Abstract
We present a bacterial genome computational analysis pipeline, called GenVar. The pipeline, based on the program GeneWise, is designed to analyze an annotated genome and automatically identify missed gene calls and sequence variants such as genes with disrupted reading frames (split genes) and those with insertions and deletions (indels). For a given genome to be analyzed, GenVar relies on a database containing closely related genomes (such as other species or strains) as well as a few additional reference genomes. GenVar also helps identify gene disruptions probably caused by sequencing errors. We exemplify GenVar's capabilities by presenting results from the analysis of four Brucella genomes. Brucella is an important human pathogen and zoonotic agent. The analysis revealed hundreds of missed gene calls, new split genes and indels, several of which are species specific and hence provide valuable clues to the understanding of the genome basis of Brucella pathogenicity and host specificity.
INTRODUCTION
We describe ‘GenVar’, a comparative genomics analysis computational pipeline, whose aim is to improve existing bacterial genome annotations as well as reveal indel polymorphisms in already-annotated protein-coding genes. GenVar is based on GeneWise, a program to analyze DNA and protein sequences that helps eukaryotic gene structure analysis (1). Even though GeneWise is aimed primarily at eukaryotic genomes, we have found it useful and effective as a platform upon which to develop GenVar.
Manual annotation is the currently agreed upon ‘gold standard’ to provide quality genome annotation (2). This gold standard, however, does not scale or keep up with the increasing pace of microbial genome sequencing. Among the main challenges in the manual annotation process are accurately identifying missed gene calls and split genes in existing genome annotations. These are the two main features addressed by GenVar. There are two possible causes of split genes: sequencing errors or mutations. Both can cause ORF truncations or over-extensions, thus creating annotation errors if left uncorrected. Comparative analysis of closely related genomes can provide important clues to help distinguish these two cases, and GenVar also provides such clues when possible.
A brief description of the GeneWise program (1) is necessary for a better understanding of GenVar. GeneWise combines hidden Markov models for gene prediction and for alignment, thereby making it possible to compare a single protein sequence directly to genomic DNA. The models take into account known statistical properties of genes as well as the possible presence of ‘sequencing errors’ or problems in translation. GeneWise will take genomic sequence and compare it to target protein sequences (assumed to be homologous) considering all possible ‘intermediate’ predicted sequences given by the gene prediction part of the combined model.
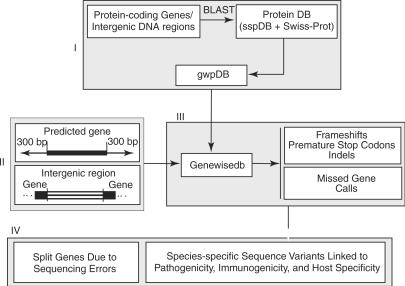
The GenVar pipeline is composed of three conceptual steps. The first two steps generate GeneWise-required inputs: a set of protein database inputs (gwpDBs) and a set of genomic DNA inputs. Each gwpDB contains orthologous proteins from a limited number of closely related species. The genomic DNA inputs are selected to represent extended regions of both predicted genes and putative intergenic regions. By breaking up the genome to be studied and the reference protein sequences into small units, we decrease the computational cost that would be incurred if GeneWise were to be used starting from the entire genome and comparing it to a general set of protein sequences. The third step comparatively analyzes missed gene calls and sequence variants among closely related species. Sequence variants are defined as genes with frameshifts, premature stop codons, insertions and deletions. Missed gene calls are DNA regions described as intergenic in the original genome annotation that can be fully aligned with gene calls in closely related or otherwise well-annotated genomes. Once the sequence variants are identified, the variants within the context of closely related genomes are classified using a simple classification scheme. This scheme facilitates the correlation of sequence variants with phenotypic differences in the species studied; it also identifies a list of frameshifts and premature stops that may be sequencing errors.
We are especially interested in pathogenic bacteria. Our underlying assumption is that gene disruptions (true split genes) and indel polymorphisms play a key role in host–pathogen evolution. The literature provides several cases of this connection. For example, split genes of the major surface protein 2 (MSP2) determine antigenic variation in the tick-transmitted pathogen Anaplasma marginale (3). An array of variable proteins is the source of diversity in host tropism and disease causation in the obligate intracellular bacterial pathogen Chlamydophila abortus (4). Gene inactivation, loss and acquisition are hypothesized to be the main mechanisms that contribute to Yersinia pestis fitness and promote its adaptive microevolution (5).
For the development and testing of GenVar, we chose the bacterial pathogen Brucella. Our motivation was as follows: three out of the six recognized taxonomic Brucella species have been sequenced, including one B. melitensis, one B. suis and two B. abortus strains, which represent the most virulent Brucella species to humans (6–9). In addition, Brucella is one of the world's major zoonotic pathogens for which there is no human vaccine (10). Although highly similar in terms of gene content (11), the six Brucella species have preferential host specificity: goats (B. melitensis), cattle (B. abortus), swine (B. suis), dogs (B. canis), sheep (B. ovis) and desert mice (B. neotomae) (12). Thus the choice of Brucella offers a unique opportunity to assay and improve the quality of current genome annotations and to identify unique genetic factors that may help explain the pathogen's niche as a facultative intracellular pathogen (13). Finally, Brucella is a priority pathogen of the National Institute of Allergic and Infectious Diseases (http://www3.niaid.nih.gov/Biodefense/PDF/cat.htm). We participate in the development of a Web resource (http://patric.vbi.vt.edu) devoted in part to Brucella (14), and GenVar is being used in that project.
We have analyzed four genomes: B. melitensis 16M (7), B. suis 1330 (9), B. abortus 9-941 (8) and B. abortus 2308 (6). Each of these genomes has two chromosomes (2.1 and 1.2 Mbp approximately). Our results indicate that GenVar was able to improve the existing annotations of these genomes. The analysis revealed hundreds of Brucella missed gene calls and dozens of new, probable bona fide split genes. Please note, however, that the results presented are meant to demonstrate the versatility of this new tool, rather than being an exhaustive list of every possible missed gene call, split gene or polymorphic indel in the Brucella genomes studied. Obtaining such results would require a much larger input database than the one we used.
MATERIALS AND METHODS
Genome sequence data
Four Brucella genomes including B. melitensis 16M, B. suis 1330, B. abortus 9-941 and B. abortus 2308 were downloaded on 4 December 2005 from the National Center for Biotechnology Information (NCBI) (ftp://ftp.ncbi.nih.gov/genomes/Bacteria). The published genome assembly fold coverage for these genomes are as follows: B. melitensis 16M: 9X (7); B. abortus 9-941: 10X (8); and B. abortus 2308: 7X (6). B. suis 1330 does not have a published fold coverage, but we assume that it is 7X or 8X based on other genomes published by The Institute for Genomic Research. The genomes of Agrobacterium tumefaciens C58 (Cereon) and Mesorhizobium loti MAFF303099 (both are alphaproteobacteria, like Brucella) and Escherichia coli K12 (generally regarded as the best annotated bacterial genome) were also used by GenVar and downloaded from the same source. Also used was the Swiss-Prot protein database (UniProt Knowledgebase Release 7.3), downloaded from http://www.pir.uniprot.org. Gene functional assignments reported are those obtained from the mentioned sources.
GenVar steps
GenVar is based on the GeneWise program (1,2). It is partitioned into three conceptual steps, as described in the Introduction section and detailed below. The GeneWise program was downloaded from http://www.sanger.ac.uk/Software/Wise2. A GenVar run is entirely automated. A run of GenVar on the 1.2 Mbp Brucella chromosome takes ∼6 h on a 500 MB RAM Pentium 4 computer running Linux. Running time increases linearly with sequence length.
Input protein databases
The first step is designed to establish, for each query genome feature (QGF), a gene-specific protein database (gwpDB), the first input for GeneWise (Figure 1, panel I). A QGF is either a protein-coding gene from the existing genome annotation or a DNA region between two immediately adjacent protein-coding genes on the chromosomes (intergenic DNA regions). The gwpDB is constructed from BLAST (15) analysis of the QGF on a species-specific protein database. The protein database consists of proteins from closely related genomes and also those that are well annotated (see above). Consequently, the gwpDB of the QGF would include a small number of proteins yet cover all its paralog and orthologs from closely related genomes as well as from well-annotated protein sequences.
Figure 1.
Data flow in GenVar showing its three constitutive conceptual steps. sspDB: species-specific database; gwpDB: gene-specific database.
Input DNA sequences
The second step is devised to generate meaningful searchable DNA regions (SDR), the second data input for the GeneWise program (Figure 1, panel II). By ‘meaningful’ we mean that an SDR needs to cover sequence variants involved in the protein-coding genes and missed gene calls so that they can be detected. For this purpose, the SDR is defined as a DNA region of a predicted protein-coding gene with 300 bp extensions both upstream and downstream (needed to identify split genes and indels) or an intergenic DNA region between two adjacent protein-encoding genes (needed to identify missed gene calls, besides sequence variants).
Identifying sequence variants and missed gene calls
The next step actually runs GeneWise and parses GeneWise outputs to identify the sequence variants and missed gene calls (Figure 1, panel III). In this study, a protein-coding gene or an intergenic DNA region is considered to contain a sequence variant if and only if such variant is detected when compared to its orthologs. The orthologs are determined by the best BLAST hits from each closely related genome. Furthermore, frameshifts and premature stop codons detected in the sequence variants are further mapped at specific chromosomal locations; indels are mapped on the proteins coded by the orthologs.
Classification scheme for comparative genomics studies
GenVar's output is then interpreted and linked to different organismal properties such as host specificity, host–pathogen interactions and other pathogenicity-related traits found in Brucella species (Figure 1, panel IV). Classification schemes were designed to determine whether the identified split genes resulted from sequencing errors or genuine mutation and to discover species-specific/selective gene disruption and their possible patterns. Such patterns can suggest the specific association between occurrences of sequence variants and pathogenicity properties. For genes with indels, the classification scheme is simple, relying on the genome association. For split genes, the classification scheme is more complex, depending on the length of the alignments of split genes with their orthologs, the status of assigned biological functions of the orthologs and genome associations. Different genomes will have different associations. For example, the split genes from B. abortus 2308 were classified into six groups (Table 1). The occurrences of these sequence variants among closely related genomes in particular protein complexes were then compared to reveal possible patterns in which genes are selectively disrupted or modified.
Table 1.
Classification of split genes detected in B. abortus 2308
| Group | Alignment length (AA) | Biological function of orthologs | Species association |
|---|---|---|---|
| G1 | <=100 | Any | Any |
| >100 | Unknown | ||
| G2 | >100 | Assigned | Detected in two Brucella species: both B. melitensis and B. abortus or both B. suis and B. abortus but not others |
| G3 | >100 | Assigned | Detected in both B. abortus genomes but not others |
| G4 | >100 | Assigned | Detected in B. abortus 2308 only |
| G5 | >100 | Assigned | Detected in all four Brucella genomes but not in genes from non Brucella genomes |
| G6 | >100 | Assigned | Detected in any other combination of Brucella genomes |
Program and data availability
GenVar is publicly available to noncommercial users at http://patric.vbi.vt.edu/downloads/software/GenVar. GenVar results are being used to reannotate all four Brucella genomes by the PATRIC project (14); some of these reannotations already are publicly available through the PATRIC website (http://patric.vbi.vt.edu); eventually all results will be incorporated into the reannotations and deposited in GenBank.
DNA resequencing
For each split gene predicted in the B. abortus S19 genome, we obtained by PCR 60 bp around the predicted disruption, resequenced this fragment, and compared to the original sequence.
RESULTS AND DISCUSSION
Results presented here are a sample of our total results, chosen primarily based on their likely roles in host specificity and other Brucella pathogenicity-related properties.
Missed gene calls
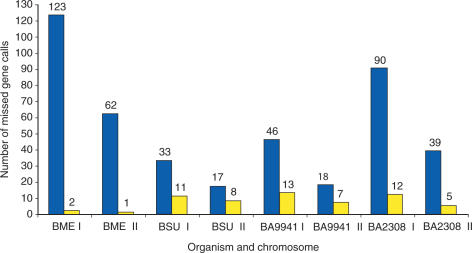
Many missed gene calls were detected and their numbers vary from genome to genome (Figure 2). For example, B. melitensis 16M has about 185 missed gene calls whereas B. suis 1330 has 50. About 77% of all missed gene calls have lengths that are less than or equal to 100 amino acids (AA). This result is consistent with previous findings that differences in gene number among completely sequenced Brucella genomes are mainly caused by annotation discrepancies in the number of small genes (9). GenVar did find several missed genes longer than 100 AA, some with orthologs having assigned biological functions (Figure 2).
Figure 2.
Missed gene calls revealed in the intergenic DNA regions from the four Brucella genomes. The bars show the total number of missed gene calls (blue) and the number of missed gene calls that are larger than 100 AA and have orthologs with assigned biological functions (yellow). BME stands for B. melitensis 16M; BSU for B. suis 1330; BA9941 for B. abortus 9-941; and BA2308 for B. abortus 2308. The letters I and II stand for chromosomes I and II, respectively.
Missed genes that have orthologs with functional assignments in other organisms encode a broad category of biological functions; some of the functions are critically important for cellular processes in Brucella. For example, missed gene calls in B. suis 1330 include an ABC transporter ATP-binding protein, a dihydroxyacetone kinase, a lipid A-myristate beta-hydroxylase, an outer membrane protein, a transcriptional regulator, a thioredoxin reductase and an RNA pseudouridylate synthase family protein. Missed gene calls in B. abortus 9-941 include a flagellar motor switch protein FliG, a bacterial regulatory protein, MarR family, a transcriptional regulatory protein, LysR family, a bicyclomycin resistance protein, a phage minor tail protein and many transposases. Missed gene calls in B. abortus 2308 further include an outer membrane protein, a thioredoxin reductase and many iron-related proteins such as a zinc protease, a calcium- or iron-binding protein, and a cobalt–zinc–cadmium resistance protein, Czc. Some of the missed gene calls are specific to just one species. For example, the gene coding for the flagellar motor switch protein FliG is missing only in B. abortus 9-941; the iron-related proteins are missing only in B. abortus 2308. In contrast, others are missing in more than one Brucella genome. The gene for outer membrane protein E is absent from the annotations in both B. suis 1330 and in B. abortus 2308. The gene coding for lipid A-myristate beta-hydroxylase is absent from the annotations in B. suis 1330, B. abortus 2308 and B. abortus 9-941.
Split genes detected in intergenic DNA regions
Table 2 presents the numbers of split genes detected in the four Brucella genomes. Supplementary Table 1 presents a sample of the split genes discovered in B. abortus 2308. We have observed that some of the sequence variants are consistently detected while others vary depending on the protein sequences to which the intergenic sequence is compared. For example, intergenic region 336 on chromosome I from B. abortus 2308, harboring a gene for a transcriptional regulatory protein in the LysR family, has a single frameshift when compared with all its orthologs (hence the frameshift is a candidate for being a sequencing error, see below). On the other hand, intergenic region 1244 on the same chromosome, covering a gene for urease accessory protein UreE, has no frameshift when compared to its ortholog from B. abortus 9-941, but two frameshifts when compared to its orthologs from B. suis 1330 and B. melitensis 16M (Figure 3), and one frameshift when compared to Yersinia pestis (note: there is a 17-residue insertion in the Yersinia ureE gene right before what would have been the second frameshift).
Table 2.
The number of split genes detected in the four Brucella genomes
| Organism | Premature stop codon | Frameshift | Both | Total |
|---|---|---|---|---|
| Intergenic regions | ||||
| B. melitensis 16M | 13 | 44 | 2 | 59 |
| B. suis 1330 | 28 | 86 | 6 | 120 |
| B. abortus 9-941 | 44 | 124 | 7 | 175 |
| B. abortus 2308 | 33 | 122 | 10 | 165 |
| Protein-coding regions | ||||
| B. melitensis 16M | 81 | 247 | 11 | 339 |
| B. suis 1330 | 19 | 91 | 1 | 111 |
| B. abortus 9-941 | 11 | 92 | 0 | 103 |
| B. abortus 2308 | 10 | 41 | 1 | 52 |
Figure 3.
Pairwise alignments of the urease accessory gene ureE-2 in B. abortus 9-941 (panel I, gi|62290255) and in B. melitensis 16M (panel II, gi|17986929) against intergenic region 1244 from B. abortus 2308. Panel I shows that intergenic region 1244 contains a missed gene call for ureE-2 in B. abortus 2308. Furthermore, both B. abortus strains present two frameshifts (FS1 and FS2) when compared to ureE-2 in B. melitensis 16M (panel II). These frameshifts cause the absence of the multi-histidine Ni2+ chelating center at the C-terminal.
Sequence variants in predicted protein-coding regions
GenVar is also able to discover split genes in already annotated protein-coding gene sequences. These can occur because the original annotators failed to see an extension of the protein-coding region downstream of the assigned stop (which therefore is a premature stop), or an extension upstream of the assigned start due to a frameshift, or both. Table 2 presents the numbers of such split genes discovered. It is noteworthy that B. melitensis 16M has a total of 339 of these split genes, three times those found in B. suis 1330 and six times more than those in B. abortus 2308. This observation is in contrast to the fact that B. melitensis 16M has the smallest number of split genes detected in intergenic DNA regions. We believe this difference is primarily due to sequencing errors in the genome of B. melitensis 16M; see below. Supplementary Table 2 contains examples of split genes in already annotated protein-coding regions in B. melitensis 16M.
GenVar analysis of protein-coding regions also revealed polymorphic indels. A total of 142 such genes were discovered in B. melitensis 16M and a comparable number in the other three Brucella genomes. Supplementary Table 3 presents a sample of these results for B. melitensis 16M.
Table 3.
Example of split gene groups in chromosome I of B. abortus 2308. The intergenic DNA regions are numbered according to their order on the chromosome; thus, Intergenic_129 is the 129th intergenic DNA region on chromosome I
| Interval name | Split gene group | Frameshift position | Premature stop position | Ortholog | Gene source | RefSeq annotation |
|---|---|---|---|---|---|---|
| Intergenic_129 | G1 | 163662 | – | 23500355 | BS1330 (II) | Hypothetical protein |
| 163662 | – | 83269513 | BA2308 (II) | Conserved hypothetical protein | ||
| – | 163653 | 23502245 | BS1330 (I) | Hypothetical protein | ||
| – | 163653 | 82700191 | BA2308 (I) | Hypothetical protein | ||
| Intergenic_560 | G2 | 608947 | 607381 | 17987624 | BM16M (I) | Phage host specificity protein |
| – | – | 17987623 | BM16M (I) | Phage host specificity protein | ||
| Intergenic_516 | G2 | 564045 | – | 23501439 | BS1330 (I) | Transcriptional regulator, AraC family |
| – | – | 17987666 | BM16M (I) | Transcriptional regulator, AraC family | ||
| – | – | 17987667 | BM16M (I) | Transcriptional regulator, AraC family | ||
| Intergenic_973 | G3 | – | – | Missed gene | BA9-941 (I) | Unannotated |
| – | 1044923 | 23501937 | BS1330 (I) | Drug resistance transporter, EmrB/QacA family | ||
| – | 1044923 | 17987210 | BM16M (I) | Drug resistance transporter, EmrB/QacA family | ||
| Intergenic_1236 | G3 | – | – | Missed gene | BA9-941 (I) | Unannotated |
| 1328845 | – | 23502222 | BS1330 (I) | ABC transporter, ATP binding/ permease protein | ||
| 1327018 | – | 17986937 | BM16M (I) | ABC transporter ATP-binding protein | ||
| 1328815 | – | YHIH_ECOLI | Swiss-Prot | Hypothetical ABC transporter ATP-binding protein yhiH | ||
| Intergenic_1244 | G3 | – | – | 62290255 | BA9-941 (I) | Urease accessory protein UreE |
| 1335216 and 1335291 | – | 17986929 | BM16M (I) | Urease accessory protein UreE | ||
| 1335216 and 1335291 | – | 23502231 | BS1330 (I) | Urease accessory protein UreE | ||
| Intergenic_336 | G4 | 381948 | – | 17987856 | BM16M (I) | Transcriptional regulator, LysR family |
| 381948 | – | 62289344 | BA9-941 (I) | Transcriptional regulator, LysR family | ||
| 381948 | – | 23501257 | BS1330 (I) | Transcriptional regulator, LysR family |
‘Orthologs’ are defined as the best BLAST hits in the custom database relied on by GenVar; the identifiers given are GenBank accession numbers or Swiss-Prot identifiers. ‘Gene source’ gives the organism or database where the ortholog was found; in this column the acronyms used are as follows: BM16M stands for B. melitensis 16M; BS1330 for B. suis 1330; BA9-941 for B. abortus 9-941 and BA2308 for B. abortus 2308. (I) stands for chromosome I and (II) for chromosome II. The split gene groups are as described in Table 1.
Classification of split genes
In order to facilitate the interpretation of GenVar results, we have created a classification scheme for split genes. This scheme uses three characteristics: lengths of alignment with orthologs; whether or not orthologs have assigned biological functions; and occurrence within the set of four Brucella genomes studied (more details in the Materials and Methods section). For instance, the split genes in the intergenic regions of chromosome I of B. abortus 2308 were classified into six groups, among which eight occur in two of the three Brucella species (group 2, or G2), 56 in both strains of B. abortus (G3) and 12 in B. abortus 2308 only (G4); see Table 3 for group samples. The split genes from groups G2 to G6 are the most interesting because their orthologs have assigned biological functions. The split genes in G3 are especially important in that they are found in both B. abortus genomes but not on the other two Brucellae, therefore being specific to this Brucella species. Such is the case of the ureE gene already mentioned.
A natural question to ask is whether some of the frameshifts and premature stops detected are the results of sequencing errors. Our classification scheme provides clues to help answer this question. Split genes that fall in groups G2, G3, G5 and G6 show disruptions that are shared by at least two genomes. The probability is very low (estimated at less than 0.01; see Supplementary Data) that the same sequencing error would occur on two or more genomes derived from independent sequencing projects. We conclude therefore that those split genes are most likely real. The disruptions observed in split genes in group G4, having been observed on only one genome, are sequencing error candidates. We conducted laboratory experiments (see DNA resequencing in the Materials and Methods section) to verify this hypothesis. These were done on a vaccine strain, Brucella abortus S19 (sequenced to 20X coverage; unpublished data). GenVar detected 138 split genes in this genome, and each putative split gene region was resequenced for confirmation. The verification showed that all split genes from groups where disruptions are shared by more than one genome had correct sequences (and hence the disruptions are very likely due to natural mutations), while all those that are S19 specific were due to sequencing errors (Table 4). In B. melitensis 16M 179 split genes were specific to this genome; based on the results presented in this paragraph, we hypothesize that a large fraction of these split genes are sequencing errors.
Table 4.
Results of experimental verification of GenVar-detected gene disruptions (frameshifts and premature stop codons) in split genes in B. abortus S19
| Genomes in which the same B. abortus S19 disruptions are found | Chromosome | Number of Genes | Sequence verification | |
|---|---|---|---|---|
| True | False | |||
| B. melitensis 16M OR B. suis 1330 OR B. abortus 2308 OR B. abortus 9-941 | I | 9 | √ | |
| II | 3 | √ | ||
| B. abortus 2308 AND B. abortus 9-941 | I | 7 | √ | |
| II | 10 | √ | ||
| No other genomes | I | 57 | √ | |
| II | 52 | √ | ||
Genes with indels
Comparative analysis of indels focused on genes from existing genome annotations. Like split genes, we can classify genes with indels but have to rely only on their occurrence in specific Brucella genomes. Supplementary Table 3 presents result for B. melitensis 16M; here we describe a few examples.
A 4-AA deletion was detected in the gene coding for primosomal protein N when compared to its ortholog from B. abortus 9-941. A 12-AA insertion in the gene coding for a 25 kDa outer-membrane immunogenic protein precursor was detected when compared to the B. ovis gene OM25_BRUOV. It is worth noting that the corresponding regions of omp genes in other Brucella species, which contain two 8-bp direct repeats and two 4-bp inverted repeats, may be related to the ‘slipped mispairing’ mechanism and an antigenic shift (16).
The gene coding for a sensor histidine kinase is especially interesting. The peptide RNVG appears in this gene in multiple and varying copies in the genomes studied: three copies in B. abortus 2308, two copies in B. abortus 9-941 and B. melitensis 16M and one copy in B. suis 1330. Other indels are specific to one Brucella species or shared by multiple Brucella genomes. For instance, a 2-residue deletion in the urease accessory protein UreD-2 (ureD gene in urease operon 2) is associated with both B. abortus genomes. The same is true for indels in the gene coding for a multidrug resistance efflux protein.
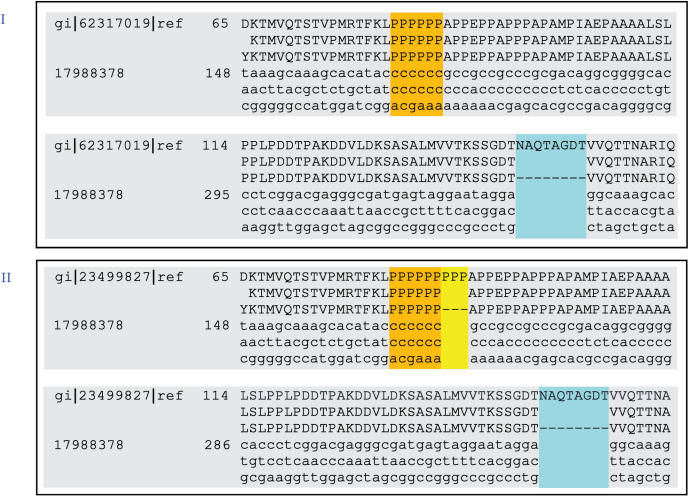
An 8-residue deletion was found in the gene coding for type IV secretion system protein VirB10 when compared to B. abortus 9-941, B. abortus 2308 and B. suis 1330; the same gene in B. suis 1330 has a 3-residue insertion unique to this species, among those compared (Figure 4). VirB10, as an energy-sensing bridge between the inner and outer membranes, is essential for the transfer of substrates from the inner to the outer membrane (17). While almost all type IV genes are highly conserved among the four Brucella genomes, virB10 is the only one that has indel polymorphisms. The first is a three-proline insertion specific to B. suis 1330 (Figure 4), part of a proline-rich region, which is a predicted extended structure in the periplasm (18). The second is an 8-residue insertion specific to both B. abortus and to B. suis 1330. Although the biological significance of such indels has yet to be investigated, their importance could not be over-emphasized considering the nature of this gene and its associated protein complex. The type IV secretion system is used by many Gram-negative bacteria to translocate virulence factors into eukaryotic cells, to mediate conjugative transfer of broad-host-range plasmids, and to facilitate host–pathogen interactions that enable bacterial survival in widely different habitats (19). Thus, experiments based on the results of this analysis may provide experimental data that would help determine the role these variants play in host specificity and other pathogenesis-related functions.
Figure 4.
Pairwise alignments of the Type IV secretion system protein VirB10 gene sequence. Panel I shows the alignment between the B. abortus 9-941 gene (gi|62317019) against that of B. melitensis 16M (gi|17988378), and panel II shows the alignment between the B. suis 1330 gene (gi|23499827) also against that of B. melitensis 16M. The two alignments show that B. melintensis 16M has an 8-residue deletion with respect to its orthologs in B. abortus 9-941 and B. suis 1330 (blue sections in panels I and II). B. suis 1330 has a 3-residue insertion (yellow part of panel II).
Split gene/indel occurrence patterns
Using GenVar, we have also discovered possible patterns of split gene and indel occurrences in the four genomes for some protein complexes. Here we present two examples. It has been reported that all the genes necessary to assemble a functional flagellum except for the chemotactic system are found in Brucella species (13). In our analysis, we found patterns of species-specific gene disruption in the flagellum protein complex, shown in Supplementary Table 5. We have also found that drug resistance genes are selectively disrupted in three Brucella species. Genes encoding multidrug resistance protein B, a Na+-driven multidrug efflux pump, and a drug resistance transporter (EmrB/QacA family) are disrupted in both B. abortus genomes, while a gene encoding a multi-drug resistance efflux protein is disrupted in B. suis 1330 only.
Additional sequence variants in virulence-related genes
Brucella abortus-specific frameshifts were identified for genes coding for an intimin/invasin family protein, an outer membrane autotransporter, and a flagellar motor switch protein. All of them are critically important virulence factors for bacterial pathogenesis (13,20). The intimin/invasin family protein mediates the internalization of the pathogen into cultured epithelial cells and is located in a pathogenicity island in Yersinia enterocolitica (28). The outer membrane autotransporter protein influences the survival of B. suis 1330 in BALB/c mice (20). Lastly, flagella subunit expression is essential for Brucella to successfully infect and replicate in macrophages (13). Gene disruptions in either the fliF (MS ring monomer), flgI (P ring monomer) or fliC (flagellin monomer) genes (Supplementary Table 5) resulted in attenuated B. melitensis 16M in BALB/c mice as measured by splenic clearance (13).
Urease, a multi-subunit enzyme whose genes are organized as an operon, is an important colonization factor in a number of bacterial species (21– 24) including B. abortus (26). Among the subunits of urease, in Proteus mirabilis the accessory protein UreE acts as a Ni2+ chelator through the seven histidine residues located among the last eight C-terminal residues (His-His-His-His-Asp-His-His-His), an essential protein active site for the urease (25). A pair of frameshifts in ureE-2 of urease operon 2 was discovered in the B. abortus genomes, which results in a shortened version of the gene in B. abortus 9-941 (in B. abortus 2308 this gene call was missed); a gene coding for a nickel ABC transporter is similarly disrupted. As a consequence, the ureE-2 subunits from both genomes are deficient in the multi-histidine Ni2+ chelating centers (Figure 3). The lack of urease activity encoded by ure-2 was reported for a ureC-1 mutant of B. abortus 2308 by Sangari et al. (26). We have observed the same lack of urease activity encoded by ure-2 for a ureC-1 mutant of B. suis 1330 (Boyle, S.M., unpublished data). Thus the lack of detectable urease activity in a ure-1 mutant of either B. abortus or B. suis correlates well with the predicted disruption of the ureE-2 subunit by GenVar. Furthermore, a 9-AA insertion was located at the nickel-chelating center of the nickel-binding accessory protein E on urease operon 1 (ureE-1) in B. melitensis 16M, B. abortus 2308 and B. abortus 9-941 but not in B. suis 1330, therefore potentially inactivating or decreasing the functionality of the urease subunits. In fact, we have found that the urease activity of B. suis 1330 is much stronger than that of B. melitensis 16M or B. abortus 2308 in urease test broth (Boyle, S.M., unpublished data). The lower urease activity in B. abortus and B. melitensis is consistent with the predicted inability of the species to bind nickel that is required for optimal urease activity (27).
CONCLUSION
We believe results presented demonstrate the usefulness of GenVar in helping enrich genome annotation. GenVar's targets, namely missed gene calls, split genes and indel polymorphisms have the best potential in taking genome annotations to a level where significant new biological insights can be gained from genome sequences. The results and interpretation we present provide information about the molecular basis underlying pathogenic variations and other species-specific genome properties among Brucella species that should help in further investigations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGMENTS
This work was funded through NIAID contract HHSN266200400035C to Bruno Sobral. Funding to pay the Open Access publication charges for this article was provided by NIH/NIAID.
Conflict of interest statement. None declared.
REFERENCES
- 1.Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curwen V, Eyras E, Andrews TD, Clarke L, Mongin E, Searle SM, Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez JL, Palmer GH, Knowles DP, Jr, Brayton KA. Distinctly different msp2 pseudogene repertoires in Anaplasma marginale strains that are capable of superinfection. Gene. 2005;361:127–132. doi: 10.1016/j.gene.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 4.Thomson NR, Yeats C, Bell K, Holden MT, Bentley SD, Livingstone M, Cerdeno-Tarraga AM, Harris B, Doggett J, et al. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 2005;15:629–640. doi: 10.1101/gr.3684805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tong Z, Zhou D, Song Y, Zhang L, Pei D, Han Y, Pang X, Li M, Cui B, et al. Pseudogene accumulation might promote the adaptive microevolution of Yersinia pestis. J. Med. Microbiol. 2005;54:259–268. doi: 10.1099/jmm.0.45752-0. [DOI] [PubMed] [Google Scholar]
- 6.Chain PS, Comerci DJ, Tolmasky ME, Larimer FW, Malfatti SA, Vergez LM, Aguero F, Land ML, Ugalde RA, et al. Whole-genome analyses of speciation events in pathogenic Brucellae. Infect. Immun. 2005;73:8353–8361. doi: 10.1128/IAI.73.12.8353-8361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DelVecchio VG, Kapatral V, Redkar RJ, Patra G, Mujer C, Los T, Ivanova N, Anderson I, Bhattacharyya A, et al. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl Acad. Sci. USA. 2002;99:443–448. doi: 10.1073/pnas.221575398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halling SM, Peterson-Burch BD, Bricker BJ, Zuerner RL, Qing Z, Li LL, Kapur V, Alt DP, Olsen SC. Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J. Bacteriol. 2005;187:2715–2726. doi: 10.1128/JB.187.8.2715-2726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulsen IT, Seshadri R, Nelson KE, Eisen JA, Heidelberg JF, Read TD, Dodson RJ, Umayam L, Brinkac LM, et al. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl Acad. Sci. USA. 2002;99:13148–13153. doi: 10.1073/pnas.192319099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cutler SJ, Whatmore AM, Commander NJ. Brucellosis – new aspects of an old disease. J. Appl. Microbiol. 2005;98:1270–1281. doi: 10.1111/j.1365-2672.2005.02622.x. [DOI] [PubMed] [Google Scholar]
- 11.Ratushna VG, Sturgill DM, Ramamoorthy S, Reichow SA, He Y, Lathigra R, Sriranganathan N, Halling SM, Boyle SM, et al. Molecular targets for rapid identification of Brucella spp. BMC Microbiol. 2006;6:13. doi: 10.1186/1471-2180-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko J, Splitter GA. Molecular host-pathogen interaction in brucellosis: current understanding and future approaches to vaccine development for mice and humans. Clin. Microbiol. Rev. 2003;16:65–78. doi: 10.1128/CMR.16.1.65-78.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fretin D, Fauconnier A, Kohler S, Halling S, Leonard S, Nijskens C, Ferooz J, Lestrate P, Delrue RM, et al. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell Microbiol. 2005;7:687–698. doi: 10.1111/j.1462-5822.2005.00502.x. [DOI] [PubMed] [Google Scholar]
- 14.Snyder EE, Kampanya N, Lu J, Nordberg EK, Karur HR, Shukla M, Soneja J, Tian Y, Xue T, et al. PATRIC: the VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 2007;35:D401–D406. doi: 10.1093/nar/gkl858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cloeckaert A, Verger JM, Grayon M, Vizcaino N. Molecular and immunological characterization of the major outer membrane proteins of Brucella. FEMS Microbiol. Lett. 1996;145:1–8. doi: 10.1111/j.1574-6968.1996.tb08547.x. [DOI] [PubMed] [Google Scholar]
- 17.Binns AN, Beaupre CE, Dale EM. Inhibition of VirB-mediated transfer of diverse substrates from Agrobacterium tumefaciens by the IncQ plasmid RSF1010. J. Bacteriol. 1995;177:4890–4899. doi: 10.1128/jb.177.17.4890-4899.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cascales E, Christie PJ. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc. Natl Acad. Sci. USA. 2004;101:17228–17233. doi: 10.1073/pnas.0405843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoppner C, Carle A, Sivanesan D, Hoeppner S, Baron C. The putative lytic transglycosylase VirB1 from Brucella suis interacts with the type IV secretion system core components VirB8, VirB9 and VirB11. Microbiology. 2005;151:3469–3482. doi: 10.1099/mic.0.28326-0. [DOI] [PubMed] [Google Scholar]
- 20.Bandara AB, Sriranganathan N, Schurig GG, Boyle SM. Putative outer membrane autotransporter protein influences survival of Brucella suis in BALB/c mice. Vet. Microbiol. 2005;109:95–104. doi: 10.1016/j.vetmic.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 21.Andrutis KA, Fox JG, Schauer DB, Marini RP, Murphy JC, Yan L, Solnick JV. Inability of an isogenic urease-negative mutant stain of Helicobacter mustelae to colonize the ferret stomach. Infect. Immun. 1995;63:3722–3725. doi: 10.1128/iai.63.9.3722-3725.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belzer C, Stoof J, Beckwith CS, Kuipers EJ, Kusters JG, van Vliet AH. Differential regulation of urease activity in Helicobacter hepaticus and Helicobacter pylori. Microbiology. 2005;151:3989–3995. doi: 10.1099/mic.0.28188-0. [DOI] [PubMed] [Google Scholar]
- 23.Tsuda M, Karita M, Mizote T, Morshed MG, Okita K, Nakazawa T. Essential role of Helicobacter pylori urease in gastric colonization: definite proof using a urease-negative mutant constructed by gene replacement. Eur. J. Gastroenterol. Hepatol. 1994;6(Suppl. 1):S49–S52. [PubMed] [Google Scholar]
- 24.Tsuda M, Karita M, Morshed MG, Okita K, Nakazawa T. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect. Immun. 1994;62:3586–3589. doi: 10.1128/iai.62.8.3586-3589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sriwanthana B, Island MD, Maneval D, Mobley HL. Single-step purification of Proteus mirabilis urease accessory protein UreE, a protein with a naturally occurring histidine tail, by nickel chelate affinity chromatography. J. Bacteriol. 1994;176:6836–6841. doi: 10.1128/jb.176.22.6836-6841.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sangari FJ, Seoane A, Rodriguez MC, Aguero J, Garcia Lobo JM. Characterization of the urease operon of Brucella abortus, and assessment of its role in the virulence of the bacteria. Infect. Immun. 2007;75:774–780. doi: 10.1128/IAI.01244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulrooney SB, Ward SK, Hausinger RP. Purification and properties of the Klebsiella aerogenes UreE metal-binding domain, a functional metallochaperone of urease. J. Bacteriol. 2005;187:3581–3585. doi: 10.1128/JB.187.10.3581-3585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fauconnier A, Allaoui A, Campos A, Van Elsen A, Cornelis GR, Bollen A. Flagellar flhA, flhB and flhE genes, organized in an operon, cluster upstream from the inv locus in Yersinia enterocolitica. Microbiology. 1997;143:3461–3471. doi: 10.1099/00221287-143-11-3461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.