Abstract
Previous work suggested that phosphorylation of DNA-PKcs at several serine/threonine (S/T) residues at positions 2609–2647 promotes DNA-PK-dependent end joining. In an attempt to clarify the role of such phosphorylation, end joining was examined in extracts of DNA-PKcs-deficient M059J cells. Joining of ends requiring gap filling prior to ligation was completely dependent on complementation of these extracts with exogenous DNA-PKcs. DNA-PKcs with either S/T → A or S/T → D substitutions at all six sites in the 2609–2647 cluster also supported end joining, but with markedly lower efficiency than wild-type protein. The residual end joining was greater with the S/T → D-substituted than with the S/T → A-substituted protein. A specific inhibitor of the kinase activity of DNA-PK, KU57788, completely blocked end joining promoted by wild type as well as both mutant forms of DNA-PK, while inhibition of ATM kinase did not. The fidelity of end joining was not affected by the mutant DNA-PKcs alleles or the inhibitors. Overall, the results support a role for autophosphorylation of the 2609–2647 cluster in promoting end joining and controlling the accessibility of DNA ends, but suggest that DNA-PK-mediated phosphorylation at other sites, on either DNA-PKcs or other proteins, is at least as important as the 2609–2647 cluster in regulating end joining.
INTRODUCTION
DNA-dependent protein kinase, consisting of the catalytic subunit DNA-PKcs and the end-binding heterodimer Ku, is a core component of the nonhomologous end-joining pathway of DNA double-strand break repair. The primary function of DNA-PK appears to be regulating the end-joining process, both by sequestering DNA ends and by catalyzing serine/threonine phosphorylation of itself as well as other proteins (1,2). In order to assess the biological significance of specific phosphorylation events, radiosensitivity and end joining were previously examined in CHO variants harboring nonphosphorylatable S/T → A substitutions at several sites shown to be targets of DNA-PK-mediated phosphorylation, either in vivo or in vitro. Data obtained thus far suggest that whereas phosphorylation of Ku and XRCC4 is dispensable (3), phosphorylation of DNA-PK itself, particularly at T2609, S2612, T2620, S2624, T2638 and T2647 (the ‘ABCDE cluster’) is critical for efficient DSB repair. For example, single S/T → A substitutions at several of these sites increase radiosensitivity of cells harboring the mutant proteins, and tandem substitutions at multiple sites in the cluster inhibit repair to a greater degree than mutations at any single site, and confer greater radiosensitivity (4,5).
Particularly intriguing is a DNA-PKcs allele with S/T → D substitutions at all six sites in the ABCDE cluster, designed to mimic constitutive phosphorylation at these sites. In an end-joining assay with purified Ku and XRCC4/DNA ligase IV complex (X4L4), this mutant (hereafter designated DNA-PKcs-D6) promotes ligation of a cohesive-ended substrate, albeit less efficiently than the wild-type protein. Moreover, whereas wortmannin, an inhibitor of DNA-PK kinase activity, completely blocks the in vitro end joining promoted by wild-type DNA-PKcs, wortmannin has no effect on the end joining promoted by the D6 mutant (6). Thus, in this assay, DNA-PKcs-D6 appears to substitute for autophosphorylated DNA-PKcs, suggesting that phosphorylation of the ABCDE cluster may be the primary means by which DNA-PK regulates the end-joining process. In DNA-PKcs-deficient CHO-V3 cells, ectopic expression of DNA-PKcs-D6 partially restores radioresistance and DSB rejoining (5,7), although not as robustly as might be expected from the in vitro studies.
In an attempt to further elucidate the role of phosphorylation of the ABCDE cluster, end joining was examined in whole-cell extracts of DNA-PKcs-deficient M059J cells supplemented with wild-type and mutant DNA-PKcs proteins. This experimental system allows direct examination of DNA end processing and end joining, in the presence of all the proteins required to carry out joining of both cohesive and incompatible DNA ends. The results confirm the importance of the ABCDE cluster as a DNA-PK phosphorylation target, but suggest that there are other target phosphorylation sites of equal if not greater importance.
MATERIALS AND METHODS
Materials
Purification of DNA-PKcs from CHO-V3 cell lines harboring wild-type and mutant human DNA-PKcs alleles has been described (6). Because the yield of DNA-PKcs from hamster cells is much lower than from human cells, DNA-PKcs from HeLa cells (8) was used for experiments that did not involve direct comparison of wild-type and mutant alleles. Nevertheless, the properties of HeLa DNA-PKcs did not appear to differ significantly from those of wild-type DNA-PKcs purified from V3 cells. To ensure reproducibility between experiments, initial frozen aliquots of DNA-PKcs were subdivided into 1 μl aliquots and flash-frozen in the presence of 1.6 mg/ml BSA. Thus, most experiments were performed with DNA-PKcs that had undergone no more than two freeze–thaw cycles. Concentrations of DNA-PKcs from V3 cells were determined by Bradford assay, and the purity and relative concentrations of the three recombinant alleles were verified by quantitation on gels stained with SYPRO orange (Supplementary Figure 1).
Previous end-joining experiments employed an SV40-based shuttle plasmid (pSV56) designed to introduce a DSB in a polylinker in the intron of the T-antigen gene (9). However, because the presence of the SV40 origin reduces the yield of the plasmid from bacterial cultures, the polylinker region of pSV56 was excised as a 625-bp PflMI/AvrII fragment, and cloned into pBR322 between the EcoRV and NheI sites (the PflMI cut was blunt-ended with T4 polymerase). This 5-kb plasmid, designated pRZ56, was cut at its unique MluI site, and then subjected to controlled 3′ → 5′ exonucleolytic digestion with T4 polymerase in the presence of dTTP. This procedure resects each 3′-terminal strand to the first thymine in the sequence, resulting in a 10-base 5′ overhang at one end and an 11-base 5′ overhang at the other. An unlabeled 13-mer and a 5′-32P-labeled 14-mer, each complementary to one of the 5′ overhangs, were successively ligated into the overhangs, to yield site-specifically labeled substrates with partially complementary (-ACG) overhangs, as described previously (9,10). Control experiments showed that the oligomers were ligated onto at least 90% of the overhangs (data not shown). The labeled plasmid was purified by agarose gel electrophoresis, electroeluted, concentrated (Amicon Centricon 100) and precipitated. Alternatively, the MluI-cut plasmid was dephosphorylated and 5′-end-labeled with polynucleotide kinase and [32P]ATP.
DNA-PKcs-deficient M059J cells were obtained from Dr Joan Allalunis-Turner (Cross Cancer Institute, Edmonton, Alberta, Canada), and were grown in MEM alpha (Gibco) containing 10% fetal bovine serum. One day after reaching confluence, cells were trypsinized, and washed extensively with serum-containing medium and PBS. Whole-cell extracts were prepared by hypotonic swelling and Dounce homogenization as described (10,11), except that instead of a 3 × hypertonic extraction buffer, 0.25 volume of a 5 × buffer (83.5 mM Tris-HCl, 1.65 M KCl, 3.3 mM EDTA, 1 mM dithiothreitol) was added prior to high-speed centrifugation, to yield a slightly higher protein concentration. Extracts consistently had a protein concentration of ∼12 mg/ml (Pierce BCA protein assay), and extracts from confluent cultures showed greater end-joining efficiency than those from subconfluent cultures. Dignam nuclear extracts, prepared as described previously (12,13), had a concentration of ∼5 mg/ml.
Kinase inhibitors KU57788 and KU55933 were provided by Dr Graeme Smith, KUDOS Inc., and were dissolved in DMSO at concentrations of 2.5 and 10 mM, respectively, and stored at −20°C. Inhibitors were further diluted in DMSO and added directly to cell extracts within 1 h.
End-joining assay
Reactions contained 50 mM triethanolamine–KOH pH 7.5, 1 mM ATP, 1 mM dithiothreitol, 1.3 mM magnesium acetate and dNTPs (or ddNTPs) at 100 μM each. Typically, a 16-μl reaction contained 10 μl of extract, resulting in a final concentration of 66 mM potassium acetate and 16% glycerol, and an effective Mg++ concentration of 1 mM (taking into account ∼0.3 mM EDTA from the extract). Buffer components were first mixed with cell extract at 22°C. DNA-PKcs and/or kinase inhibitors were then added and the solution mixed by pipeting. Finally, 10 ng substrate was added and the reaction again mixed by pipeting, and placed in a 37°C water bath, usually for 1 or 6 h. Samples were deproteinized as described (10) and either loaded directly onto 0.8% agarose gels, or cut with BstXI and TaqI and analyzed on 20% polyacrylamide DNA sequencing gels. Storage phosphor screens were exposed to frozen polyacrylamide or dried agarose gels, and images were analyzed with ImageQuant 3.3 software.
RESULTS
Accurate joining of incompatible or cohesive DNA ends in M059J extracts requires catalytically active DNA-PKcs but not ATM
As demonstrated previously, human whole-cell extracts contain robust activity for joining of both cohesive and incompatible DNA ends (10,11,14). When end-joining reactions are performed in the presence of relatively low Mg++ concentrations, the joining is largely dependent on core end-joining factors such as DNA-PKcs, XRCC4, DNA ligase IV (11,15) and, in the case of incompatible ends, DNA polymerase λ (12).
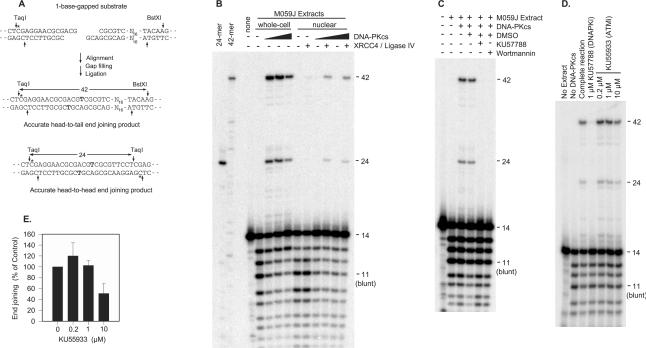
To assess the dependence of end joining on DNA-PKcs, and specifically on DNA-PKcs kinase activity, whole-cell extracts were prepared from DNA-PKcs-deficient M059J glioma cells (16). Western blotting (not shown) confirmed absence of DNA-PKcs in these extracts. End joining of an internally labeled substrate with partially complementary (-ACG) 3′ overhangs was then examined in these extracts either with or without addition of purified DNA-PKcs from HeLa cells (Figure 1).
Figure 1.
Requirement for catalytically active DNA-PK in end joining of a substrate with partially complementary 3′ overhangs. The internally labeled (asterisk) substrate shown (10 ng) was incubated for 6 h in whole-cell extracts (120 μg) or nuclear extracts (30 μg) from DNA-PKcs-deficient M059J cells. Extracts were supplemented with purified human DNA-PKcs from HeLa cells and/or recombinant X4L4 as indicated. DNA was then cut with BstXI and TaqI and analyzed on denaturing sequencing gels. (A) Internally labeled substrate and expected end-joining products. The bolded ‘T’ indicates fill-in of the 1 base gap in each strand of the aligned ends. (B) End joining by whole-cell extracts supplemented with 0, 3, 6 or 10 nM DNA-PKcs, or nuclear extracts supplemented with 0, 3 or 6 nM DNA-PKcs plus 20 nM X4L4 as indicated. (C) Inhibition of end joining in the presence of 6 nM DNA-PKcs by the DNA-PK inhibitors KU57788 (1 μM) and wortmannin (3 μM). Samples contained 1.2% DMSO as indicated. (D) Effect of the ATM inhibitor KU55933; same conditions as (C) except that all samples contained 1.2% DMSO. (E) Quantitative data from experiments similar to that shown in (D); the extent of accurate end joining was calculated as the sum of the 24- and 42-base fragments divided by the total phosphorimage intensity in each lane, then normalized to the end joining seen in the presence of DNA-PKcs and DMSO without any inhibitor. Error bars indicate SEM of four measurements, each from a separate experiment except the 10 μM point, which was from two measurements.
When this substrate was incubated in M059J extracts, and then cut with BstXI and TaqI to release short fragments from opposite ends of the plasmid, there was some 3′ → 5′ exonucleolytic digestion, but no ligation products were detectable (Figure 1B). Supplementation of the extracts with DNA-PKcs resulted in appearance of prominent 42- and 24-base labeled products. As demonstrated previously (10), these products reflect ‘accurate’ head-to-tail (42-mer) and head-to-head (24-mer) end joining by a mechanism involving annealing of the overhangs, fill-in of the resulting one-base gap, and ligation (Figure 1A). Although traces of 34- to 38-base products, corresponding to small deletions in the repair joints, were detected, the accurate products consistently accounted for at least 90% of the end joining. Using various batches of cell extract, 4–20% of the initial DNA ends were accurately joined. The specific DNA-PK inhibitor KU57788 (aka NU7441) (17) completely blocked formation of both accurate products at a concentration of 1 μM, as did the less specific DNA-PKcs inhibitor wortmannin at a concentration of 3 μM (Figure 1C). A specific inhibitor of ataxia-telangiectasia-mutated (ATM) protein, KU55933 (18), had little effect at concentrations expected to completely inhibit ATM kinase activity (0.2–1 μM, Figure 1D and E). However, it did partially inhibit end joining at 10 μM, a concentration expected to affect DNA-PK as well (18). DMSO (solvent for all these inhibitors) had only a slight inhibitory effect (<20%) on the efficiency of end joining. These results confirm the requirement for DNA-PKcs, and for its kinase activity, in the joining of incompatible ends by human whole-cell extracts. Similar DNA-PKcs dependence was seen in nuclear extracts, except that, as reported previously (19), significant end joining was only seen when extracts were also supplemented with purified X4L4 (Figure 1B). However, whole-cell extracts were used for all subsequent experiments because supplemented nuclear extracts gave lower yields (2–3%) of end-joined products, and showed a trace of X4L4-dependent end joining in the absence of DNA-PKcs. This latter result, which is reminiscent of studies showing accurate end joining of gapped DSB substrates in extracts of DNA-PKcs-deficient CHO strains (10,20), could be explained either by the presence in human whole-cell extracts of factors whose inhibitory effect is abrogated by DNA-PK, or the presence in nuclear extracts of factors that can substitute for DNA-PK in promoting end-to-end synapsis.
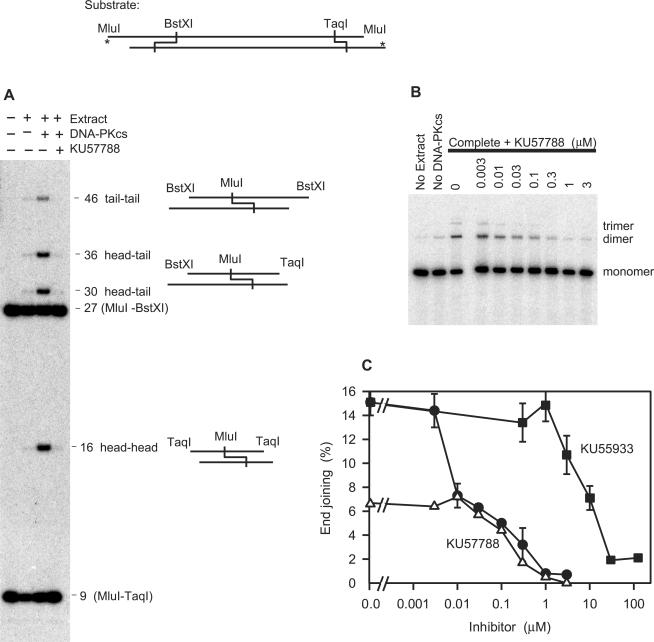
DNA-PKcs-supplemented M059J extracts were also capable of joining a 5′-end labeled cohesive-end substrate (Figure 2A). Consistent with simple cohesive-end ligation, such joining yielded exclusively 16-base (head-head), 46-base (tail-tail), 30-base and 36-base (both head-tail) labeled fragments, which comigrated with products generated by treatment with T4 ligase (not shown). For this substrate, the incubation time was reduced to 1 h, which avoided significant loss of 5′ label but did not significantly decrease the extent of joining (data not shown). Although a trace of cohesive end joining was sometimes detected in unsupplemented extracts, DNA-PKcs addition consistently increased end joining by at least 15-fold. As with the gapped substrate, KU57788 blocked end joining at concentrations expected to inhibit DNA-PKcs, with half-maximal inhibition at ∼0.02 μM and complete inhibition at 1 μM (Figure 2B and C). Again, the ATM inhibitor KU55933 had no effect at concentrations expected to inhibit ATM [Ki ∼ 0.013 μM, ref. (18)], but did block end joining at the extremely high concentrations expected to inhibit DNA-PK as well (Ki ∼ 2.5 μM) (Figure 2C). These results indicate that joining of DSBs with cohesive as well as incompatible ends in human cell extracts is completely dependent on catalytically active DNA-PK but not ATM.
Figure 2.
Requirement for catalytically active DNA-PK in end joining of a substrate with cohesive 5′ overhangs. MluI-cut pRZ56 was 5′-32P-end-labeled (asterisk) and incubated for 1 h in whole-cell extracts containing 6 nM purified DNA-PKcs and inhibitors as indicated. (A) Denaturing sequencing gel showing products formed. (B) Agarose gel showing titration of inhibition by KU57788. (C) Quantitation of inhibition of end joining of the cohesive end substrate by KU57788 (filled circle) or by the ATM inhibitor KU55933 (filled square). Error bars show range of values obtained in two independent experiments. A single titration for KU57788 with the partially complementary substrate (see Figure 1) is shown for comparison (open triangle). Note that since a single joining event results in both the joined and unjoined ends of a single plasmid migrating as a dimer, the agarose gel assay results in higher apparent levels of end joining than the sequencing gel assay.
DNA-PKcs proteins with mutations at autophosphorylation target sites show reduced proficiency in end joining
To determine whether the observed requirement for DNA-PKcs kinase activity reflects a requirement for autophosphorylation of the S/T 2609–2647 cluster, end joining was examined in M059J cells supplemented with either wild-type DNA-PKcs or DNA-PKcs proteins having either S/T → A (A6) or S/T → D (D6) substitutions at all six sites in the ABCDE cluster. To ensure consistency, all three proteins were expressed in and purified from CHO-V3 cells, and previous work showed that they all have comparable kinase activity (6,7).
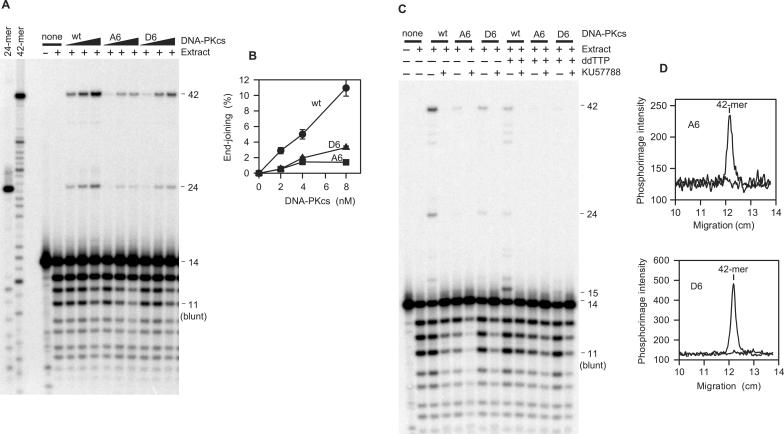
Figure 3 shows end joining of the 1-base-gapped substrate in extracts supplemented with these wild-type and mutant DNA-PKcs proteins. DNA-PKcs-D6, designed to mimic constitutive phosphorylation of the ABCDE cluster, supported end joining, but with markedly lower efficiency than the wild-type protein (Figure 3A and B). The S/T → D mutations did not affect the fidelity of end joining, as the accurate 42- and 24-base products were formed almost exclusively. DNA-PKcs-A6 also supported end joining, but even less efficiently than the D6 mutant. Nevertheless, even at the lowest concentration of DNA-PKcs-A6, joining was significantly greater than that seen with no DNA-PKcs (0.5% versus <0.05%). Moreover, in three replicate experiments, the difference between the D6 and A6 alleles was consistently more pronounced at 8 nM than at 2–4 nM (Figure 3B).
Figure 3.
Promotion of end joining of a substrate with partially complementary 3′ overhangs by wild-type and mutant alleles of DNA-PKcs. The substrate shown in Figure 1 was incubated for 6 h in whole-cell extracts supplemented with wild-type DNA-PKcs (wt), or DNA-PKcs with 6 S/T → A (A6) or S/T → D (D6) substitutions in the ABCDE cluster. Products were then analyzed on denaturing sequencing gels. See Figure 1 for definition of substrates and products. (A) End joining in presence of 2, 4 or 8 nM of each DNA-PKcs protein. (B) Total accurate end joining (42-mer plus 24-mer product) was calculated for the wt (filled circle), A6(filled square) and D6 (filled triangle) alleles at each concentration of DNA-PKcs. Error bars represent range of values in two independent experiments. (C) Effect of KU57788 (1 μM) on end joining and gap filling by each DNA-PKcs allele (4 nM). Some samples contained 100 μM ddTTP in place of dTTP, as indicated. The labeled single-stranded 15-mer fragment (released from the top strand of the plasmid by TaqI) represents addition of an unligatable ddTMP nucleotide to the -AGC overhang in the labeled top strand. (D) Phosphorimage intensity profiles from the gel shown in (C), in the area of the 42-base end-joining product, for the lanes containing samples with the A6 and D6 mutant alleles, without ddTTP. The profiles for the samples with and without KU57788 have been superimposed. In both cases, there is clearly a peak of 42-base product that is completely eliminated by KU57788.
The formation of accurate repair products from the gapped substrate requires two end-processing steps, gap filling and ligation. To determine the effect of the phosphorylation site mutations specifically on the gap-filling step, end-joining reactions were performed in the presence of ddTTP rather than dTTP. As expected, in the presence of wild-type DNA-PKcs, ddTTP resulted in the generation of a 15-mer, corresponding to a single-base-elongated but unligated intermediate, accompanied by a decrease in intensity of the 42- and 24-base ligation products (Figure 3C). The residual 42- and 24-mer products presumably reflect gap filling by residual dTTP in the cell extracts (extracts were dialyzed for only 3 h (11)). With the D6 and A6 mutant forms of DNA-PKcs, there was little if any apparent gap filling. If the extent of gap filling were comparable to that of overall end joining, the 15-mer intermediate might not be detected due to its proximity to the dominant 14-mer band corresponding to the initial substrate. In any case, however, the mutant proteins are clearly much less efficient in promoting gap filling on aligned DSB ends than is wild-type DNA-PKcs.
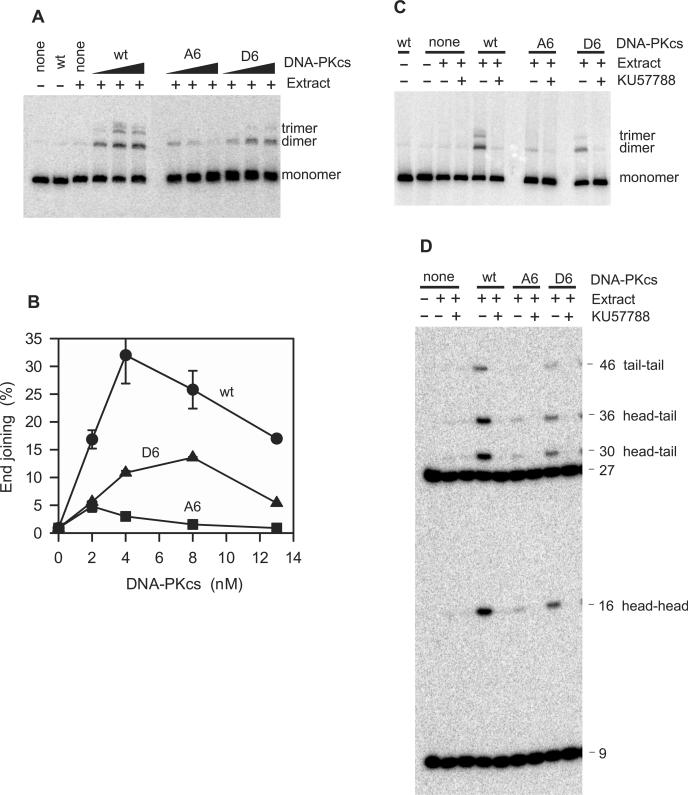
To examine the effect of phosphorylation site mutations specifically on the ligation step, similar end-joining assays were performed with a simple 5′-end-labeled substrate bearing cohesive 4-base (CGCG-) 5′ overhangs (Figure 4). The results were very similar to those obtained with the substrate requiring gap filling; that is, the D6 mutant was markedly less efficient in promoting ligation than wild-type DNA-PKcs, and the A6 mutant was even less efficient than the D6 mutant (Figure 4A and B). Thus, end joining of a substrate requiring only simple religation appears to be just as dependent on the availability of phosphorylation sites in the ABCDE cluster, as end joining of a substrate requiring more complex processing. Thus, gap filling, ligation and end joining overall, show roughly the same relative dependence on ABCDE phosphorylation, consistent with the model previously proposed (6,7) in which phosphorylation of the ABCDE cluster promotes some conformational or other change that increases accessibility of DNA ends and thereby facilitates both gap filling and ligation.
Figure 4.
Promotion of end joining of a cohesive-end substrate by wild-type and mutant alleles of DNA-PKcs. The substrate shown in Figure 2 was endlabeled and incubated for 1 h in extracts supplemented with the wt, A6 and D6 DNA-PKcs alleles. (A) Agarose gel analysis of end joining in the presence of each DNA-PKcs allele at 0, 2, 4 or 8 nM. (B) Concentration dependence of end joining with the wild-type (filled circle), A6(filled square) and D6 (filled triangle) alleles. Error bars show range of values obtained in two independent experiments when larger than the symbols; the 13 nM point was from a single experiment due to the large amount of DNA-PKcs required. (C) Effect of KU57788 (1 μM) on end joining with each DNA-PKcs allele (4 nM). (D) Same as C., except analysis was by BstXI/TaqI cleavage and denaturing sequencing gel electrophoresis.
End joining promoted by mutant DNA-PKcs proteins is still dependent on DNA-PK-catalyzed phosphorylation
The finding that the D6 mutant promotes end joining more efficiently than the A6 mutant, is consistent with a model wherein the S/T → D mutations mimic phosphorylation of the ABCDE cluster (albeit imperfectly) and promote a similar conformational change in DNA-PKcs. To assess whether this was the only essential DNA-PK-mediated phosphorylation event, joining of both cohesive and incompatible DNA ends was examined in the presence of M059J extracts, mutant DNA-PKcs proteins and the DNA-PKcs inhibitor KU57788. With the gapped DSB substrate, KU57788 completely blocked the end joining promoted by wild-type DNA-PKcs as well as both the A6 and the D6 mutants (Figure 3C and D). Similarly, KU57788 reduced end joining of the cohesive-end substrate in the presence of either mutant to the extremely low level seen in the absence of DNA-PKcs (Figure 4C and D). This complete inhibition by KU57788 was confirmed in at least three independent experiments with each substrate and each of the three DNA-PKcs alleles. Thus, mimicking ABCDE phosphorylation with S/T → D substitutions was not sufficient to promote end joining in the absence of DNA-PKcs activity. These results suggest that there are target sites for DNA-PKcs-mediated phosphorylation, other than the ABCDE cluster, that are important if not essential for end joining.
In assays with the substrate bearing 3′ overhangs, 3′ → 5′ exonucleolytic processing in cell extracts is also apparent. Although the nuclease responsible for this digestion has not been identified, it nevertheless provides an independent measure of DNA end accessibility. Despite some variability between different batches of cell extract, exonucleolytic processing was consistently greater with the D6 than with the A6 mutant, consistent with phosphorylation of the ABCDE cluster playing an important role in controlling DNA end accessibility (Figure 3C). Nevertheless, KU57788 reduced exonucleolytic processing with all three DNA-PKcs alleles, suggesting that DNA-PK-mediated phosphorylation of sites outside the ABCDE cluster contribute to regulation of end accessibility.
DISCUSSION
Several lines of evidence suggest that DNA-PKcs autophosphorylation in the ABCDE cluster plays an important role in promoting nonhomologous end joining. When nonphosphorylatable S/T → A substitutions are introduced at all six sites in the cluster, DNA-PKcs no longer stimulates ligation of cohesive ends by X4L4 (6). Moreover, CHO cells harboring the same DNA-PKcs-A6 allele are even more radiosensitive than cells lacking DNA-PK entirely (5). Both these results are consistent with the proposed model wherein autophosphorylation of these sites induces a conformational change in DNA-PKcs that increases accessibility of DNA ends to specific end-processing enzymes such as polymerase λ and ligase IV, thus allowing end joining to proceed to completion. However, the A6 mutant does support a low level of Artemis-mediated hairpin opening and V(D)J coding joint formation, suggesting some residual activity (though perhaps only in the hairpin cleavage step) (5). The coding joints formed in cells expressing this mutant are more similar to those formed in wild-type than in DNA-PKcs-deficient cells, but show even less nucleotide loss from the ends (consistent with constitutive end sequestration), and completely lack any microhomology-based joining.
Conversely, a mutant with S/T → D substitutions at the same six sites promotes ligation of cohesive ends by purified X4L4, albeit less efficiently than the wild-type DNA-PKcs, even when DNA-PK kinase activity is blocked (6). When expressed in DNA-PKcs-deficient CHO cells, DNA-PKcs-D6 largely restores normal V(D)J recombination, and partially rescues radioresistance (5). Both these results suggest that the S/T → D substitutions mimic DNA-PKcs autophosphorylation to a considerable extent, though perhaps not perfectly.
While all of these studies strongly suggest an important role for phosphorylation of the ABCDE cluster in both V(D)J recombination and DSB repair, they do not address the issue of whether such phosphorylation is sufficient to promote these processes, or whether other sites of DNA-PK-mediated phosphorylation exist that are of equal or greater importance. These results also do not indicate whether quantitative differences between DNA-PKcs alleles in restoring radioresistance reflect qualitative differences in the types of DSBs that can be effectively processed by each allele. In intact CHO cells, complete DNA-PKcs deficiency reduces DSB rejoining by only ∼40%, whereas replacement of DNA-PKcs with DNA-PKcs-A6 reduces rejoining by ∼50% (5). These results suggest that some DSBs are repaired by alternative DNA-PK-independent mechanism(s) (21) that are partially inhibited by DNA-PKcs-A6, probably because it fails to dissociate from unrepaired DSBs (7). In the whole-cell extract assay, however, the low Mg++ concentration restricts end joining to the ligase IV-mediated pathway (15), and thus accurate joining of the substrate requiring gap filling is absolutely dependent on DNA-PKcs (Figure 1). This stringency allowed the detection of a small amount of residual accurate joining promoted by DNA-PKcs-A6 (Figure 3), reminiscent of the low level of restored V(D)J recombination in cells expressing this allele (5). Thus, at least in cell extracts, DNA-PKcs does have some residual activity in promoting end joining, even when phosphorylation of the ABCDE cluster is completely abolished. This residual activity, like that seen with wild-type DNA-PKcs, is still blocked by KU57788, and therefore must require DNA-PK-mediated phosphorylation of sites outside the ABCDE cluster, either on DNA-PKcs itself or on other proteins. The results suggest that once these phosphorylations occur, DNA-PKcs-A6 either dissociates from some DNA ends (5), or can adopt a conformation that permits some end processing.
The level of end joining promoted by DNA-PKcs-D6 in cell extracts, greater than DNA-PKcs-A6 but typically 2- to 5-fold lower than wild type (Figure 3), is in reasonable agreement with its 2-fold lower activity in promoting ligation by purified X4L4 (6), as well as with its ability to partially restore radioresistance (∼50%) and DSB rejoining (∼20%) to DNA-PKcs-deficient V3 cells (5,7). However, in previous work wortmannin had no effect on the ability of the D6 allele to promote X4L4-mediated ligation (6). Thus, the finding that the more specific inhibitor KU57788 completely blocks the ability of DNA-PKcs-D6 to promote end joining in cell extracts (Figures 3 and 4), was unexpected. The simplest explanation for these conflicting results is that the presence of other proteins at DNA ends in cell extracts (e.g. XLF, Artemis and/or the Mre11/Rad50/NBS complex) confers a requirement for additional phosphorylation events, besides DNA-PKcs phosphorylation in the ABCDE cluster—events that are apparently dispensable in the simpler reaction with DNA-PK and X4L4 only. On the other hand, the failure of the D6 mutant to fully substitute for the wild-type protein in both in vitro and in vivo assays is not particularly surprising, and is consistent with a model wherein optimum end joining requires properly timed interconversion between phosphorylated and nonphosphorylated forms of DNA-PKcs. (There is, however, the formal possibility that S/T → D substitutions do not fully mimic phosphorylated S/T residues, and that if fully ABCDE-phosphorylated DNA-PKcs could be generated, it would promote some level of end joining in the absence of other phosphorylations.) Overall, the data confirm the importance of ABCDE phosphorylation in promoting DNA-PK-mediated end joining, but suggest that there are additional DNA-PK phosphorylation targets that are as important as ABCDE cluster, if not moreso. Histone H1 may be one such target (22).
Because DNA-PKcs is a large and complex protein, any comparisons among the wild-type and mutant forms of the enzyme are potentially complicated by the possibility that differences in protein stability rather than in phosphorylation status could account for the observed differences in end-joining efficiency. However, we have not seen any indication of differences in stability, and all three alleles have similar levels of kinase activity. In fact, in some assays, such as DNA end protection and X4L4 recruitment, the A6 protein (which is least active in end joining) is more active than the D6 protein (6), consistent with the proposal that it more strongly binds to and sequesters the DNA end. Extreme care was taken that the three alleles of DNA-PKcs were purified, stored and aliquotted identically. Moreover, although a single batch of each form of DNA-PKcs was used in all experiments in Figures 1–4, the previously reported differences in biochemical activities were seen consistently with at least two batches of each allele (Ramsden,D.A., unpublished data). Nevertheless, the possibility that one or both of the mutant proteins tends to spontaneously degrade in a way that compromises its end-joining activity but not its kinase activity, cannot be definitively excluded.
Recently, it has been reported that in irradiated cells (10 Gy) the ABCDE cluster is phosphorylated by ATM rather than DNA-PK (23). However, if such ATM-mediated phosphorylation is required for DNA repair, it is difficult to reconcile the small defect in DSB rejoining seen in ATM-deficient cells (24–26) with the much larger deficit seen in cells harboring DNA-PKcs-A6 (5). The distinct lack of an inhibitory effect of KU55933 on end joining at concentrations known to block ATM kinase activity (Figures 1 and 2) suggests that, at least in cell extracts, the ABCDE phosphorylation that is functionally relevant for repair is carried out by DNA-PK rather than ATM. At survivable levels of damage in intact cells, DNA-PK autophosphorylation might be transient and involve only an extremely small number of DNA-PKcs molecules actually bound to DSB ends, and so might not be detected in any direct phosphorylation assays.
A second ‘PQR’ cluster of five phosphorylated serines (2023, 2029, 2041, 2053, 2056) has been identified in DNA-PKcs, and in contrast to ABCDE, phosphorylation of these sites appears to decrease accessibility of DNA ends (27). However, even simultaneous S → A substitutions at all five PQR sites confers only slight radiosensitivity. A threonine → alanine substitution at a newly identified C-terminal site, T3950, confers radiosensitivity intermediate between normal and DNA-PKcs-deficient cells (28), but while in vivo phosphorylation at this site has been demonstrated it is not known whether the phosphorylation is catalyzed by DNA-PK. Overall, the available data suggest that functionally relevant phosphorylation occurs at a large number of sites on DNA-PKcs, and that while no single site is essential, some are more important than others. Thus, while elimination of phosphorylation throughout the ABCDE cluster (as in DNA-PKcs-A6) reduces end joining to a low level, elimination of phosphorylation at all non-ABCDE sites (DNA-PKcs-D6 plus KU57788) reduces end joining to an undetectable level (Figures 3 and 4). Interpretation of results with the D6 mutant is complicated by the possibility that optimal end joining may require not only that these sites be phosphorylated, but that they be phosphorylated/dephosphorylated in a specific order or at a specific stage of the end-joining pathway. Although the assays with the gapped substrate can potentially discriminate between the gap-filling step and the final ligation step, mutations in the ABCDE cluster as well as DNA-PK/ATM inhibitors appear to affect both steps about equally, consistent with a model where ABCDE phosphorylation precedes both steps.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr Graeme Smith and KUDOS for providing KU57788 and KU55933, and Dr Kathy Meek for providing cell lines expressing mutant DNA-PKcs alleles. This work was supported by Grants CA40615, CA72955, AG023783 and CA84442 from the National Institutes of Health, DHHS. Funding to pay the Open Access publication charges for this article was provided by CA40615.
Conflict of interest statement. None declared.
REFERENCES
- 1.Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–5812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- 2.Meek K, Gupta S, Ramsden DA, Lees-Miller SP. The DNA-dependent protein kinase: the director at the end. Immunol. Rev. 2004;200:132–141. doi: 10.1111/j.0105-2896.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- 3.Douglas P, Gupta S, Morrice N, Meek K, Lees-Miller SP. DNA-PK-dependent phosphorylation of Ku70/80 is not required for non-homologous end joining. DNA Repair. 2005;4:1006–1018. doi: 10.1016/j.dnarep.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, Chen DJ. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, Lees-Miller SP, Meek K. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol. Cell. Biol. 2003;23:5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy YV, Ding Q, Lees-Miller SP, Meek K, Ramsden DA. Non-homologous end joining requires that the DNA-PK complex undergo an autophosphorylation-dependent rearrangement at DNA ends. J. Biol. Chem. 2004;279:39408–39413. doi: 10.1074/jbc.M406432200. [DOI] [PubMed] [Google Scholar]
- 7.Block WD, Yu Y, Merkle D, Gifford JL, Ding Q, Meek K, Lees-Miller SP. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic Acids Res. 2004;32:4351–4357. doi: 10.1093/nar/gkh761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodarzi AA, Lees-Miller SP. Biochemical characterization of the ataxia-telangiectasia mutated (ATM) protein from human cells. DNA Repair. 2004;3:753–767. doi: 10.1016/j.dnarep.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 9.Bennett RAO, Gu X-Y, Povirk LF. Construction of a vector containing a site-specific DNA double-strand break with 3′-phosphoglycolate termini and analysis of the products of end-joining in CV-1 cells. Intl. J. Radiat. Biol. 1996;70:623–636. doi: 10.1080/095530096144509. [DOI] [PubMed] [Google Scholar]
- 10.Chen S, Inamdar KV, Pfeiffer P, Feldmann E, Hannah MF, Yu Y, Lee JW, Zhou T, Lees-Miller SP, et al. Accurate in vitro end-joining of a DNA double-strand break with partially cohesive 3′-overhangs and 3′-phosphoglycolate termini: Effect of Ku on repair fidelity. J. Biol. Chem. 2001;276:24323–24330. doi: 10.1074/jbc.M010544200. [DOI] [PubMed] [Google Scholar]
- 11.Baumann P, West SC. DNA end-joining catalyzed by human cell-free extracts. Proc. Natl Acad. Sci. USA. 1998;95:14066–14070. doi: 10.1073/pnas.95.24.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JW, Blanco L, Zhou T, Bebenek K, Garcia-Diaz M, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 13.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daza P, Reichenberger S, Göttlich B, Hagmann M, Feldmann E, Pfeiffer P. Mechanisms of nonhomologous DNA end-joining in frogs, mice and men. Biol. Chem. 1996;377:775–786. doi: 10.1515/bchm3.1996.377.12.775. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Zeng ZC, Perrault AR, Cheng X, Qin W, Iliakis G. Genetic evidence for the involvement of DNA ligase IV in the DNA-PK-dependent pathway of non-homologous end joining in mammalian cells. Nucleic Acids Res. 2001;29:1653–1660. doi: 10.1093/nar/29.8.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lees-Miller SP, Godbout R, Chan DW, Weinfeld M, Day RS, III, Barron GM, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 17.Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, Smith GC. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg. Med. Chem. Lett. 2004;14:6083–6087. doi: 10.1016/j.bmcl.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 18.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 19.Huang J, Dynan WS. Reconstitution of the mammalian DNA double-strand break end-joining reaction reveals a requirement for an Mre11/Rad50/NBS1-containing fraction. Nucleic Acids Res. 2002;30:667–674. doi: 10.1093/nar/30.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhfittig-Kulle S, Feldmann E, Odersky A, Kuliczkowska A, Goedecke W, Eggert A, Pfeiffer P. The mutagenic potential of non-homologous end joining in the absence of the NHEJ core factors Ku70/80, DNA-PKcs and XRCC4-LigIV. Mutagenesis. 2007;22:217–233. doi: 10.1093/mutage/gem007. [DOI] [PubMed] [Google Scholar]
- 21.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 22.Kysela B, Chovanec M, Jeggo PA. Phosphorylation of linker histones by DNA-dependent protein kinase is required for DNA ligase IV-dependent ligation in the presence of histone H1. Proc. Natl Acad. Sci. USA. 2005;102:1877–1882. doi: 10.1073/pnas.0401179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Yajima H, Löbrich M, Chen DJ. ATM is essential for DNA-PKcs phosphorylations at T2609 cluster upon DNA double-strand break. J. Biol. Chem. 2007;282:6582–6587. doi: 10.1074/jbc.M611605200. [DOI] [PubMed] [Google Scholar]
- 24.Foray N, Priestley A, Alsbeih G, Badie C, Capulas EP, Arlett CF, Malaise EP. Hypersensitivity of ataxia telangiectasia fibroblasts to ionizing radiation is associated with a repair deficiency of DNA double-strand breaks. Int. J. Radiat. Biol. 1997;72:271–283. doi: 10.1080/095530097143266. [DOI] [PubMed] [Google Scholar]
- 25.Kuhne M, Riballo E, Rief N, Rothkamm K, Jeggo PA, Lobrich M. A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res. 2004;64:500–508. doi: 10.1158/0008-5472.can-03-2384. [DOI] [PubMed] [Google Scholar]
- 26.Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 27.Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol. Cell Biol. 2005;25:10842–10852. doi: 10.1128/MCB.25.24.10842-10852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Douglas P, Cui X, Block WD, Yu Y, Gupta S, Ding Q, Ye R, Morrice N, Lees-Miller SP, et al. The DNA-dependent protein kinase catalytic subunit (DNA-PKcs) is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol. Cell Biol. 2007;27:1581–1591. doi: 10.1128/MCB.01962-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.