11A; (iii) interaction(s) only to −11A, with other amino acids also interacting with this base; (iv) interactions to both −11A, and to another group of the DNA; (v) novel interactions with the DNA due to adaptation of the protein (43,44) to the DNA site that lacks the −11A. Our results with DNAs that are abasic for −11 in the non-template strand rule out (ii) and (iii), above: If the only roles of Y430 and W433 were to recognize and flip the −11A, in the absence of this base the actual residue present at positions 430 or 433 of σ70 would not matter, and a constant (low) level of strand separation would be expected regardless of the particular amino acid side chain at these positions. The simplest explanations for the results with the abasic DNA are possibilities (i) or (iv), but we cannot exclude (v) based on the available data. The adaptabilty of σ70 may in fact be of crucial importance for the recognition of a variety of non-consensus promoters by RNAP.
For comparison we have also investigated the effect of an alanine substitution for R588 of σ70, which is involved in recognition of the −35 region (45) and would be a highly unlikely interacting partner of the −11A. It is found (data from Figures 2 and 3, and data not shown) that RNAP containing WT and R588A σ70, bind the following percentage of input DNA, respectively, at room temperature subsequent to a 200 μg/ml heparin challenge: Duplex, 66 ± 7 and 46 ± 1; −11 2AP Duplex, 28 ± 7 and 3 ± 0.4; −11Ab Duplex, 51 ± 2 and 22 ± 2. These numbers for R588A are very similar to those for W433L with the same DNAs, and also qualitatively similar to those for Y430F, consistent with the possibility that Y430 and W433, like R588, would not interact with the −11A either.
The finding that with the short Duplex DNA at position 430 small (A) or medium sized (L) aliphatic side chains are preferred, but at position 433, aromatic amino acids [this work and also (15)], must reflect the different roles these two residues play in the process of open complex formation. Perhaps the W433 is involved in stacking onto another aromatic residue. Consistent with the conclusion that amino acids 430 and 433 of σ70 have other (or additional) roles than interacting with the −11 base, it is found that substitutions at 430 and 433, and at −11 in the DNA (e.g. the 2AP or the G) exacerbate each others’ deleterious effects, indicating that different processes may be affected by the DNA—and σ70 substitutions. Also, a mismatch at −11, expected to facilitate removal of the −11 base out of the helix as shown here (Figures 2 and 3) and elsewhere (22,27) to improve open complex formation on Duplex DNA, results in only a small improvement of stable complex formation with the templates for which the −11 A has been substituted by 2AP. This suggests that there is sequence recognition subsequent to removal of the −11 base out of the helix and thus constitutes another demonstration that nucleation of strand separation is, at least, a two-step process.
The steps in the kinetic scheme affected by base analog substitutions at position −11 of Duplex DNA and amino acid substitutions at positions 430 and 433 of σ70
Our results show that substitutions at −11 in Duplex DNA, and Y430 and W433 of σ70 both increase the Kd and the koff for the stable complex. Ten-fold effects on the off-rate were seen for the substitution of the −11A with 2AP, for the WT RNAP. However, for amino acid substitutions in σ70 the effects on the koff of complexes formed with either Duplex DNA or −11 2AP Duplex are more modest (up to 4 fold). Interestingly, the Y430A (and L) RNAP tolerates substitutions at −11A better than the WT RNAP (see also Figure 2). Both the −11 2AP substitution in Duplex DNA, and the three amino acid substitutions investigated had little, if any, effect on the kon for stable complex formation (Figure 6B). In a previous study (11) using a promoter with non-consensus −10 and −35 regions and on a longer piece of DNA with extensions in both directions, we found order of magnitude effects of single substitutions in σ70 on the rates of formation of stable complexes, monitored just as in the experiments described here, by using EMSA subsequent to a heparin challenge. Here we did not detect such effects on kobs, even though our experimental conditions should have readily allowed detection of both increases and decreases in its value (Figure 5B). A recent study (22), as well as countless prior ones [e.g. (46)] also found significant effects on kobs of base substitutions in promoter DNA. A possible explanation for the discrepancy between our data and other work is the difference in length (Duplex is truncated at +1) and the fact that A and its base analog, 2AP, both have an unsubstituted N1 that may be important in the strand separation process (21). Therefore, non-consensus −35 and −10 elements, as well as substitution of −11A with another base or base analog that changes the hydrogen bonding capabilities of the N1 position could affect on-rate. With regard to an abasic site at the −11 position, our work confirms prior data suggesting that such a site causes a local disruption of the DNA helix in the surrounding area, thus aiding in RNAP-induced strand separation (17,22,39). In any case, effects on the ‘on’ and ‘off ’ rates would both be exerted at the rate limiting step, which is the I1 to I2 conversion in either direction. As I1 is in rapid equilibrium with RPc and I2 with RPo (47), it is also possible that the mutations in either DNA or RNAP skew these equilibria towards RPc, thus decreasing the kobs on the one hand, or towards I2, thereby increasing the koff, on the other.
We have no information on the equilibrium between RPc and I1 (although the DNA bending step may not occur for our short Duplex promoter lacking DNA downstream of the +1 site), but our experiments did address whether the substitutions might lead to increased formation of stable complexes that are not strand separated (e.g. I2). In view of the good correlation between the EMSA (determination of stable complexes; Figures 2 and 3) and the KMnO4 (determination of strand opening; Figure 7) studies, there is no evidence for accumulation of I2, and consequently not for inhibition of the I2 to RPo step, due to any substitutions in σ70 or promoters. This is in contrast to published data, which showed that stable complexes of WT RNAP and a −11 2AP containing promoter could still form, but that no strand separation could be detected (19). Likely the actual promoter sequence plays a role as our group previously showed that strand separation could occur at another −11 2AP-substituted promoter (36) as well.
CONCLUSIONS
Substitutions at both −11A of promoter DNA, and Y430 and W433 of σ70 affect the I1 to I2, and possibly also the RPc to I1 step. This conclusion agrees with published work concerning the effects of the substitutions at Y430 and W433 (12,13). Our study of the roles of the Y253 and W256 residues of Taq σA (orthologs of Y430 and W433, respectively) also led to the conclusion that these amino acid residues participated in a step beyond the closed complex (16). We have additionally shown that the effects of substitutions in σ70 (for Y430 and W433) and promoter DNA (for −11A) are cumulative, and that the Y430 and W433 may not participate in interactions with the −11A. While these conclusions are pertinent to the particular promoter used here, they are likely to be relevant to other promoters as well, although it is probable that exceptions will be encountered. The next task is to determine which amino acids interact with the −11A, and which DNA bases, if any, interact with the Y430 and W433 residues of σ70. Establishing the contacts between σ70 and promoter DNA that are necessary for open complex formation will be an important step towards understanding the mechanism of transcription initiation.
ACKNOWLEDGEMENTS
We thank members of the deHaseth laboratory for advice and Dr Vernon Anderson, Dr Mark Caprara and Dr Mike Harris for their comments and suggestions. We also thank Kate Donatto for help with site-directed mutagenesis of some of the sigma factors. This work was supported by NIH grant GM31808 to PLH. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM 31808 to PLH.
Conflict of interest statement. None declared.
REFERENCES
- 1.deHaseth PL, Zupancic M, Record MT., Jr RNA polymerase-promoter interaction: the comings and goings of RNA polymerase. J. Bacteriol. 1998;180:3019–3025. doi: 10.1128/jb.180.12.3019-3025.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saecker RM, Tsodikov OV, McQuade KL, Schlax PE, Capp MW, Record MT., Jr Kinetic studies and structural models of the association of E. coli σ70 RNA polymerase with the λPR promoter: large scale conformational changes in forming the kinetically significant intermediates. J. Mol. Biol. 2002;319:649–671. doi: 10.1016/S0022-2836(02)00293-0. [DOI] [PubMed] [Google Scholar]
- 3.Craig ML, Tsodikov OV, McQuade KL, Schlax PE, Jr, Capp MW, Saecker RM, Record MT., Jr DNA footprints of the two kinetically significant intermediates in formation of an RNA polymerase-promoter open complex: evidence that interactions with the start site and downstream DNA induce sequential conformational changes in polymerase and DNA. J. Mol. Biol. 1998;283:741–756. doi: 10.1006/jmbi.1998.2129. [DOI] [PubMed] [Google Scholar]
- 4.Sen R, Dasgupta D. Simple fluorescence assays probing conformational changes of Escherichia coli RNA polymerase during transcription initiation. Meth. Enzymol. 2003;370:598–605. doi: 10.1016/S0076-6879(03)70050-0. [DOI] [PubMed] [Google Scholar]
- 5.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 6.Malhotra A, Severinova E, Darst SA. Crystal structure of a σ70 subunit fragment from E. coli RNA polymerase. Cell. 1996;87:127–136. doi: 10.1016/s0092-8674(00)81329-x. [DOI] [PubMed] [Google Scholar]
- 7.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 8.Vassylyev DG, Sekine S-I, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 9.Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. Structure of the bacterial RNA polymerase promoter specificity σ subunit. Mol. Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 10.Juang Y-L, Helmann JD. A promoter melting region in the primary sigma factor of Bacillus subtilis: identification of functionally important aromatic amino acids. J. Mol. Biol. 1994;235:1470–1488. doi: 10.1006/jmbi.1994.1102. [DOI] [PubMed] [Google Scholar]
- 11.Panaghie G, Aiyar SE, Bobb KL, Hayward RS, deHaseth PL. Aromatic amino acids in region 2.3 of Escherichia coli sigma 70 participate collectively in the formation of an RNA polymerase-promoter open complex. J. Mol. Biol. 2000;299:1217–1230. doi: 10.1006/jmbi.2000.3808. [DOI] [PubMed] [Google Scholar]
- 12.Tomsic M, Tsujikawa L, Panaghie G, Wang Y, Azok J, deHaseth PL. Different roles for basic and aromatic amino acids in conserved region 2 of Escherichia coli σ70 in the nucleation and maintenance of the single-stranded DNA bubble in open RNA polymerase-promoter complexes. J. Biol. Chem. 2001;276:31891–31896. doi: 10.1074/jbc.M105027200. [DOI] [PubMed] [Google Scholar]
- 13.Fenton MS, Lee HJ, Gralla JD. Escherichia coli promoter opening and −10 recognition: mutational analysis of σ70. EMBO J. 2000;19:1130–1137. doi: 10.1093/emboj/19.5.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsujikawa L, Tsodikov OV, deHaseth PL. Interaction of RNA polymerase with forked DNA: evidence for two kinetically significant intermediates on the pathway to the final complex. Proc. Natl Acad. Sci. (USA) 2002;99:3493–3498. doi: 10.1073/pnas.062487299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenton MS, Gralla JD. Roles for inhibitory interactions in the use of the −10 promoter element by σ70 holoenzyme. J. Biol. Chem. 2003;278:39669–39674. doi: 10.1074/jbc.M307412200. [DOI] [PubMed] [Google Scholar]
- 16.Schroeder LA, deHaseth PL. Mechanistic differences in promoter DNA melting by Thermus aquaticus and Escherichia coli RNA polymerases. J. Biol. Chem. 2005;280:17422–17429. doi: 10.1074/jbc.M501281200. [DOI] [PubMed] [Google Scholar]
- 17.Fenton MS, Gralla JD. Effect of DNA bases and backbone on σ70 holoenzyme binding and isomerization using fork junction probes. Nucleic Acids Res. 2003;31:2745–2750. doi: 10.1093/nar/gkg400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helmann JD, deHaseth PL. Protein-nucleic acid interactions during open complex formation investigated by systematic alteration of the protein and DNA binding partners. Biochemistry. 1999;37:5959–5967. doi: 10.1021/bi990206g. [DOI] [PubMed] [Google Scholar]
- 19.Lim HM, Lee HJ, Roy S, Adhya S. A “master” in base unpairing during isomerization of a promoter upon RNA polymerase binding. Proc. Natl Acad. Sci. USA. 2001;98:14849–14852. doi: 10.1073/pnas.261517398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee HJ, Lim HM, Adhya S. An unsubstituted C2 hydrogen of Adenine is critical and sufficient at the −11 position of a promoter to signal base pair deformation. J. Biol. Chem. 2004;279:16899–16902. doi: 10.1074/jbc.C400054200. [DOI] [PubMed] [Google Scholar]
- 21.Matlock DL, Heyduk T. Sequence determinants for the recognition of the fork junction DNA containing the −10 region of promoter DNA by E. coli RNA polymerase. Biochemistry. 2000;39:12274–12283. doi: 10.1021/bi001433h. [DOI] [PubMed] [Google Scholar]
- 22.Heyduk E, Kuznedelov K, Severinov K, Heyduk T. A consensus adenine at position −11 of the nontemplate strand of bacterial promoter is important for nucleation of promoter melting. J. Biol. Chem. 2006;281:12362–12369. doi: 10.1074/jbc.M601364200. [DOI] [PubMed] [Google Scholar]
- 23.Fenton MS, Gralla JD. Function of the bacterial TATAAT −10 element as single-stranded DNA during RNA polymerase isomerization. Proc. Natl Acad. Sci. USA. 2001;98:9020–9025. doi: 10.1073/pnas.161085798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helmann JD, Chamberlin J. Structure and function of bacterial sigma factors. Annu. Rev. Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 25.deHaseth PL, Nilsen TW. When a part is as good as the whole. Science. 2004;303:1307–1308. doi: 10.1126/science.1095483. [DOI] [PubMed] [Google Scholar]
- 26.Young BA, Gruber TM, Gross CA. Minimal machinery of RNA polymerase holoenzyme sufficient for promoter melting. Science. 2004;303:1382–1384. doi: 10.1126/science.1092462. [DOI] [PubMed] [Google Scholar]
- 27.Roberts JW, Roberts CW. Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase. Cell. 1996;86:495–501. doi: 10.1016/s0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 28.Zhi H, Jin DJ. Purification of highly-active and soluble Escherichia coli σ70 polypeptide overproduced at low temperature. Methods in Enzymol. 2003;370:174–180. doi: 10.1016/S0076-6879(03)70015-9. [DOI] [PubMed] [Google Scholar]
- 29.deHaseth PL, Tsujikawa L. Probing the role of region 2 of Escherichia coli σ70 in nucleation and maintenance of the single stranded bubble in RNA polymerase-promoter open complexes. Methods in Enzymol. 2003;370:553–567. doi: 10.1016/S0076-6879(03)70047-0. [DOI] [PubMed] [Google Scholar]
- 30.Lozinski T, Wierzchowski KL. Inactivation and destruction of Escherichia coli RNA polymerase open transcription complex: recommendations for footprinting experiments. Anal. Biochem. 2003;320:239–251. doi: 10.1016/s0003-2697(03)00381-6. [DOI] [PubMed] [Google Scholar]
- 31.Burr T, Mitchell J, Kolb A, Minchin S, Busby S. DNA sequence elements located immediately upstream of the −10 hexamer in Escherichia coli promoters: a systematic study. Nucleic Acids Res. 2000;28:1864–1870. doi: 10.1093/nar/28.9.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanderson A, Mitchell JE, Minchin SD, Busby SJW. Substitutions in the Escherichia coli RNA polymerase σ70 factor that affect recognition of extended −10 elements at promoters. FEBS Lett. 2003;544:199–205. doi: 10.1016/s0014-5793(03)00500-3. [DOI] [PubMed] [Google Scholar]
- 33.Szoke PA, Allen TA, deHaseth PL. Promoter recognition by Escherichia coli RNA polymerase: Effects of base substitution in the −10 and −35 regions. Biochemistry. 1987;26:6188–6194. doi: 10.1021/bi00393a035. [DOI] [PubMed] [Google Scholar]
- 34.Roy S. On the use of 2-aminopurine as a probe for base pair opening during transcription initiation. Methods in Enzymol. 2003;370:568–576. doi: 10.1016/S0076-6879(03)70048-2. [DOI] [PubMed] [Google Scholar]
- 35.Ujvari A, Martin CT. Thermodynamic and kinetic measurements of promoter binding by T7 RNA polymerase. Biochemistry. 1996;35:14574–14582. doi: 10.1021/bi961165g. [DOI] [PubMed] [Google Scholar]
- 36.Tsujikawa L, Strainic MG, Watrob H, Barkley MD, deHaseth PL. RNA polymerase alters the mobility of an A-residue crucial to polymerase-induced melting of promoter DNA. Biochemistry. 2002;41:15334–15341. doi: 10.1021/bi026539m. [DOI] [PubMed] [Google Scholar]
- 37.Tang G-Q, Patel SS. Rapid binding of T7 RNA polymerase is followed by simultaneous bending and opening of the promoter DNA. Biochemistry. 2006;45:4947–4956. doi: 10.1021/bi052292s. [DOI] [PubMed] [Google Scholar]
- 38.Li X-Y, McClure WR. Stimulation of open complex formation by nicks and apurinic sites suggests a role for nucleation of DNA melting in Escherichia coli promoter function. J. Biol. Chem. 1998;273:23558–23566. doi: 10.1074/jbc.273.36.23558. [DOI] [PubMed] [Google Scholar]
- 39.Gelfand CG, Plum GE, Grollman AP, Johnson F, Breslauer KJ. Thermodynamic consequences of an abasic lesion in duplex DNA are strongly dependent on base sequence. Biochemistry. 1998;37:7321–7327. doi: 10.1021/bi9803372. [DOI] [PubMed] [Google Scholar]
- 40.Lisser S, Margalit H. Compilation of E.coli mRNA promoter sequences. Nucleic Acids Res. 1993;21:1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niedziela-Majka A, Heyduk T. Escherichia coli RNA polymerase contacts outside the −10 promoter element are not essential for promoter melting. J. Biol. Chem. 2005;280:38219–38227. doi: 10.1074/jbc.M507984200. [DOI] [PubMed] [Google Scholar]
- 42.Sasse-Dwight S, Gralla JD. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J. Biol. Chem. 1989;264:8074–8081. [PubMed] [Google Scholar]
- 43.Jen-Jacobson JJ. Protein-DNA recognition complexes: conservation of structure and binding energy in the transition state. Biopolymers. 1997;44:153–180. doi: 10.1002/(SICI)1097-0282(1997)44:2<153::AID-BIP4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 44.Mossing MM, Record MT., Jr Thermodynamic origins of specificity in the lac Repressor-Operator interaction. J. Mol. Biol. 1985;186:295–305. doi: 10.1016/0022-2836(85)90106-8. [DOI] [PubMed] [Google Scholar]
- 45.Gardella T, Moyle H, Susskind MM. A mutant Escherichia coli σ70 subunit of RNA polymerase with altered promoter specificity. J. Mol. Biol. 1989;206:579–590. [Google Scholar]
- 46.McClure WR. Mechanism and control of transcription initiation in prokaryotes. Annu. Rev. Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 47.Tsodikov OV, Record MT., Jr General method of analysis of kinetic equations for multistep reversible mechanisms in the single-exponential regime: application to kinetics of open complex formation between Eσ70 RNA polymerase and λPR promoter DNA. Biophys. J. 1999;76:1320–1329. doi: 10.1016/S0006-3495(99)77294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]