Abstract
Protein–protein interactions are an important element of signal transfer within and between organisms. They are mainly mediated by short oligopeptide motifs and represent a widely used alternative to small, organic molecules for conveying information. The transcription factor TetR, a regulator of tetracycline resistance in Gram-negative bacteria, is naturally induced by tetracycline or its derivatives. The oligopeptide Tip (Transcription inducing peptide) fused to either N- or C-terminus of Thioredoxin A (TrxA) has been isolated as an artificial inducer for TetR in Escherichia coli. This inducing property can be exploited to monitor the in vivo expression of a protein of interest by fusing Tip to its C-terminus. We improve the induction efficiency of Tip by adding an aromatic amino acid before residue 1 of Tip in C-terminal fusions to TrxA. The induction efficiency of that modified TrxA-Tip fusion is further enhanced when the effector-binding pocket of TetR is enlarged by the N82A and F86A mutations. The double mutant is also insensitive to induction by tetracyclines. Thus, Tip is an exclusive inducer of this TetR variant, representing the first example of fully converting a small molecular weight effector-dependent transcription factor into one depending solely on protein–protein recognition.
INTRODUCTION
Peptides are involved in many essential processes in signal transduction and cell-to-cell communication. Several prominent examples are the peptide hormones that bind to cell surface receptors triggering a signal cascade within the cell (1), peptides involved in innate immunity (2), or the quorum sensing peptides found in Gram-positive bacteria (3). Since methods like phage display or mRNA display facilitate the isolation of peptides that recognize protein targets, the number of synthetic peptides with novel biological functions is steadily and rapidly growing. These include enzyme and toxin inhibitors (4,5), peptides with antiviral activity (6,7) or with the ability to specifically detect tissues with histological changes associated with diseases (8,9). Peptides selected in this manner mostly exert their effect by binding to a specific target site.
Recently, a 16-mer peptide termed Tip (Transcription inducing peptide; Figure 1A) was isolated via phage display. Tip is not only able to bind specifically to its target, the bacterial transcription factor Tet repressor (TetR), but also triggers a conformational change in vivo normally induced by tetracyclines, the natural effectors of TetR (10). TetR is widely used to control regulation of gene expression in prokaryotes and, as a fusion protein with an activation domain, in eukaryotes (11), because it combines high specificity for its cognate DNA sequence (tetO) with extremely sensitive induction by tetracyclines (tc), especially the potent analogues doxycycline (dox) and anhydrotetracycline (atc) (12,13). Non-tc inducers of TetR, like Tip, may lead to alternative inducers when used as a scaffold for peptidomimetics to generate novel, small-molecule effectors. The recently published crystal structure of the TetR–Tip complex provides a molecular basis for such efforts (14). Another application of Tip is to fuse it to a protein of interest and use it to analyse target protein expression in vivo by monitoring a TetR-controlled reporter (15). Although a wide variety of tag-systems suitable for diverse applications exist, Tip is the only protein tag that allows quantitative in vivo analysis of protein expression. Mandatory for this application is a high efficiency of induction to ensure that low levels of Tip fusion proteins can be detected. We demonstrate here that addition of an aromatic residue between C-terminally fused Tip and TrxA along with mutations in the effector-binding pocket of TetR lead to a strong increase of reporter induction. Furthermore, the introduced TetR mutations render it insensitive to induction by tetracyclines, thereby creating exclusive specificity for Tip.
Figure 1.
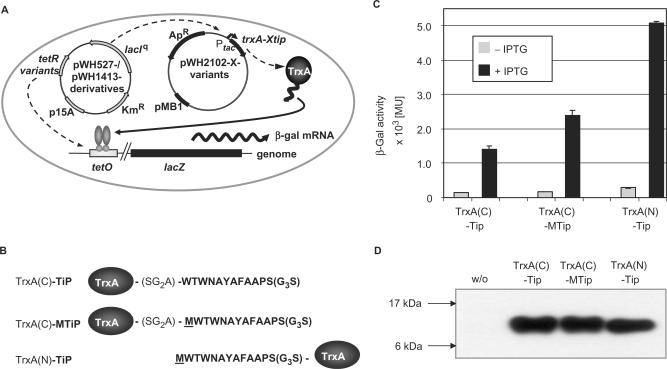
Induction activity of the C-terminal Met insertion mutant TrxA-Mtip. (A) A schematic presentation of the E. coli screening strain for TetR inducing peptides is shown (10). Escherichia coli DH5α(λtet50) contains lacZ under Tet control on the chromosome. Derivatives of pWH527 or pWH1413 constitutively express LacI and the TetR variants. The TetR dimer is depicted by light gray ovals with circular DNA-binding domains. TetR controls transcription of lacZ. The peptide-expressing plasmids encode either TrxA (pWH2200) or SbmC (pWH2300) or the respective fusion protein (e.g. TrxA(C)-XTip is encoded by pWH2102-X) under control of Ptac. A TetR inducing thioredoxin-peptide fusion leads to β-Gal expression as indicated by the arrow. The respective resistance genes and origins of replication of the plasmids are indicated. (B) Sequences of Tip peptides (bold) fused C-terminal or N-terminal to TrxA and the linker between TrxA and Tip are shown in one-letter abbreviations. (C) β-Gal activities obtained with the indicated TrxA-Tip fusions in the presence and absence of IPTG are shown. Gray bars show the repressed states in the absence of IPTG. Expression of the Tip-fusion proteins was induced with 60 µM IPTG (black bars). (D) Western blots indicating the steady-state levels of Tip-fusion proteins in the presence of 60 µM IPTG. Molecular weights of marker proteins (not shown) are indicated on the left side.
MATERIALS AND METHODS
Materials and general methods
Chemicals were obtained from Merck (Darmstadt), Sigma (Munich) or Roth (Karlsruhe) and were of the highest purity available. Media, buffers and solutions were prepared with Millipore water or deionized water and autoclaved. Heat labile substances were dissolved and filtered with a sterile filter (0.2 µm). Enzymes for DNA restriction and modification were obtained from New England Biolabs (Frankfurt/M.), Roche Diagnostics (Mannheim), Invitrogen (Karlsruhe) or Fermentas (St. Leon-Rot). Sequencing was carried out according to the protocol provided by PE Applied Biosystems (Weiterstadt) for cycle sequencing.
Bacterial strains, plasmids and phage
All bacterial strains are derived from Escherichia coli K12. Strains DH5α (hsdR17 (rK− mK+), recA1, endA1, gyrA96, thi, relA1, supE44, ϕ80dlacZΔM15, Δ(lacZYA-argF)U169) (16) and RB791 (IN[rrnD-rrnE]1, lacIqL8) (17) were used for general cloning procedures. Strain DH5α(λtet50) (Tn10 tetA-lacZ transcriptional fusion) (10,18) served as host strain for β-galactosidase assays. The plasmids pWH1200 (19), pWH527- and pWH1413-derivatives for TetR variant expression (10), pWH806 (20), pWH2100-, pWH2200- and pWH2300-derivatives for C- and N-terminal TrxA/SbmC-XTip variant expression (10) were used in the in vivo studies. Class B TetR (21) was used in all experiments.
Insertion of Met in C-terminal TrxA-Tip fusion
Insertion of Met into the C-terminal TrxA-Tip fusion was done by standard 2-step PCR using pWH2101 as template, the mutagenic primer 5′-GTCGGGTGGAGCTATGTGGACTTGGAATG-3′ to introduce the Met codon (underlined) and the flanking oligonucleotides 5′-TGACAATTAATCATCGGCTCG-3′ and 5′-AAGGAATGGTGCATGCCTGC-3′. The product obtained was restricted with HindIII and PstI and cloned into likewise-digested pWH2101 (10). The resulting plasmid was named pWH2102.
Randomization of Met at position −1 of TrxA-MTip
Randomization of the Met at position −1 in TrxA(C)-MTip was performed by combined chain reaction (22). The phosphorylated oligonucleotide 5′-GTCGGGTGGAGCT(NNK)TGGACTTGGAATG-3′ introducing the randomized codon (NNK; N = A, C, G or T; K = G or T) and the flanking oligonucleotides 5′-TGACAATTAATCATCGGCTCG-3′ and 5′-AAGGAATGGTGCATGCCTGC-3′ were used in a standard PCR reaction with pWH2102 as template. The resulting DNA was restricted with HindIII and PstI and cloned into likewise-digested pWH2102. Candidates were identified by sequencing and named pWH2102-X with X denoting the amino acid replacing the M.
Construction of the C-terminal SbmC(C)-Tip fusion
The construction of a plasmid-encoded SbmC(C)-Tip fusion was done by amplification of the fragment encoding the fusion from genomic DNA from E. coli BW25113(λtet50)sbmC(C)-tip, a strain containing the linker and Tip-encoding sequence as used in the TrxA fusion fused to the 3′ end of the sbmC gene at its position in the E. coli genome (unpublished data) with the primers 5′-ATGATGTCTAGAAACTACGAGATTAAGCAGGAAGAGAAACG-3′ introducing a restriction site for XbaI (underlined) and 5′-GTAGTTGCATGCTTACGAACCTCCACCACTAGGAGC-3′ introducing a restriction site for SphI (underlined). The product was restricted with XbaI and SphI and cloned into likewise-digested pWH610 (23). The resulting plasmid was named pWH2301.
Insertion of Met in the C-terminal SbmC(C)-Tip fusion
Insertion of Met (underlined) into the C-terminal SbmC-Tip fusion protein was done genetically by standard 2-step PCR using pWH2301 as template, a mutagenic primer 5′-CTCGGGTGGAGCTATGTGGACTTGGAATG-3′ and the flanking primers 5′-ATGATGTCTAGAAACTACGAGATTAAGCAGGAAGAGAAACG-3′ and 5′-GTAGTTGCATGCTTACGAACCTCCACCACTAGGAGC-3′, introducing restriction sites (underlined) for XbaI and SphI, respectively. The product obtained was cleaved with both enzymes and cloned into likewise-digested pWH610 (10). The resulting plasmid was named pWH2301-Met.
Construction of tetR-N82A-F86A
The F86A mutation was introduced into tetR-N82A encoded by pWH527-N82A via standard 2-step PCR using a mutagenic primer 5′-CGTAATAACGCTAAAAGTGCTAGATGTGCTTT-3′ and the two flanking primers 5′-CTCGACATCTTGGTTACCG-3′ and 5′-CGCCGTACTGCCCGCTTGG-3′. The resulting double mutant was cloned via restriction with NcoI and XbaI into pWH527 resulting in low-level constitutive expression of the TetR mutant (10).
β-Galactosidase (β-Gal) activity measurements
Repression and inducibility of TetR was assayed in E. coli DH5α(λtet50) (10). The strain was transformed with pWH527-derivatives expressing TetR variants at a low level or pWH1413-derivatives expressing TetR at a higher level and with plasmids from the pWH2100/2200/2300 series encoding the different C- and N-terminal Tip fusions to TrxA or SbmC. Overnight cultures and log-phase cultures were grown at 28°C in LB medium supplemented with 100 µg/ml ampicillin and either 60 µg/ml kanamycin for pWH527-derivatives or 25 µg/ml chloramphenicol for pWH1413-derivatives. For this purpose, stationary phase cultures were diluted 1:100 in fresh medium and expression of the fusion proteins induced using 60 µM IPTG, if not indicated otherwise. The cells were then grown to an OD600 of ∼0.4 and their β-Gal activities determined as described by Miller (24). Three independent clones were assayed for each combination of constructs and experiments repeated at least twice. The values obtained were normalized to the maximal β-Gal activity in the absence of TetR which was set to 100% and typically varied from 6500 to 7500 Miller units in the individual experiments.
Western blot analysis of the TrxA-XTip fusion variants and the TetR variants
Escherichia coli DH5α(λtet50) was transformed with the derivatives listed in the respective β-Gal assays and grown under the same conditions as stated there. Cells were harvested at an OD600 of 0.4. The crude lysates were prepared by sonication and centrifugation. Ten microgram of crude lysate of each construct was loaded either on a 14% (TrxA-XTip fusions) or a 10% SDS-PAA gel (TetR variants) and electrophoresed. Proteins were transferred (120 mA, overnight at 4°C) to a PVDF membrane (BIORAD) in a Mini V8.10 blotting apparatus (Gibco-BRL) using 10 mM NaHCO3, 3 mM Na2CO3 and 20% (v/v) methanol as transfer buffer. After blocking with 0.2% I-Block™ (TROPIX) in phosphate-buffered saline (75 mM Na-phosphate; 68 mM NaCl; pH 7.8) with 0.1% Tween 20, membranes were treated either with a monoclonal anti-TrxA antibody (TrxA-XTip fusions, Anti-Thio™ antibody, Invitrogen) or a polyclonal TetR-specific antibody (SA1851; lab stock). Signals were detected with anti-mouse IgG (TrxA-XTip fusions) or anti-rabbit IgG (TetR variants) conjugated to horseradish peroxidase (Amersham Pharmacia) and the ECL+ kit (Amersham Pharmacia) following the manufacturer's instructions.
RESULTS
Induction of TetR by C-terminal TrxA-XTip variants
Figure 1A shows the relevant features of the E. coli screening strain used in this study to score the efficiency of TetR induction by TrxA fusions with Tip. This strain has been described in detail (10). It contains one plasmid encoding TetR or its variants and LacI. TetR represses a chromosomally located lacZ, while LacI controls expression of the respective TrxA-Tip fusion protein. The latter is encoded on a second, compatible plasmid giving rise to IPTG-inducible β-Gal expression mediated by TrxA-Tip. For our studies, Tip was fused C- or N-terminally to TrxA, and it had been observed that the latter is a much better inducer of TetR than the former (10) (see Figure 1B and C). A potential reason might be a sequence difference in Tip resulting from the failure of cleaving the initiator amino acid M from the N-terminus in E. coli when the following residue is W (see Figure 1B) (25,26). This M residue is missing in the C-terminal fusion. To examine this possibility we inserted an M residue into the C-terminal TrxA(C)-Tip fusion at the corresponding position termed −1 (see Figure 1B). This insertion leads to a nearly 2-fold increase in β-Gal activity, but still reaches only half of the β-Gal activity obtained with the N-terminal TrxA(C)-Tip fusion (see Figure 1C). The steady-state protein levels of all three TrxA fusion proteins are similar as concluded from Western blots (see Figure 1D) confirming that the induction levels are intrinsic properties of the fusions.
We next asked if M is the optimal amino acid at position −1 for induction of TetR by randomizing this residue. Western blot analyses were carried out with the resulting 19 TrxA(C)-XTip variants to make sure their expression levels are the same. Only TrxA(C)-PTip showed a reduced steady-state level while all other variants were present in similar amounts as TrxA(C)-MTip and TrxA(C)-Tip (data not shown). The TrxA(C)-XTip variants were then scored for their induction efficiencies in the E. coli screening strain (see Figure 1A). The results are summarized in Table 1. All activities of the TrxA(C)-XTip fusions are presented relative to 100% β-Gal activity defined by a strain lacking TetR. TrxA(C)-XTip variants with P, charged residues like D, E, K or R or with aliphatic, hydrophobic residues like V, L or I display either no or only marginal induction of TetR. All other amino acids at this position yield fusion proteins that induce TetR. They are either less active (Y), as active (S, T, C, F) or more active (G, A, N, Q, H, W) than the one with M.
Table 1.
Induction of TetR by all 20 possible variants of TrxA(C)-Xtip
| TetR-inducing protein | β-Gal activity (%) | |
|---|---|---|
| Repression | Induction | |
| TrxA(C)-Tip | 2.3 ± 0.1 | 24 ± 2 |
| TrxA(C)-MTip | 2.6 ± 0.1 | 37 ± 3 |
| TrxA(C)-XTip | ||
| X = Glycine | 3.5 ± 0.4 | 57 ± 5 |
| Alanine | 3.7 ± 0.2 | 63 ± 4 |
| Serine | 2.7 ± 0.1 | 41 ± 3 |
| Threonine | 2.4 ± 0.2 | 34 ± 3 |
| Cysteine | 2.4 ± 0.2 | 36 ± 3 |
| Valine | 2.2 ± 0.1 | 6.9 ± 0.5 |
| Isoleucine | 2.6 ± 0.2 | 8.7 ± 1.2 |
| Leucine | 2.1 ± 0.1 | 6.3 ± 0.6 |
| Aspartic Acid | 2.2 ± 0.1 | 2.0 ± 0.1 |
| Asparagine | 3.1 ± 0.1 | 46 ± 4 |
| Glutamic Acid | 1.9 ± 0.2 | 2.3 ± 0.2 |
| Glutamine | 3.8 ± 0.1 | 64 ± 3 |
| Lysine | 2.1 ± 0.1 | 2.2 ± 0.1 |
| Arginine | 2.0 ± 0.1 | 2.2 ± 0.1 |
| Histidine | 3.4 ± 0.1 | 57 ± 4 |
| Phenylalanine | 3.3 ± 0.2 | 38 ± 1 |
| Tyrosine | 2.3 ± 0.1 | 21 ± 1 |
| Tryptophan | 2.7 ± 0.1 | 55 ± 1 |
| Proline | 2.0 ± 0.1 | 1.5 ± 0.4 |
β-Gal activities were determined in E. coli DH5α(λtet50) expressing lacZ under Tet-control. tetR is encoded by pWH527. Expression of the C-terminal TrxA-Tip fusions was induced with 60 µM IPTG to determine peptide-based induction of TetR.
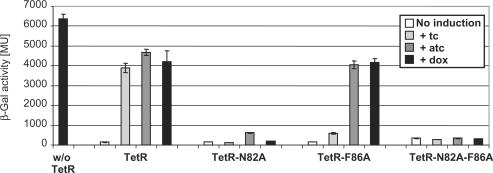
Induction of TetR-N82A, -F86A and -N82A-F86A by Tip variants
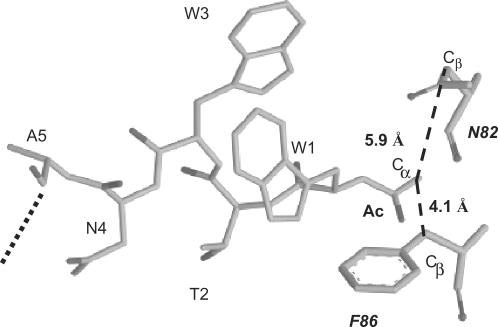
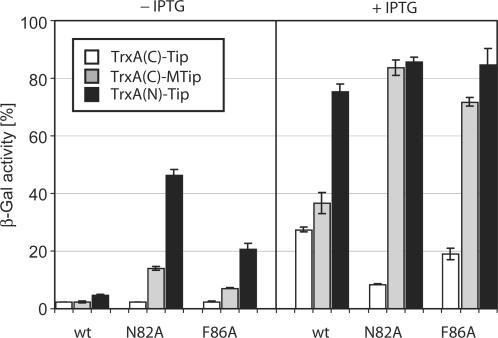
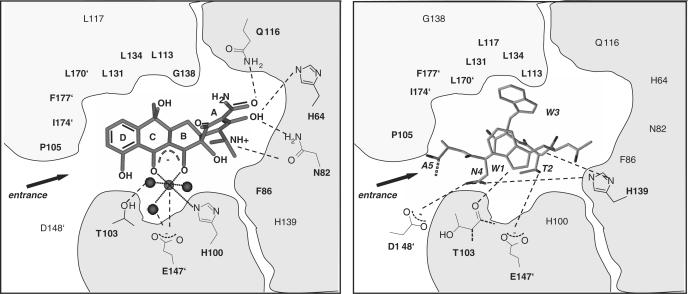
X-ray crystallography of TetR complexed with a 16-mer Tip oligopeptide acetylated at the N-terminus revealed details of the location of Tip inside the tc-binding pocket. The Cα atom of the acetyl moiety is in close proximity to residues N82 and F86 of TetR (14). Figure 2 shows an excerpt of the structure highlighting this proximity. We assumed that a large residue at this position, as found in several active C-terminal TrxA(C)-XTip variants, might lead to sterical hindrance. Therefore, we replaced the bulky N82 and F86 residues with a smaller A residue and scored the induction properties of these mutants with TrxA(C)-MTip. The results are shown in Figure 3. Even in the absence of IPTG, in which only basal expression of TrxA(C)-MTip from the leaky tac promoter occurs, we observed induction of TetR-N82A and TetR-F86A. However, TrxA(N)-Tip is still the more efficient inducer under these conditions. At the higher, IPTG-induced, expression level of TrxA(C)-MTip, induction of TetR-N82A reaches the same level as with TrxA(N)-Tip and wt TetR. A slightly smaller maximal induction by TrxA(C)-MTip is observed for TetR-F86A. As expected, altering the size of the binding pocket in TetR-N82A and TetR-F86A does not lead to increased induction by TrxA(C)-Tip lacking the additional M residue.
Figure 2.
Location of Tip in the TetR–Tip complex structure. Residues W1 to A5 of Tip and the residues N82 and F86 of TetR are displayed as gray stick models. Ac indicates the acetyl moiety at the N-terminus of Tip present in the complex. The distance (dashed lines) between the Cα of the acetyl and the Cβ atoms of N82 or F86 is indicated. The dotted line indicates the continuation of Tip. The coordinates were taken from the PDB-file 2NS8 (14).
Figure 3.
Induction of TetR, TetR-N82A and TetR-F86A by C- and N-terminal TrxA-Tip fusions. The TetR variants are indicated below the bars and encoded by pWH527-derivatives. White bars show the activities induced by the C-terminal TrxA(C)-Tip fusion, gray bars those of TrxA(C)-MTip and black bars those of the N-terminal TrxA(N)-Tip fusion. The left side of the figure (labeled − IPTG) shows induction by basal expression levels while the right side (labeled + IPTG) shows the effects of induced (60 µM IPTG) expression of the Tip protein fusions.
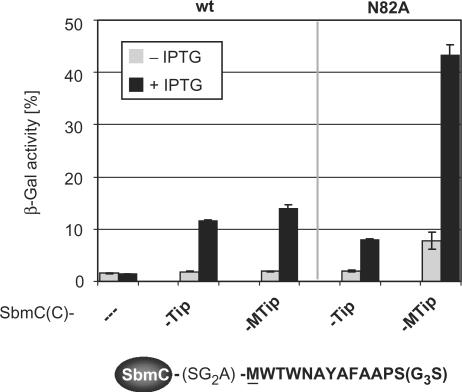
The induction properties of TrxA-Tip could depend in part on the TrxA portion of the fusion protein. To address this question, we constructed Tip and MTip fusions to the C-terminus of the E. coli protein SbmC (27) and replaced the TrxA-Tip fusion in our screening strain by them. SbmC is a small, soluble protein of 157 residues with solvent-exposed and flexible N- and C-termini (28). It is part of the SOS regulon and involved in reducing DNA damage (29). Induction of TetR by SbmC(C)-MTip is slightly more efficient than induction by SbmC(C)-Tip. In contrast, TetR-N82A is induced to a 5-fold higher level of β-Gal activity by SbmC(C)-MTip compared to SbmC(C)-Tip as shown in Figure 4. This result establishes an only marginal influence of the carrier protein on Tip activity. Hence, the improved induction of the TetR variants by the elongated Tip results solely from their effects on Tip–TetR interaction.
Figure 4.
Induction of TetR and TetR-N82A by C-terminal SbmC-Tip fusions. Induction of the TetR variants indicated above the plot and encoded by pWH527-derivatives is given by the β-Gal activities expressed by the screening strain. Gray bars represent basal expression levels of SbmC(C)-Tip fusions in the absence of IPTG, while induced (60 µM IPTG) expression levels are represented by black bars. The sequence of Tip fused to SbmC is illustrated below the chart. The residues of Tip fused to SbmC are shown in bold, and the linker between SbmC and Tip is shown in light print.
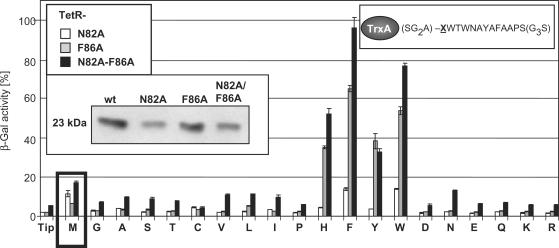
We wondered if the induction efficiency profiles of the TrxA(C)-XTip variants depend on the shape of the inducer-binding pocket. In addition to the TetR-N82A and -F86A mutants, we constructed the double mutant TetR-N82A-F86A and introduced that also into the screening strain. The Western blot shown as insert in Figure 5 indicates that TetR and TetR-F86A are expressed to about the same level, while TetR-N82A and TetR-N82A-F86A are present in roughly half the amount of the former variants. The induction activities were scored without IPTG at the basal expression levels of the TrxA(C)-XTip variants to increase the sensitivity and are shown in Figure 5. TrxA(C)-Tip shows about the same induction for TetR-N82A and TetR-F86A (∼2% β-Gal activity), but the double mutant appears slightly de-repressed under these conditions (∼4%). No other TrxA(C)-XTip variant induces TetR-N82A to a significantly higher degree than TrxA-MTip, and only TrxA(C)-FTip and TrxA(C)-WTip reach about the same induction efficiency (see the white columns in Figure 5). In contrast, TetR-F86A is very efficiently induced by TrxA-XTip variants with an aromatic residue. While TrxA-MTip shows only ∼6% β-Gal activity, TrxA(C)-HTip and TrxA(C)-YTip lead to ∼35% β-Gal activity, TrxA(C)-WTip gives rise to 53% and TrxA-FTip even to 60% β-Gal activity. The latter variant is hence the most efficient inducer of TetR-F86A. Similar to TetR-F86A, we observed the highest induction activities for TetR-N82A-F86A by TrxA-XTip derivatives with aromatic residues, albeit in a slightly changed order: TrxA-YTip exhibits the same induction efficiency for both TetR variants, while TrxA-HTip, TrxA-FTip and TrxA-WTip are better inducers for the double than for the single TetR mutant. Taken together, it appears that TrxA(C)-FTip and TrxA(C)-WTip are the best inducers for all three TetR variants, but their efficiencies increase from TetR-N82A via TetR-F86A to TetR-N82A-F86A. It is also noteworthy to mention that these induction profiles do not correlate generally to the differences in expression levels established for these three TetR variants.
Figure 5.
Induction of TetR mutants by Tip variants. Induction of TetR mutants by basal expression levels (absence of IPTG) of TrxA-XTip variants is shown. X represents any of the 20 amino acids as depicted in one-letter abbreviations (see upper right insert for the sequence). TetR variants are encoded by pWH527-derivatives. Induction of TetR variants is indicated by the corresponding β-Gal activities expressed by the screening strain. White bars show the induction values of TetR-N82A, gray bars those of TetR-F86A and black bars those of TetR-N82A-F86A. Induction by the initially analysed M insertion is highlighted by a black box. Tip indicates induction by basal expression levels of TrxA(C)-Tip and the one-letter abbreviations indicate the inserted residue. The insert in the upper left corner shows the coding of the bars. The central insert shows Western blots indicating the steady-state protein levels of TetR, TetR-N82A, TetR-F86A and TetR-N82A-F86A. The molecular mass of the TetR variants is indicated on the left.
The TetR residues N82 and F86 contact tc as derived from the crystal structure of the TetR-tc complex (30). Hence, we tested whether the three TetR mutants in which one or both of these residues are replaced by A are inducible by tc, atc or dox, three widely used inducers of TetR. The results are shown in Figure 6. TetR-F86A is not induced by tc, but atc and dox are very efficient inducers. In contrast, TetR-N82A and TetR-N82A-F86A are not induced by any of these compounds. Since these two are best induced by Tip we have created functionally novel TetR mutants, which will not respond to the most commonly used inducers atc and dox anymore, but instead are highly sensitive to Tip.
Figure 6.
Induction of the TetR variants by tetracycline (tc), anhydro-tetracycline (atc) and doxycycline (dox). β-Gal activity was determined in the E. coli screening strain with TetR variants encoded by pWH527-derivatives. β-Gal activities are given in Miller Units (MU) with the maximal expression in the absence of TetR (w/o TetR, black bar). White bars show the expression levels obtained with the TetR variant indicated at the bottom. Light gray bars indicate the β-Gal activities after induction with 0.2 μg/ml tc, gray bars after induction with 0.2 μg/ml atc and dark gray bars after induction with 0.2 μg/ml dox.
DISCUSSION
An additional Met preceding Tip increases its induction efficiency
TrxA(N)-Tip exhibits a much higher activity as inducer of TetR than TrxA(C)-Tip (10). We show here that part but not all of this activity difference is due to the fact that an additional M residue is present when Tip is N-terminal, owing to the fact that removal of the initiator M residue in E. coli is very inefficient before a W residue (26). The crystal structure of the TetR–Tip complex (14) reveals that residues W1 to N4 of Tip are located in the tc-binding pocket, thereby piercing the TetR core. We assume that an N-terminally fused Tip may enter the inducer-binding pocket more easily because the interacting residues can enter in a head first orientation. This is not possible for a C-terminal fusion of Tip, and hence, may contribute to the lower induction efficiency.
Random mutation of position −1 in TrxA(C)-XTip yields more efficient inducers of TetR
The M residue in position −1 of Tip is not the only, nor the optimal residue for increased induction of TetR. Indeed, randomizing the residue at this position in TrxA(C)-XTip revealed that several other amino acids also lead to an increase in TetR induction. The better induction seen with small residues like G, A or S could simply be interpreted as an extension of the linker sequence separating Tip from the TrxA scaffold, thereby increasing the flexibility of Tip. This simple explanation does not hold true for the increased induction seen when aromatic residues occupy position −1, as TrxA(C)-FTip is as active as TrxA(C)-MTip, and TrxA(C)-WTip and TrxA(C)-HTip are even slightly more active. Furthermore, TrxA(C)-DTip, TrxA(C)-ETip, TrxA(C)-KTip or TrxA(C)-RTip carrying charged residues exhibit no induction of TetR, while the uncharged, but similarly sized residues TrxA(C)-NTip and TrxA(C)-QTip are very good inducers of TetR. According to the crystal structure of the Tip–TetR complex, the N-terminus of Tip is located in a predominantly hydrophobic environment formed by residues L60, F67, F86, L90 and L142 of the tc-binding pocket. Placing a charged residue in the hydrophobic core of a protein should be very unfavourable (31,32). Hence, we conclude that the residue at position −1 must exert its function via contacts with the inducer-binding pocket of TetR.
There is a clear distinction among the hydrophobic amino acids between aromatic and aliphatic residues. C-terminal TrxA(C)-XTip variants with V, L or I show a decrease in activity compared to M, while variants with H, F, Y or W induce TetR more efficiently. Comparison of the contacts in the TetR inducer-binding pocket to tc and the first five residues of Tip (Figure 7) shows that most of the key interactions for tc (H64, N82, F86, H100, P105) are not formed with Tip (14). In particular, the space occupied by the A-ring of tc appears to be empty in the Tip–TetR complex. It is thus conceivable that the aromatic residues mimic the A-ring structure of tc best, followed by the flexible M residue with the easily polarizable sulphur, while the large hydrophobic residues fit least well.
Figure 7.
Schematic illustration of contacts made by tc and the first five residues of Tip to residues located in the TetR inducer-binding pocket. Tc (left) and the first five residues of Tip (right, one-letter abbreviations in italics) are shown as gray stick models inside the TetR inducer-binding pocket. The hydrophobic contact region of the TetR-binding pocket is shown in light gray and the two hydrophilic contact regions in gray. W1 of Tip is acetylated and the continuation of the Tip backbone is indicated as a dotted line. Mg2+ is shown as a gray ball and the H2O molecules involved in Mg2+-coordination as black balls. Residues of TetR indicated with an apostrophe belong to the second TetR monomer in the dimer. Relevant hydrophilic interactions are indicated by dashed lines and the side chains of respective TetR amino acids are shown schematically. Residues of TetR shown solely as bold letters are positioned to make hydrophobic interactions with the inducer. Residues of TetR in normal lettering make no contact to the inducer. The PDB-files 2TRT (30) for the TetR–tc complex and 2NS8 (14) for the TetR–Tip complex were used for this presentation.
TetR mutants with altered binding pocket are more efficiently induced by TrxA-XTip
Figures 2 and 7 display the proximity of the N-terminus of Tip to the residues N82 and F86 in the inducer-binding pocket of TetR. The replacement of these bulky residues by the much smaller A in single and double exchange mutants of TetR results in a dramatic increase of the induction activities of several TrxA(C)-XTip variants. These gain of function mutants reinforce the hypothesis that the residue at position −1 in TrxA(C)-XTip is located in the inducer-binding pocket facing the altered residues of the TetR variants. This conclusion is highlighted by the observation that TrxA(C)-XTip variants show different activity profiles for induction of the TetR-N82A, -F86A and -N82A-F86A mutants. We assume that this reflects the slightly different interactions of the various residues with the mutated tc-binding pockets.
The increase of in vivo induction efficiency of the TrxA(C)-XTip TetR variant pairs is indeed dramatic because nearly complete induction is obtained without having to relieve the LacI-mediated repression of TrxA(C)-XTip expression. Previous studies of the expression levels of TrxA(C)-Tip have revealed that the fusion protein is barely detectable in Western blots in the absence of IPTG (10). Since we also demonstrate that the induction efficiencies are not confined to TrxA fusions, we have created much more sensitive tools for monitoring the expression levels of proteins in general as compared to the one used previously (15). Furthermore, two of the TetR mutants are not inducible by tc, atc or dox anymore, allowing their use in conjunction with tc inducible expression systems (11). Taken together, the data presented here suggest that proteins carrying a modified Tip as a C-terminal protein tag in combination with a reporter system under control of a TetR mutant allows highly sensitive detection of their expression levels. Inserting such a tag at the C-terminus of a protein is of great advantage over the N-terminus as one can expect less interference with translation initiation (33).
ACKNOWLEDGEMENTS
We want to thank Janko Daam for helpful discussions and Dr Yves A. Muller for providing us with the coordinates of the TetR–Tip complex prior to their publication. This work was supported by the Deutsche Forschungsgemeinschaft through SFB 473, the Volkswagen-Stiftung through grant AZ I/81 614 and the Fonds der Chemischen Industrie. Funding to pay the Open Access publication charges for this article was provided by the Deutsche Forschungsgemeinschaft through SFB 473.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ogawa H, Qiu Y, Ogata CM, Misono KS. Crystal structure of hormone-bound atrial natriuretic peptide receptor extracellular domain: rotation mechanism for transmembrane signal transduction. J. Biol. Chem. 2004;279:28625–28631. doi: 10.1074/jbc.M313222200. [DOI] [PubMed] [Google Scholar]
- 2.Rosenfeld Y, Papo N, Shai Y. Endotoxin (lipopolysaccharide) neutralization by innate immunity host-defense peptides. Peptide properties and plausible modes of action. J. Biol. Chem. 2006;281:1636–1643. doi: 10.1074/jbc.M504327200. [DOI] [PubMed] [Google Scholar]
- 3.Rohde BH, Quadri LE. Functional characterization of a three-component regulatory system involved in quorum sensing-based regulation of peptide antibiotic production in Carnobacterium maltaromaticum. BMC Microbiol. 2006;6:93. doi: 10.1186/1471-2180-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mourez M, Kane RS, Mogridge J, Metallo S, Deschatelets P, Sellman BR, Whitesides GM, Collier RJ. Designing a polyvalent inhibitor of anthrax toxin. Nat. Biotechnol. 2001;19:958–961. doi: 10.1038/nbt1001-958. [DOI] [PubMed] [Google Scholar]
- 5.Desjobert C, de Soultrait VR, Faure A, Parissi V, Litvak S, Tarrago-Litvak L, Fournier M. Identification by phage display selection of a short peptide able to inhibit only the strand transfer reaction catalyzed by human immunodeficiency virus type 1 integrase. Biochemistry. 2004;43:13097–13105. doi: 10.1021/bi049385e. [DOI] [PubMed] [Google Scholar]
- 6.Ho KL, Yusoff K, Seow HF, Tan WS. Selection of high affinity ligands to hepatitis B core antigen from a phage-displayed cyclic peptide library. J. Med. Virol. 2003;69:27–32. doi: 10.1002/jmv.10266. [DOI] [PubMed] [Google Scholar]
- 7.Sticht J, Humbert M, Findlow S, Bodem J, Müller B, Dietrich U, Werner J, Kräusslich HG. A peptide inhibitor of HIV-1 assembly in vitro. Nat. Struct. Mol. Biol. 2005;12:671–677. doi: 10.1038/nsmb964. [DOI] [PubMed] [Google Scholar]
- 8.Wiesehan K, Buder K, Linke RP, Patt S, Stoldt M, Unger E, Schmitt B, Bucci E, Willbold D. Selection of D-amino-acid peptides that bind to Alzheimer's disease amyloid peptide Aβ1-42 by mirror image phage display. Chembiochem. 2003;4:748–753. doi: 10.1002/cbic.200300631. [DOI] [PubMed] [Google Scholar]
- 9.McGuire MJ, Samli KN, Johnston SA, Brown KC. In vitro selection of a peptide with high selectivity for cardiomyocytes in vivo. J. Mol. Biol. 2004;342:171–182. doi: 10.1016/j.jmb.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 10.Klotzsche M, Berens C, Hillen W. A peptide triggers allostery in Tet repressor by binding to a unique site. J. Biol. Chem. 2005;280:24591–24599. doi: 10.1074/jbc.M501872200. [DOI] [PubMed] [Google Scholar]
- 11.Berens C, Hillen W. Gene regulation by tetracyclines. Genet. Eng. (N.Y.) 2004;26:255–277. doi: 10.1007/978-0-306-48573-2_13. [DOI] [PubMed] [Google Scholar]
- 12.Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 13.Lederer T, Kintrup M, Takahashi M, Sum PE, Ellestad GA, Hillen W. Tetracycline analogs affecting binding to Tn10-encoded Tet repressor trigger the same mechanism of induction. Biochemistry. 1996;35:7439–7446. doi: 10.1021/bi952683e. [DOI] [PubMed] [Google Scholar]
- 14.Luckner SR, Klotzsche M, Berens C, Hillen W, Muller YA. How an agonist peptide mimics the antibiotic tetracycline to induce Tet-repressor. J. Mol. Biol. 2007;368:780–790. doi: 10.1016/j.jmb.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 15.Schlicht M, Berens C, Daam J, Hillen W. Random insertion of a TetR-inducing peptide tag into Escherichia coli proteins allows analysis of protein levels by induction of reporter gene expression. Appl. Environ. Microbiol. 2006;72:5637–5642. doi: 10.1128/AEM.00739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 17.Brent R, Ptashne M. Mechanism of action of the lexA gene product. Proc. Natl Acad. Sci. USA. 1981;78:4204–4208. doi: 10.1073/pnas.78.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith LD, Bertrand KP. Mutations in the Tn10 tet repressor that interfere with induction. Location of the tetracycline-binding domain. J. Mol. Biol. 1988;203:949–959. doi: 10.1016/0022-2836(88)90120-9. [DOI] [PubMed] [Google Scholar]
- 19.Altschmied L, Baumeister R, Pfleiderer K, Hillen W. A threonine to alanine exchange at position 40 of Tet repressor alters the recognition of the sixth base pair of tet operator from GC to AT. EMBO J. 1988;7:4011–4017. doi: 10.1002/j.1460-2075.1988.tb03290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wissmann A, Wray LV, Jr, Somaggio U, Baumeister R, Geissendörfer M, Hillen W. Selection for Tn10 Tet repressor binding to tet operator in Escherichia coli: isolation of temperature-sensitive mutants and combinatorial mutagenesis in the DNA binding motif. Genetics. 1991;128:225–232. doi: 10.1093/genetics/128.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy SB, McMurry LM, Barbosa TM, Burdett V, Courvalin P, Hillen W, Roberts MC, Rood JI, Taylor DE. Nomenclature for new tetracycline resistance determinants. Antimicrob. Agents Chemother. 1999;43:1523–1524. doi: 10.1128/aac.43.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bi W, Stambrook PJ. CCR: a rapid and simple approach for mutation detection. Nucleic Acids Res. 1997;25:2949–2951. doi: 10.1093/nar/25.14.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schnappinger D, Schubert P, Berens C, Pfleiderer K, Hillen W. Solvent-exposed residues in the Tet repressor (TetR) four-helix bundle contribute to subunit recognition and dimer stability. J. Biol. Chem. 1999;274:6405–6410. doi: 10.1074/jbc.274.10.6405. [DOI] [PubMed] [Google Scholar]
- 24.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 25.Ben-Bassat A, Bauer K, Chang SY, Myambo K, Boosman A, Chang S. Processing of the initiation methionine from proteins: properties of the Escherichia coli methionine aminopeptidase and its gene structure. J. Bacteriol. 1987;169:751–757. doi: 10.1128/jb.169.2.751-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirel PH, Schmitter MJ, Dessen P, Fayat G, Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl Acad. Sci. USA. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baquero MR, Bouzon M, Varea J, Moreno F. sbmC, a stationary-phase induced SOS Escherichia coli gene, whose product protects cells from the DNA replication inhibitor microcin B17. Mol. Microbiol. 1995;18:301–311. doi: 10.1111/j.1365-2958.1995.mmi_18020301.x. [DOI] [PubMed] [Google Scholar]
- 28.Romanowski MJ, Gibney SA, Burley SK. Crystal structure of the Escherichia coli SbmC protein that protects cells from the DNA replication inhibitor microcin B17. Proteins. 2002;47:403–407. doi: 10.1002/prot.10102. [DOI] [PubMed] [Google Scholar]
- 29.Gupta S, Chatterji D. Bimodal protection of DNA by Mycobacterium smegmatis DNA-binding protein from stationary phase cells. J. Biol. Chem. 2003;278:5235–5241. doi: 10.1074/jbc.M208825200. [DOI] [PubMed] [Google Scholar]
- 30.Hinrichs W, Kisker C, Düvel M, Müller A, Tovar K, Hillen W, Saenger W. Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science. 1994;264:418–420. doi: 10.1126/science.8153629. [DOI] [PubMed] [Google Scholar]
- 31.Ladbury JE, Wynn R, Thomson JA, Sturtevant JM. Substitution of charged residues into the hydrophobic core of Escherichia coli thioredoxin results in a change in heat capacity of the native protein. Biochemistry. 1995;34:2148–2152. doi: 10.1021/bi00007a007. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura H. Roles of electrostatic interaction in proteins. Q. Rev. Biophys. 1996;29:1–90. doi: 10.1017/s0033583500005746. [DOI] [PubMed] [Google Scholar]
- 33.Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]