Abstract
Since the proposal of ‘in-line attack’ conformation as a possibly important intermediate in RNA cleavage, its structure has been captured in various protein and RNA enzymes; these structures strengthen the belief that this conformation plays an essential role in the catalysis of RNA cleavage. As generally discussed, this intermediate structure can be involved in energy barrier reduction in two possible ways, e.g. through either conformational effect or electrostatic effect. In order to quantitatively elucidate the contribution of conformational effect in this type of enzyme catalysis, free energy simulations were performed on the RNA structures both in a splicing endonuclease complex and in the aqueous solution. Our free energy simulation results revealed that the ‘in-line attack’ conformational effect plays a modest role in facilitating the reaction rate enhancement (∼12-fold) compared with the overall 1012-fold rate increase. The close agreement between the present computational estimation and an experimental measurement on the spontaneous RNA cleavage in an in vitro evolved ATP aptamer motives us to realize that the conformation distribution of an enzyme substrate prior to rather than after its binding determines the upper bound of the rate enhancement ability through the conformational strategy.
INTRODUCTION
Regardless of catalysis strategies in various biomolecular processes, many RNA cleavage reactions undergo an intra-molecular phosphoester transfer (1). This mechanism involves a nucleophilic attack by the 2′ oxygen on the adjacent phosphorus center, followed by the formation of a pentacovalent phosphate intermediate and the subsequent departure of the 5′-oxyanion group (Scheme 1B). Both protein enzymes, as typified by Ribonuclease A (RNase A) (2) and RNA enzymes, as represented by hammerhead ribozyme (3–5), can facilitate this RNA cleavage reaction. The mechanism on how these enzymes exert their catalytic powers is of intense interest (1–6).
Scheme 1.
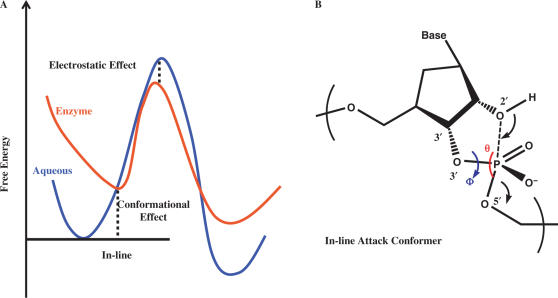
The illustration of the ‘in-line attack’ conformation in RNA cleavage reaction (B) and its conformation effect in enzyme catalysis (A).
RNA cleaving enzymes can bring up to 1012-fold rate enhancement, as compared with the uncatalyzed reaction in the aqueous solution (2). This large rate acceleration can be attributed to various factors such as conformational effect and electrostatic transition state stabilization effect (7). Recently, conformational effect has been invoked to understand catalytic strategies in various enzymes (8–11), such as in chorismate mutase, where this effect alone can contribute up to 103-fold rate enhancement (12). Upon the substrate binding, a tight conformation, which is structurally close to the corresponding transition state, is usually formed within the bound substrate prior to any chemical steps. Specifically, in the RNA intra-molecular phosphoester transfer reactions, such tight conformation is referred as an ‘in-line attack’ conformation (13,14). Here, the attacking atom O2′ is placed opposite to the leaving O5′ atom and aligned in the direction of the broken P–O5′ bond (Scheme 1B). This ‘in-line attack’ conformation has been observed in the structures of a hammerhead ribozyme (6) and two RNA cleaving enzymes bound to their RNA substrates (15,16) (Figure 1B). These structural data are consistent with the previous speculation that ‘in-line attack’ conformation can be important for RNA cleavage (14). In the present work, we wish to quantitatively evaluate how much the conformational effect of this ‘in-line attack’ structure contributes to the rate acceleration in an enzyme-catalyzed RNA cleavage reaction.
Figure 1.
‘In-line attack’ conformation of a pre-cleaved RNA substrate in splicing endonuclease (A) comparison of a regular RNA conformation (upper) and an ‘in-line attack’ conformation (lower) in term of dihedral angle Φ(C3′–O3′–P–O5′); (B) illustration of ‘in-line attack’ conformer of RNA substrate inside enzyme (gray surface) binding pocket.
We achieved this goal by taking the advantage of our recently determined co-crystal structure of a splicing endonuclease and its RNA substrate (15). The RNA splicing endonuclease is responsible for the removal of the intervening sequences in nuclear tRNA and all archaeal RNAs (16). It recognizes a small RNA structure, called the bulge–helix–bulge (BHB) motif. The BHB motif comprises two three-nucleotide-bulges separated by four base pairs. The endonuclease forms a homodimer and cleaves two phosphodiester bonds located after the second bulge nucleotide in these two bulges. In this co-crystal structure, a similar conformation of the RNA substrate is captured in both pre- and post-cleavage states, suggesting that the substrate conformations closely mimic that of the transition state intermediate. To our knowledge, this structure and the structure of sarcin/ricin loop RNA bound to restrictocin (16) [with the sarcin/ricin RNA fortuitously trapped at a minor cleavage site rather than its canonical site (17)] are the only crystal structures that have trapped the ‘in-line attack’ conformation of a pre-cleaved RNA substrate in a protein-catalyzed intra-molecular phosphoester transfer reaction (Figure 1B). In the present work, we employed free energy simulations based on our crystal structure in order to quantitatively dissect the contribution of the ‘in-line attack’ conformational effect to the overall catalysis.
COMPUTATIONAL DETAILS
As discussed in the Results and Discussions section, two free energy simulations were comparatively performed. One is on the complex between a splicing endonuclease and a RNA; the other is on a free RNA with the same sequence.
For the bound state simulation, the crystal structure with PDB code 2GJW was used as the initial input structure (15). During this set-up, the protonation states of the charged residues were determined based on their local environments. Then a stochastic boundary condition (18) was set up with this complex structure overlapped with a 25 Å water sphere, centered at one of the scissile phosphates. During molecular dynamics simulations, the atoms within the sphere of the radius of 22 Å around the same center were treated as the dynamic region; the atoms in this region was propagated with regular Newtonian dynamics using the leapfrog integrator, and 1 fs time step was used. The atoms in the layer between the radii of 22 and 25 Å were treated as the buffer region; the heavy atoms of this biopolymer complex in this region were harmonically restrained with the force constants scaled linearly with their distances from the sphere center, and the force constants around the boundary of this 25 Å sphere were set effectively the same as crystal B factor implies. In the buffer region, Langevin dynamics was applied with the friction coefficients set also linearly scaled with their distances from the sphere center and the friction coefficients around the boundary 25 Å sphere were set as 60. The atoms beyond 25 Å sphere were fixed throughout the simulations and their charges were scaled based on the electrostatic equations. CHARMM 27 forces fields (19) were utilized as the potential in these simulations, and the water molecules were described using the TIP3P model. For the non-bonded interactions, an extended electrostatic treatment was applied with the electrostatic interactions within 12 Å described by the group based coulomb interaction and these interactions beyond 12 Å described by the multipole expansion (20). The simulations were performed with the temperature set as 298.15 K. For the unbound state simulations, the RNA BHB motif also from 2GJW structure was used as the simulation input. Then, a periodic boundary was set with this structure overlapped with a water box sized as 61 × 37 × 37 Å3 and during the dynamic propagations, constant pressure condition was employed with the pressure set as 1 atm. Dependent on the distance from RNA, the effective ionic strength ranges from 1.6 M (RNA contact region) to 0.1 M (bulk), which agrees with the study by Draper (21). The simulations were treated the PME algorithm (20). All the other simulation details are the same as those in the bound state simulations.
During the umbrella sampling simulations (22), the restraint potentials were added with the reference dihedral angle Φ incremented by 1° every 1 ns molecular dynamics simulation, starting with the ‘in-line attack’ conformation captured in the co-crystal structure for both unbound and bound states. The quadratic forms of the restraint potentials were applied and the restraint force constant was set to be 30 kcal/mol/rad2. Upon the completion of the dihedral space scanning, weighted histogram analysis method (WHAM) (23) was utilized to generate the potentials of mean forces along the reaction coordinate Φ. All the calculations were performed using the program CHARMM (24).
RESULTS AND DISCUSSIONS
In order to analyze various effects in the enzymatic catalysis of certain reaction, the free energy profile of the corresponding reaction in the aqueous solution is usually set as the reference (the curve colored in blue in Scheme 1A). As discussed in our introduction, upon binding to the enzyme, the substrate may take a tighter conformation, which is closer to the transition state. If we align the free energy profiles of the enzymatic reaction and the solution reaction by setting the free energies at this tight binding conformation to be the same, as shown in Scheme 1A, the free energy difference between the minimum conformations of the reactants in the enzymatic reaction and in the solution reaction can be the quantitative measure of the conformational effect contributing to the catalysis. This conformational effect plays a role in bringing a conformation far from the transition state, which is the preferred structure in the aqueous solution, to a closer one. Specifically, in the RNA cleavage catalysis by the splicing endonuclease, this conformational effect represents the prepaid free energy penalty required to reach the ‘in-line attack’ conformation due to the binding to the enzyme. The reduction of this energy barrier solely due to the binding of the enzyme is a conformational entropic effect and reflects a population shift from the unbound state (free in the aqueous solution) to the bound state (in the enzymatic environment). Based on the same free energy profile alignment in Scheme 1A, the free energy difference between the transition states in the solution reaction and the enzymatic reaction contributes to the rest of the barrier reduction, which is mostly electrostatic effect (7).
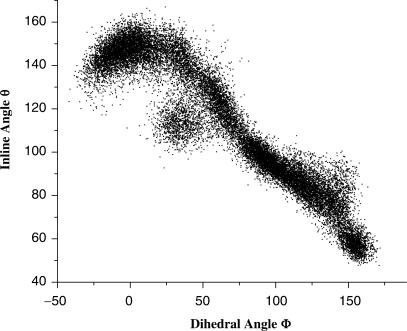
Free energy simulations were carried out on the BHB motif RNA in both the unbound and the bound states. Specifically, umbrella sampling simulations (22) were performed with the dihedral angle (C3′–O3′–P–O5′) of the second bulge nucleotide set as the reaction coordinate Φ (Scheme 1B). Changes in Φ displayed strong correlation with the changes of the in-line geometry defined by angle θ (O2′–P–O5′) (Scheme 1B), as manifested in our simulation results (Figure 2). Other dihedral angles that include (C2′–C3′–O3′–P) and (O2′–C2′–C3′–O3′) did not show any correlation with the in-line geometry angle and were not adopted as the reaction coordinates. Free energy profiles were thus generated as a function of Φ for both the bound and the unbound RNA (Figure 3).
Figure 2.
Correlation of dihedral angle Φ (C3′–O3′–P–O5′) with inline angle θ (O2′–P–O5′) of substrate nucleotide in solution BHB motif RNA. Samples in this plot are from all the molecular dynamics simulations in the present work.
Figure 3.
Difference of the free energy profiles of the substrate nucleotide of BHB motif RNA, in the enzyme binding site and in the aqueous solution, along the reaction coordinate ‘dihedral angle Φ’. (A) Computed potential of mean forces (PMF) of the substrate nucleotide of BHB motif RNA in the enzyme; (B) computed potential of mean forces (PMF) of the substrate nucleotide of BHB motif RNA in solution.
As shown in Figure 3 (red-colored curve), the global minimum of the BHB RNA in the enzymatic environment is located around Φ = −20°, which corresponds to the region of the optimal in-line geometry (θ = ∼155°−160°) (Figure 2) and the dihedral angle observed in the co-crystal structure for the 5′ cleavage site (Φ = −15°) (Figure 1A, lower). In contrast, the global minimum of the BHB RNA in the aqueous solution is shifted to Φ = 45°, which corresponds to a region far from the ‘in-line attack’ conformation (θ = ∼110°−120°) (Figure 3, blue). According to the scheme for estimating the catalytic effect of the ‘in-line attack’ conformation (Scheme 1), these free energy profiles yielded a value of 1.2 kcal/mol [GEnzyme(Φ = −20°) – GSolution(Φ = 45°)] corresponding to an 8-fold reaction rate acceleration. As shown in Figure 3, the free energy uncertainties in the conformation regions determining this value are very small (∼0.04 kcal/mol) compared with the determined catalytic effect 1.2 kcal/mol. It is noted that in the bound state simulations, the free energy profile in the range from −175° to 110° cannot be determined because they have very high free energy values caused by the structural clashes between the RNA substrate with the protein environments. Since this region is a high free energy one, it does not contribute to the catalysis.
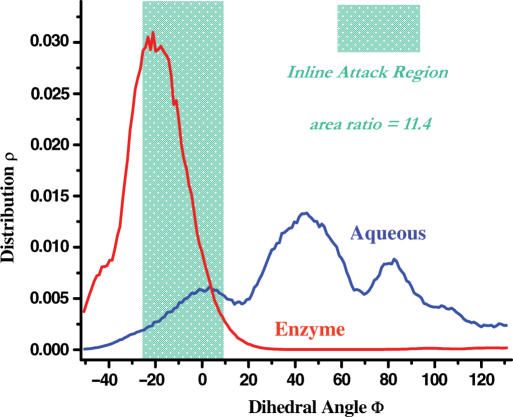
Clearly, the ‘in-line attack’ conformation corresponds to a range of the dihedral angle Φ (−25–10°) (or θ > 155°) (shaded green areas in Figures 3 and 4). A more accurate estimate of the catalytic effect of the ‘in-line attack’ conformation requires taking all the ‘in-line attack’ geometries into consideration. Based on the calculated free energy profiles (Figure 3), the normalized population (normalized in the range of 360°; for the bound structure, the region from −175° to 110° contributes nearly zero weight due to its high free energy values) curves can be obtained (Figure 4). Then, we can get the ratio of the normalized populations in the ‘in-line attack’ conformation regions for the unbound and the bound states; here, the normalized populations are equivalent to the relative populations between the ‘in-line attack’ conformation regions and the overall conformation spaces. This ratio should give us the quantitative measure of the ‘in-line attack’ conformational effect, which led to an 11.4-fold reaction rate acceleration. The rate acceleration remained at similar values when the average in-line angle was shifted to 145° (12.2-fold) or 160° (10.8-fold), suggesting that this measure is robustly insensitive to the precise definition of the in-line geometry.
Figure 4.
Free energy profile-converted conformation distributions of the substrate nucleotide of BHB motif RNA in solution (blue) and enzyme binding site (red), taking the ‘dihedral angle Φ’ as the reaction coordinate.
Interestingly, our calculated value of the reaction rate acceleration for the splicing endonuclease based on free energy simulations quantitatively agrees with that previously obtained on the spontaneous RNA transesterification solely due to the ‘in-line attack’ conformational effect. Soukup and Breaker (25) determined the rate acceleration for cleaving a near perfect in-line nucleotide to be 12-fold by comparing the spontaneous cleavage rates of an in vitro evolved ATP aptamer in the presence and absence of ATP. The agreement between our computed rate acceleration in an enzyme-catalyzed reaction and that of the spontaneous RNA transestification of an unrelated RNA underlines a generally moderate contribution of the ‘in-line attack’ conformational effect on the RNA cleavage. Based on the work on ATP aptamer (25), the Breaker group (26) speculated that the ‘in-line attack’ conformational effect (referred as α catalysis) in the enzymes is very unlikely to exceed 100-fold. Our result on splicing endonuclease strongly supports this speculation.
The ∼12-fold increase from the ‘in-line attack’ conformational effect is insignificant compared to the overall ∼1012-fold increase of the reaction rate. As shown in Figure 4, the apparently modest contribution from the ‘in-line attack’ conformational effect may be understood from the relatively shallow energy profile and the broad conformation distribution around the ‘in-line’ geometry of the unbound state. Consequently, even though the ‘in-line’ conformation of the substrate has a narrow distribution in its bound state, achieving this conformation through the enzyme binding can only result in a relatively minor reduction in the free energy barrier. Quantitatively, an upper limit on the conformational effect in catalysis can be estimated using 1/ρtight, where ρtight is the probability of the tight binding conformation distribution in solution. For instance, chorismate mutase has a relatively low population of the tight conformation in solution, and correspondingly the conformational effect in chorimate mutase can provide nearly 2 × 103-fold rate enhancement (12). Hence, only when the tight conformation distribution of the substrate has very small population of the tight conformation in the unbound environment, it is possible for the enzyme to lower the free energy barrier substantially through the conformational control strategy. We can conclude that the conformation distribution of the substrate prior to, rather than after the enzyme binding, determines the upper bound of the rate enhancement through the conformational strategy than previously thought. Specifically, our simulation result appears to contradict to the general belief that the ‘in-line’ conformational effect in RNA is important for the catalysis of RNA cleavage.
The tight conformation populations for the substrate are typically high in the enzymatic environment, because the intrinsic driving force for the transition state stabilization can effectively bias the reactant conformation toward the transition state structure. So for an enzyme with multiple substrates, the conformational effects for the catalysis of the reactions involving these substrates tend to be controlled by their conformational distributions in solution, although their bound conformations are usually similar.
Results from this study suggest that other catalytic effects, such as the electrostatic stabilization of transition state, play more important roles in the RNA cleavage. Efforts in computation are currently being made to advance our understanding of the detailed path of RNA cleavage.
ACKNOWLEDGEMENTS
We would like to thank Council on Research and Creativity of Florida State University and Florida State University Research Foundation (W.Y.) and a National Science Foundation grant 00517300 (H.L.) for financial supports and K. Calvin for critical reading of the manuscript. All the calculations were carried in Anfinsen Cluster of School of Computational Science at Florida State University. Full waiver of the Open Access publication charges for this article.
Conflict of interest statement. None declared.
REFERENCES
- 1.Oivanen M, Kuusela S, Lonnberg H. Kinetics and mechanisms for the cleavage and isomerization of the phosphodiester bonds of RNA by bronsted acids and bases. Chem. Rev. 1998;98:961–990. doi: 10.1021/cr960425x. [DOI] [PubMed] [Google Scholar]
- 2.Raines RT. Ribonuclease A. Chem. Rev. 1983;3:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 3.Kuimelis RG, McLaughlin LW. Mechanisms of ribozyme-mediated RNA cleavage. Chem. Rev. 1998;98:1027–1044. doi: 10.1021/cr960426p. [DOI] [PubMed] [Google Scholar]
- 4.Blount KF, Uhlenbeck OC. The structure-function dilemma of the hammerhead ribozyme. Annu. Rev. Biophys. Biomol. Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- 5.Lilley DMJ. Structure, folding and mechanisms of ribozymes. Curr. Opin. Struc. Biol. 2005;15:313–323. doi: 10.1016/j.sbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Martick M, Scott WG. Cell 126. 2006. Tertiary contacts distant from the active site prime a ribozyme for catalysis; pp. 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warshel A, Sharma PK, Kato M, Xiang Y, Liu H, Olsson MHM. Electrostatic basis for enzyme catalysis. Chem. Rev. 2006;8:3210–3235. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- 8.Bruice TC. Computational approaches: reaction trajectories, structures, and atomic motions. Enzyme reactions and proficiency. Chem. Rev. 2006;8:3119–3139. doi: 10.1021/cr050283j. [DOI] [PubMed] [Google Scholar]
- 9.Gao JL, Byun KL, Kluger P. Catalysis by enzyme conformational change. Top. Curr. Chem. 2004;238:113–136. [Google Scholar]
- 10.Mackerell AO, Bashford D, Bellot M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. An all-atom empirical energy function for the simulation of nucleic acids. J. Am. Chem. Soc. 1995;117:11946–11975. [Google Scholar]
- 11.Guo H, Cui Q, Lipscomb WN, Karplus M. Understanding the role of active-site residues in chorismate mutase catalysis from molecular-dynamics simulations. Proc. Natl Acad. Sci. USA. 2001;98:9032–9037. [Google Scholar]
- 12.Ranaghan KE, Mulholland AJ. Conformational effects in enzyme catalysis: QM/MM free energy calculation of the “NAC” contribution in chorismate mutase. Chem. Comm. 2004;10:1238–1239. doi: 10.1039/b402388a. [DOI] [PubMed] [Google Scholar]
- 13.Lightstone FC, Bruice TC. Ground state conformations and entropic and enthalpic factor in the efficiency of intramolecular and enzymatic reactions: 1 cyclic anhydride formation by substituted glutarates, succinate, and 3,6-endoxo-δ(4)-tetrahydrophthalate monophenyl esters. J. Am. Chem. Soc. 1996;118:2595–2605. [Google Scholar]
- 14.Vantol H, Buzayan JM, Feldstein PA, Eckstein F, Bruening G. Two autolytic processing reactions of a satellite RNA proceed with inversion of configuration. Nucleic Acids Res. 1990;18:1971–1975. doi: 10.1093/nar/18.8.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song X, Calvin K, Li H. RNA recognition and cleavage by a splicing endonuclease. Science. 2006;312:906–910. doi: 10.1126/science.1126629. [DOI] [PubMed] [Google Scholar]
- 16.Abelson J, Trotta CR, Li H. T-RNA splicing. J. Biol. Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 17.Yang XJ, Gerczei T, Glover LT, Correll CC. Crystal structure of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping. Nat. Struct. Biol. 2001;8:968–973. doi: 10.1038/nsb1101-968. [DOI] [PubMed] [Google Scholar]
- 18.Brooks CL, III, Karplus M. Deformable stochastic boundaries in molecular dynamics. J. Chem. Phys. 1983;79:6312–6325. [Google Scholar]
- 19.Mackerell AD, Jr, Bashford D, Bellot M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D. An all-atom empirical energy function for the simulation of nucleic acids. J. Am. Chem. Soc. 1995;117:11946–11975. [Google Scholar]
- 20.Darden T, York D, Pedersen L. The effect of long-range electrostatic interactions in simulations of macromolecular crystals – a comparison of the ewald and truncated list methods. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 21.Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torrie GM, Valleau JP. Non-physical sampling distributions in Monte-Carlo free-energy estimation – umbrella sampling. J. Comp. Phys. 1977;23:187–199. [Google Scholar]
- 23.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram method for free-energy calculations on biomolecules. 1. The method. J. Comp. Chem. 1992;13:1011–1021. [Google Scholar]
- 24.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 1983;4:187–217. [Google Scholar]
- 25.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emilsson GM, Nakamura S, Roth A, Breaker RR. Ribozyme speed limits. RNA. 2003;9:907–918. doi: 10.1261/rna.5680603. [DOI] [PMC free article] [PubMed] [Google Scholar]