Abstract
DNA topoisomerase (topo) II modulates DNA topology and is essential for cell division. There are two isoforms of topo II (α and β) that have limited functional redundancy, although their catalytic mechanisms appear the same. Using their COOH-terminal domains (CTDs) in yeast two-hybrid analysis, we have identified phospholipid scramblase 1 (PLSCR1) as a binding partner of both topo II α and β. Although predominantly a plasma membrane protein involved in phosphatidylserine externalization, PLSCR1 can also be imported into the nucleus where it may have a tumour suppressor function. The interactions of PLSCR1 and topo II were confirmed by pull-down assays with topo II α and β CTD fusion proteins and endogenous PLSCR1, and by co-immunoprecipitation of endogenous PLSCR1 and topo II α and β from HeLa cell nuclear extracts. PLSCR1 also increased the decatenation activity of human topo IIα. A conserved basic sequence in the CTD of topo IIα was identified as being essential for binding to PLSCR1 and binding of the two proteins could be inhibited by a synthetic peptide corresponding to topo IIα amino acids 1430-1441. These studies reveal for the first time a physical and functional interaction between topo II and PLSCR1.
INTRODUCTION
The α and β isoforms of mammalian DNA topoisomerase II (topo II) decatenate double-stranded DNA and are involved in numerous cellular processes including replication, gene transcription, chromosomal segregation, differentiation and apoptosis as well as playing an important role in chromatin structure and remodelling (1). Topo IIα is found mainly in proliferating cells and its expression levels vary substantially in different phases of the cell cycle (2). In contrast, topo IIβ is expressed at constant levels throughout the cell cycle and in a broad range of cell types, with higher expression levels during embryogenesis and in tumour cells (2,3). Topo IIβ has also been implicated in cellular maturation and differentiation in the brain (4,5). The two mammalian isoforms have comparable catalytic activities in that both can complement functional defects in yeast that conditionally lack the single yeast topo II isoform (6). However, Top2a-/- mice die early in embryogenesis because nuclei fail to divide properly (7), while Top2b-/- mice die at birth due to defects in diaphragm muscle innervation and have significant abnormalities in neurogenesis (8,9). Thus, topo IIα is essential for cell division, while topo IIβ serves a critical role in development.
In addition to their roles in cellular proliferation and differentiation, topo II α and β are well-established targets in cancer chemotherapy (10–12). Clinically important and widely used topo II-targetting drugs such as doxorubicin and etoposide are considered topo II poisons because they stabilize cleaved DNA/enzyme complexes, leading to the induction of apoptosis (10,11). Other drugs are known as catalytic inhibitors and act at different stages of the topo II catalytic cycle, such as when topo II binds to DNA or ATP (12). There is a substantial body of compelling evidence indicating that changes in topo II expression, function and/or localization can play a major role in the response of cancer cells to drugs that target these enzymes (13–17).
Both topo II α and β consist of three domains: the NH2-proximal ATPase domain, the central DNA cleavage and religation domain (both of which are highly conserved), and the divergent COOH-terminal domains (CTDs) (18). Mutant topo IIα lacking its CTD retains activity in vitro, indicating that the CTD has more of a regulatory function rather than being essential for catalytic activity (19,20). The CTDs of topo II contain most of the utilized phosphorylation sites which are COOH-proximal to the enzymes’ nuclear export sequences (NES) (21–24). The CTDs also contain nuclear localization sequences (NLS) (24) and mutations in these sequences (at least in topo IIα) lead to cytosolic localization of the protein and confer drug resistance (13,14,16,17). Quite recently, Linka et al. (25) reported convincing evidence that the topo II CTDs are important determinants of the isoform-specific functions of these enzymes.
Previous studies have shown that human topo IIα and topo IIβ both participate in protein–protein interactions with a diverse range of nuclear proteins, including p53 (26), retinoblastoma protein (27), cyclic AMP-response element-binding protein and c-Jun (28), histone deacetylase (HDAC) 1 and 2 (29,30), 14-3-3ε (31) and caspase-activated DNase (CAD) nuclease (32,33). However, there is evidence that the screens for topo II protein binding partners to date have been incomplete. In the present study, by using yeast two-hybrid analysis with the CTDs of topo II α and β as ‘bait’, we have identified phospholipid scramblase 1 (PLSCR1) as a novel binding partner of both isozymes. While the majority of studies of this primarily plasma membrane localized protein have focused on its role in phosphatidylserine externalization during apoptosis, PLSCR1 is also actively imported into the nucleus, where it appears to have a role in cellular proliferation and differentiation (34,35). Since topo II is involved in proliferation (α isoform) and differentiation (β isoform), and both isoforms are known to have a role in the cellular responses to cytotoxic drugs, our findings raise the intriguing possibility that interactions between topo II and PLSCR1 may influence tumour cell growth and drug responsiveness.
MATERIALS AND METHODS
Plasmid construction
‘Bait’ plasmids pOBD2/topo IIα1152-1531 and pOBD2/topo IIβ1165-1621 were prepared by PCR amplification of pBS/hTOP2 (ATCC, Rockville, MD) and pYEShTOP2B (gift of Dr. I. Hickson, Oxford, UK), respectively, using PfuTurbo® DNA polymerase (Stratagene, LaJolla, CA, USA) and oligonucleotide primers to create Nco1 and Xma1 sites at the 5′ and 3′ ends of the PCR products, respectively. The PCR products were cloned into Nco1/Xma1 digested pOBD2. pACT2/HDAC1220-482 was supplied by Dr. B. Turner (University of Birmingham, UK) (30).
pGEX6P-1/topo IIαCTD (containing amino acids 1171-1531) and pGEX6P-1/topo IIβCTD (containing amino acids 1184-1621) were prepared by PCR amplification of pBS/hTOP2A and pYEShTOP2B, respectively, to add SmaI and XhoI restriction sites in the correct reading frame at the 5′ and 3′ ends, respectively, and to add two glycine residues as a flexible spacer between glutathione S-transferase (GST) and the topo II fragment. These PCR fragments were cloned in-frame at the 3′ end of GST using the SmaI and XhoI sites of pGEX-6P-1 (Amersham Biosciences, Baie D’Urfé, QC, Canada).
pGEX6P-1/topo IIα1258-1531 and pGEX6P-1/topo IIα1365-1531 were prepared by digesting pGEX6P-1/topo IIαCTD with Sma1/Stu1 and Sma1/SnaB1, respectively, and re-ligating the plasmid backbone to itself. Site-directed mutagenesis, as described subsequently, was then used to create the SnaB1 site and appropriate stop codons in pGEX6P-1/topo IIα1171-1531, thus generating pGEX6P-1/topo IIα1171-1431, pGEX6P-1/topo IIα1171-1441, pGEX6P-1/topo IIα1171-1461 and pGEX6P-1/topo IIα1171-1477.
pMAL-C2/PLSCR1 was prepared by PCR amplification of pCMV-SPORT6.1/PLSCR1 (ATCC) using Platinum Pfx® polymerase (Gibco-BRL) and primers to create a PCR product with 5′ EcoRV and 3′ EcoR1 sites. The PCR fragment was cloned into Xmn1/EcoR1 digested pMAL-C2. All restriction enzymes and pMAL-C2 were from New England Biolabs (Ipswich, MA, USA), primers were from IDT (Coralville, IA, USA), and the fidelity of the constructs was confirmed by sequencing.
Site-directed mutagenesis
Site-directed mutagenesis was performed using PfuTurbo® DNA polymerase and mutagenic forward and reverse complementary primers to amplify pGEX6P-1/topo IIαCTD containing the desired mutations. In all cases, mutations were confirmed by sequencing prior to subcloning appropriate fragments into parental vectors. The SnaB1 site in pGEX6P-1/topo IIαCTD was created using a primer with the sequence 5′-CCCCAAAACTTACGTACAAAGAACTGAAACC-3′ and its complement (new SnaB1 site italicized). The new stop codons in pGEX-6P-1/topo IIα CTD, which created pGEX6P-1/topo IIα 1171-1431, 1171-1441, 1171-1451, 1171-1461 and 1171-1477, were generated using the following primers and their complements (newly introduced Xma1 sites are italicized and inserted stop codons are in boldface): 5′-CC ACT ACC GGT TAG AAC CCG GGG GCT GCC CCA AAA G-3′, 5′-CCA AAA GGA ACT TAA AGC CCG GGA GCT TTG AAT TCT GG-3′, 5′-G AAT TCT GGT GTC TAA CAC CCG GGT GAT CCT GCC-3′, 5′-CC AAA ACC AAG TAG CGC CCG GGA AGG AAG CCA TCC-3′ and 5′-CT GAC TCT AAT TAG GAC CCG GGT GTT TCG AAA GC-3’.
Yeast two-hybrid analysis
Two-hybrid screening was performed using yeast strain PJ69-4A (ATCC 201540). Yeast harbouring a bait plasmid (pOBD2/topo IIα1152-1531 or pOBD2/topo IIβ1165-1621) were transformed with a human B lymphocyte library (ATCC 87003) (provided by Dr. D. LeBrun, Queen's University), and HIS3 and ADE2 reporter gene expression monitored on histidine and adenine deficient agar plates (interaction-selective plates). Plasmids were isolated, transformed into JF1754, and then grown on leucine-deficient agar plates. Plasmids isolated from JF1754 colonies were transformed into PJ69 harbouring pOBD2/topo IIα1152-1531 or pOBD2/topo IIβ1165-1621 to confirm interactions, or into PJ69 harbouring pOBD2 to identify false positives. Plasmids that encoded putative topo II α or β interacting proteins were sequenced and the partner proteins identified by comparison to nucleotide databases.
Purification of GST and MBP fusion proteins
BL21(DE3)-RIL Escherichia coli cells (Stratagene) transfected with the pGEX6P-1/topo IIα or pMAL-C2/PLSCR1 constructs were used to express GST-topo IIα or maltose-binding protein (MBP)-PLSCR1 fusion proteins, respectively. Bacteria were induced with 1.0 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) for 3 h, harvested and then sonicated in PBS containing a protease inhibitor cocktail (Roche, Laval, QC, Canada), DTT (5 mM) and benzamidine (10 μg/ml). Lysates containing GST-topo IIα fusion proteins were pre-cleared by ultra-centrifugation and batch-bound overnight to GSH-Sepharose 4B (Amersham Biosciences, Uppsala, Sweden). GST-topo IIα fusion proteins were eluted with 50 mM Tris pH 7.5 containing 15 mM GSH, 5 mM DTT and a protease inhibitor cocktail, and dialysed overnight against PBS. Lysates containing the MBP-PLSCR1 fusion protein was treated in a similar manner, except that it was bound to amylose resin (New England Biolabs), eluted with PBS containing 10 mM maltose, 5 mM DTT and a protease inhibitor cocktail, and dialysed against PBS containing 250 mM sucrose. Purified fusion proteins were stored at −80°C.
Immunoblotting
Protein samples were resolved by Tricine gel electrophoresis (binding assays involving recombinant PLSCR1) or SDS-PAGE (all other samples) and transferred to polyvinylidene fluoride membranes in 50 mM N-cyclohexyl-3-aminopropanesulfonic acid, pH 11 (for topo II immunoblots) or carbonate buffer pH 9.0 containing 20% methanol (for all other immunoblots). Membranes were incubated with primary antibodies overnight at 4°C in TBS-T (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% Tween-20) containing 4% (w/v) skim milk powder. The filters were washed in TBS-T, incubated for 1 h with the appropriate horseradish peroxidase conjugated anti-mouse or anti-rabbit secondary antibody, and then developed using chemiluminescence detection (Amersham Pharmacia Biotech).
Topo II α and β specific mAbs, 8D2 and 3H10, respectively, were supplied by Dr A. Kikuchi (Nagoya University, Japan) (36). A PLSCR1-specific polyclonal antiserum generated against the 14 COOH-terminal amino acids of PLSCR1 (CESTGSQEQKSGVW) was supplied by Dr D. Bratton (National Jewish Medical and Research Center, Denver, CO) (37). A mAb against tubulin and a polyclonal antibody against calnexin were from Sigma Diagnostics; a mAb against lysosomal-associated membrane protein (LAMP) 2 was from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Co-immunoprecipitation of topo II and PLSCR1
HeLa cell nuclear extracts were prepared by lysing approximately 100 × 106 cells in hypotonic buffer (10 mM HEPES pH 7.6, 15 mM KCl, 2 mM EDTA, 0.5 mM spermidine, 0.5 mM spermine, 0.5% NP-40) on ice for 15 min, underlaying the suspension with a 30% sucrose solution in the same buffer and centrifuging at 2200 × g for 20 min. The pellet was resuspended in nuclear lysis buffer (10 mM HEPES, 100 mM KCl, 0.1 mM EDTA, 10% glycerol, 3 mM MgCl2), and KCl added to a final concentration of 0.55 M. After incubation for 30 min, the nuclear lysate was centrifuged at 100 000 × g and the supernatant dialysed overnight against 50 mM Tris (pH 7.5)/150 mM NaCl. The dialysate was centrifuged at 15 000 × g prior to overnight incubation with antibodies against topo IIα (mAb 8D2), topo IIβ (mAb 3H10), PLSCR1, or pre-immune serum at 4°C. Gamma-Bind Sepharose™ (Amersham) was then added for 1 h prior to collecting the beads by centrifugation. Bound proteins were eluted with sample buffer and analysed by immunoblotting with topo II α and β mAbs 8D2 and 3H10 as described above.
Confocal microscopy
HeLa cells were grown on gelatin-coated glass coverslips until approximately 80% confluent, fixed in 3.7% formaldehyde and then permeabilized in 1% TX-100. After blocking for 1 h in 1% BSA, cells were incubated with topo II α or β mAbs and PLSCR1 antiserum (ICN, Aurora, OH, USA). Antibody binding was detected with Alexa546™ goat anti-mouse and Alexa488™ goat anti-rabbit conjugated secondary antibodies (Molecular Probes, Eugene, OR, USA) and fluorescent images were captured using a Leica TCS SP2 multiphoton confocal microscope.
GST-pull-down assays
GST-pull-down assays of GST-tagged topo II CTD fusion proteins and endogenous PLSCR1 were performed using NP-40 lysates prepared from HeLa cells. Cells were lysed in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, protease inhibitors and 0.5% NP-40 (lysis buffer) for 15 min on ice. The lysate was cleared by centrifugation and the resulting supernatant incubated with purified GST or GST-topo II CTD fusion proteins for 90 min at 20°C. In some cases, the lysate was treated with 40 mU/ml Turbo™ DNase 1 (Ambion, Austin, TX, USA) for 60 min at 20°C prior to the addition of GST or GST-topo II CTD fusion proteins. In some experiments, recombinant PLSCR1 was used in place of HeLa cell lysates. For these studies, the MBP tag was first removed by digesting the recombinant protein with Factor Xa and incubating with X-Arrest agarose (Novagen, San Diego, CA, USA). GSH-Sepharose 4B was then added and after 1 h, the beads were collected by centrifugation. Bound proteins were eluted with sample buffer and analysed by immunoblotting using PLSCR1 antiserum as described earlier.
Decatenation assay
To measure the effect of PLSCR1 on topo II activity, recombinant MBP-PLSCR1 was digested with Factor Xa as above and ∼2 pmol PLSCR1 was incubated with 0.4 U purified human topo IIα for 1.5 h at 20°C according to the manufacturer's instructions (TopoGEN, Inc., Port Orange, FL, USA). The protein mixture was then incubated with kinetoplast DNA (kDNA) for 15 min at 37°C. After terminating the reactions, the samples were resolved on a 1% agarose gel containing ethidium bromide.
Peptide competition assay
Dodecapeptides corresponding to residues 1430-1441 of topo IIα (TGAKKRAAPKGT) or the same amino acids in random order (KRGATAKGTAPK) were purchased from Bio-Synthesis (Lewisville, TX, USA). After removing the MBP tag with Factor Xa, recombinant PLSCR1 was incubated with increasing concentrations of peptide for 1 h at 20°C. Purified GST or GST-topo IIα1258-1531 was then added and samples were treated as described earlier for the GST- pull-down assay.
RESULTS
Identification of PLSCR1 as a topo II α and β binding partner
Vectors encoding the topo II CTDs expressed as NH2-terminal fusion proteins with the DNA binding domain of the GAL4 transcription factor (pOBD2/topo IIα1152-1531 and pOBD2/topo IIβ1165-1621) were constructed, and expression in Saccharomyces cerevisiae yeast strain PJ69 was confirmed by immunoblotting (data not shown). While the GAL4-topo IIβ construct was expressed as a single protein, several degradation products of the polypeptide encoded by the topo IIα construct were seen, suggesting that the product of this construct might be unstable or toxic to yeast. This may be a general problem in yeast two-hybrid screens using human topo IIα as bait, since a screen of a HeLa cell library for topo IIβ binding partners was successful (38), as were two screens for yeast topo II (39,40), while no yeast two-hybrid screens for topo IIα binding partners have been reported.
No growth was detected on interaction-selective plates when either pOBD2/topo IIα1152-1531 or pOBD2/topo IIβ1165-1621 were co-transformed with pACT/SNF4 (negative control) while growth was detected when co-transformed with pACT/HDAC220-482 (positive control) (29,30), suggesting the two topo II constructs were suitable candidates for yeast two-hybrid analysis (Table 1). Screening of the library, identified several clones as potential binding partners of both the topo II α and β CTDs, one of which was confirmed by sequencing as PLSCR1. Human PLSCR1 has 318 amino acids and the clone obtained, pACT/PLSCR1, encoded a polypeptide missing amino acids 1-29 and 118-143. Yeast co-transformed with pACT/PLSCR1 and pOBD2 failed to grow on selective plates, demonstrating that transcription was not induced by pACT/PLSCR1 independently of its interaction with the topo II CTDs.
Table 1.
Identification of PLSCR1 as a binding partner of the topo II α and β CTDs using yeast two-hybrid analysis
| Plasmid combination | Growtha |
|---|---|
| pOBD2/topo IIα1152-1531 + pACT/SNF4 | – |
| pOBD2/topo IIβ1165-1621 + pACT/SNF4 | – |
| pOBD2 + pACT/PLSCR1 | – |
| pOBD2/topo IIα1152-1531 + pACT/PLSCR1 | + |
| pOBD2/topo IIβ1165-1621 + pACT/PLSCR1 | + |
| pOBD2/topo IIα1152-1531 + pACT/HDAC1220-482 | −/+ |
| pOBD2/topo IIβ1165-1621 + pACT/HDAC1220-482 | + |
aYeast strain PJ69 was co-transformed with the plasmids as indicated and allowed to grow on interaction-selective plates. Growth was scored by presence of colonies after 5 days at 30°C. +, yeast growth; −, no growth.
Confirmation of interactions between PLSCR1 and topo II α and β in vitro
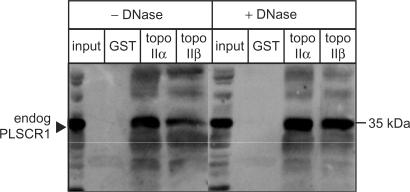
To confirm the interactions of topo IIα and topo IIβ with PLSCR1, the binding of endogenous PLSCR1 to GST-tagged topo IIα CTD and topo IIβ CTD fusion proteins was investigated using HeLa cell lysates. To eliminate the possibility that the interactions between PLSCR1 and the topo II fragments were DNA-mediated, the cell lysates were pre-treated with DNase1. However, as shown in Figure 1, addition of DNase1 had no effect on the binding between either of the topo II CTD fusion proteins and endogenous PLSCR1, indicating that the interactions are DNA independent. The input lane in the immunoblot shown represents 5% of the lysate used in the binding assay, suggesting that the interactions may be relatively weak, at least under the conditions of this assay, although other explanations are possible.
Figure 1.
GST-tagged topo IIα CTD and topo IIβ CTD interact with PLSCR1. An NP-40 soluble HeLa cell lysate (150 μg protein) was incubated sequentially with DNase1 (20 U), GST-tagged topo IIα or topo IIβ CTD fusion proteins (5 μg) and GSH-Sepharose beads (25 μl of 50% slurry) as described in Materials and Methods section. The beads were collected by centrifugation and bound proteins analysed by immunoblotting with rabbit PLSCR1 antiserum. Input lane represents 5% of lysate used in binding assay.
Of some interest, the addition of a FLAG tag to the NH2-terminus of PLSCR1 completely inhibited the interaction of the protein with the GST-tagged topo IIα CTD (results not shown). The reason for this is unknown but may be related to the highly charged nature of the FLAG-tag, since the fusion of MBP to the NH2-terminus of PLSCR1 did not affect its binding to either of the GST-topo II α or β CTD fusion proteins (data not shown).
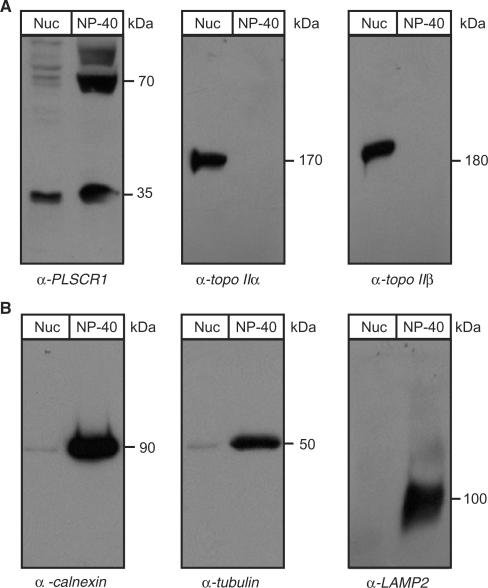
PLSCR1 co-fractionates with topo II α and β
To confirm that PLSCR1 colocalizes with topo II α and β in intact cells, HeLa cell nuclei were isolated and the relative amounts of nuclear and NP-40 soluble PLSCR1 and topo II α and β in the two cellular fractions were determined by immunoblotting. Samples were prepared from equivalent numbers of cells rather than equal protein concentrations so that the relative fraction of total cellular PLSCR1 present in the nucleus could be determined. As shown in Figure 2A, the nuclear fraction contains ∼10–20% of the amount of the 35 kDa PLSCR1 found in the NP-40 soluble fraction, an estimate that is supported by confocal microscopy (see subsequently). The identity of the 70 kDa band is unknown, but it was consistently present, albeit at variable levels. It was not detected by the pre-immune serum and may be an undissociated dimer. Topo II α and β were detected only in the nuclear fraction as expected. A lack of ER and cytoplasmic contamination of the nuclear lysates was demonstrated by immunoblotting for calnexin (integral ER membrane marker), tubulin (cytoskeletal marker) and LAMP2 (lysosomal marker) (Figure 2B).
Figure 2.
PLSCR1 co-fractionates with topo II α and β. (A) Nuclear and NP-40 soluble lysates were prepared from HeLa cells as described in Materials and Methods section for co-immunoprecipitation and GST-pull-down assays, respectively. Samples representing equivalent numbers of cells were analysed by immunoblotting with antibodies against PLSCR1, topo IIα (mAb 8D2) and topo IIβ (mAb 3H10). (B) The same samples described in Panel A were analysed for calnexin, LAMP2 and tubulin content by immunoblotting to confirm the absence of contaminating ER, lysosomal and cytoskeletal proteins, respectively. To prevent overexposure of the film due to high intensity signals, the analyses for tubulin and calnexin content used 10% of the number of cells used for other immunoblots. Nuc, nuclear lysate; NP-40, NP-40 soluble lysate; TX-100, Triton X-100 soluble lysate.
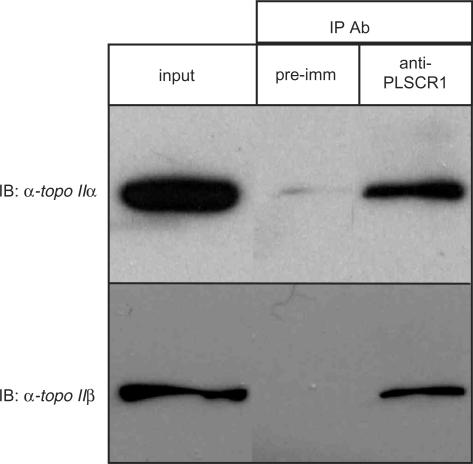
Endogenous topo II α and β co-immunoprecipitate with PLSCR1
To confirm that endogenous topo II α and β interact with endogenous PLSCR1 in intact cells, nuclear lysates were subjected to immunoprecipitation analysis using a PLSCR1 antiserum, followed by immunoblotting with topo II α and β specific mAbs. As shown in Figure 3, both topo II α and β were immunoprecipitated by the PLSCR1 antiserum, but not the pre-immune serum, demonstrating that the endogenous proteins interact in vivo. These results also show that the interaction is not an artifact caused by the use of recombinant topo II CTDs rather than the endogenous proteins, or the forced colocalization of proteins that do not normally share a compartment, as can occur in yeast two-hybrid analysis. The input lane represents 5% of the nuclear lysate used in each binding assay, suggesting that as much as 5% of nuclear topo II α and β is associated with PLSCR1. A high background associated with the PLSCR1 antiserum in co-immunoprecipitation experiments precluded consistent western blot detection of nuclear PLSCR1 immunoprecipitated by the topo II α and β specific mAbs (data not shown).
Figure 3.
Endogenous topo II α and β co-immunoprecipitate with endogenous PLSCR1. Nuclear extracts prepared from HeLa cells were subjected to co-immunoprecipitation with an anti-PLSCR1 antiserum or pre-immune serum as described in Materials and Methods section. Immunoprecipitated samples were then analysed by immunoblotting with topo II α and β specific mAbs 8D2 and 3H10, respectively. Input lanes represent 5% of sample used in the assay. Shown are results from a representative experiment and similar results were obtained in two additional experiments.
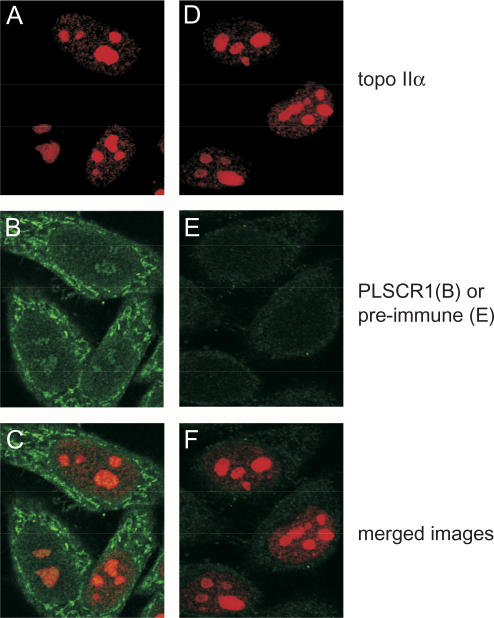
Nuclear colocalization of PLSCR1 and topo IIα by confocal microscopy
The nuclear colocalization of PLSCR1 and topo IIα in HeLa cells was confirmed by immunocytochemistry (Figure 4). We found that 1% TX-100 and incubation at 37°C was needed to permeabilize the nucleus sufficiently to detect nuclear PLSCR1. These conditions also substantially increased staining of nuclear topo IIα. This PLSCR1 staining pattern is consistent with a previous study of PLSCR1 in HeLa cells using a different primary antibody (41). It is noted that nuclear localization of PLSCR1 and its induction by interferons can vary dramatically according to cell type (34).
Figure 4.
Colocalization of endogenous topo IIα and PLSCR1. HeLa cells were cultured on gelatinized glass coverslips, fixed in formaldehyde and permeabilized with Triton X-100. Topo IIα was detected with mAb 8D2 in conjunction with Alexa546 ™ -conjugated goat anti-mouse secondary antibody (Panels A and D, red signal) and PLSCR1 was detected with rabbit PLSCR1 antiserum or the pre-immune serum (Panels B and E, respectively, green signal) in conjunction with Alexa488 ™ -conjugated goat anti-rabbit secondary antibody. Merged images are shown in Panels C (A + B) and F (D + E).
Both topo IIα and PLSCR1 were localized throughout the nucleus although, as reported by other investigators (42), topo IIα appeared more abundant in nucleoli of some cells (Figure 4, Panels A and D). Not unexpectedly, given the differences in the relative abundance of the two proteins, the signal intensity for nuclear PLSCR1 (Panel B) was very low compared to that for topo IIα, making colocalization studies difficult. However, examination of the separate images showed a small increase in PLSCR1 staining in the nucleoli relative to the nucleoplasm. This was not caused by overlap of the red channel (topo IIα) into the green channel (PLSCR1), since cells treated with the pre-immune serum in place of the PLSCR1 antiserum did not display an increased green signal in the nucleoli (Panel E). Unfortunately, comparable colocalization experiments of PLSCR1 and topo IIβ were unsuccessful since staining by the topo IIβ-specific mAb under the fixation conditions required to detect nuclear PLSCR1 was poor.
Recombinant PLSCR1 stimulates the decatenation activity of topo IIα
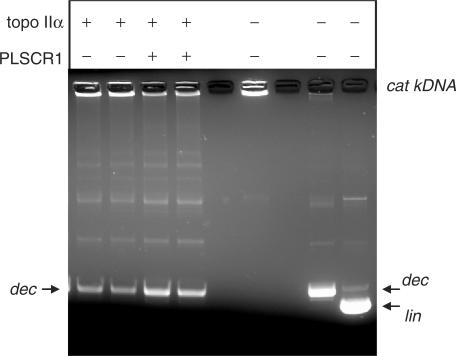
The ability of PLSCR1 to modulate topo II activity was assessed using a topo II assay that measures the decatenating activity of the enzyme. Thus purified topo IIα was preincubated with PLSCR1 for 1.5 h at 20°C prior to incubation with kDNA for 15 min at 37°C. As shown in Figure 5, the catenated kDNA did not enter the gel (lane 5) as expected, while decatenated DNA entered the gel, but its electrophoretic mobility was slightly retarded compared to linear DNA (compare lanes 6 and 7). The addition of recombinant PLSCR1 to the assay mixture significantly increased the ability of topo IIα to decatenate kDNA (compare lanes 3 and 4 versus 1 and 2).
Figure 5.
PLSCR1 stimulates the decatenation activity of topo IIα. Topo II decatenating activity was assayed using purified topo IIα enzyme in the presence and absence of untagged recombinant PLSCR1 as described in Materials and Methods section. Similar results to those shown were obtained in two additional experiments. The positions of the catenated kDNA (cat kDNA), decatenated kDNA minicircles (dec) and linear (lin) kDNA are indicated.
Topo IIα residues 1432-1441 are essential for binding to PLSCR1
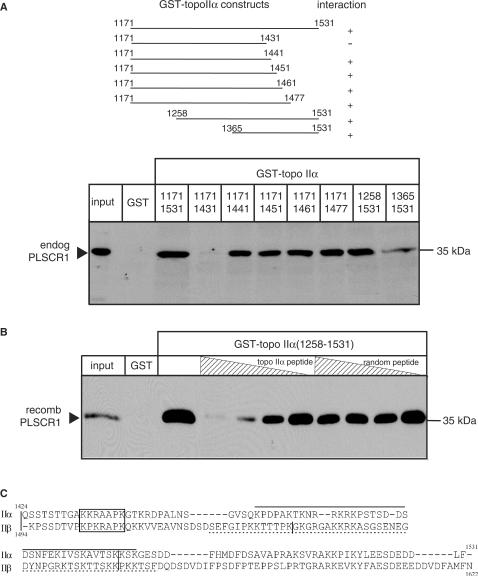
To identify the region of topo IIα that mediates binding to PLSCR1, eight GST fusion proteins encoding NH2– and COOH– truncated forms of the topo IIα CTD (Figure 6A, upper panel) were generated and tested for their ability to bind endogenous PLSCR1 in a GST-pull-down assay. Only one of these fusion proteins (topo IIα1171-1431) failed to bind to endogenous PLSCR1, indicating that topo IIα residues 1432-1441 are required for binding to PLSCR1 (Figure 6A, lower panel).
Figure 6.
Localization of PLSCR1 binding to amino acids 1432-1441 of topo IIα. (A) Upper panel, Schematic diagram showing the eight GST-tagged topo IIαCTD fusion proteins used to define the region of topo IIα involved in binding to PLSCR1. Lower panel, NP-40 solubilized HeLa cell lysates were incubated with each of the eight GST-topo IIαCTD fusion proteins and complexes pulled down with GSH-Sepharose beads as described in Materials and Methods section. The proteins were eluted and immunoblotted with PLSCR1 antiserum. (B) Untagged recombinant PLSCR1 was incubated with increasing concentrations of synthetic dodecapeptides corresponding to topo IIα residues 1430-1441 or as a control, randomly scrambled topo IIα residues 1430-1441, and GST-topo IIα1258-1531 or GST alone (control). Complexes were pulled down with GSH-Sepharose beads and bound proteins analysed by immunoblotting with PLSCR1 antiserum. The experiment shown was repeated three or more times with comparable results. (C) The sequences of the COOH-termini of topo IIα and topo IIβ were aligned using ClustalW. The basic conserved putative PLSCR1 binding motifs in topo IIα and topo IIβ are boxed; the vertical bars in the topo II sequences represent intron/exon boundaries (43), and the functional NLS sequences in this region are underlined (topo IIα, solid line; topo IIβ, broken line) (24).
To confirm that these residues were required, a dodecapeptide corresponding to topo IIα amino acids 1430-1441 (TGAKKRAAPKGT) was examined for its ability to inhibit the binding of GST-topo IIα1258-1531 to recombinant PLSCR1 in a GST-pull-down assay. A second dodecapeptide consisting of the same amino acids in random order was included as a negative control. As shown in Figure 6B, the topo IIα peptide specifically blocked the interaction between topo IIα and PLSCR1 in a concentration-dependent manner, while the random peptide had no effect. These observations provide further confirmation that residues 1432-1441 are involved in the interaction of topo IIα with PLSCR1.
The CTDs of topo II α and β are poorly conserved, making alignments of these regions difficult. Nevertheless, when residues 1424-1531 of topo IIα (exons 34-35) were aligned with residues 1493-1621 of topo IIβ (exons 34-36) (43), a basic stretch of seven amino acids was identified between topo IIα residues 1433 and 1439 (topo IIβ residues 1501-1507) (Figure 6C) that may form a positively charged patch that mediates the interaction with PLSCR1. These putative PLSCR1 interaction motifs are both approximately the same distance from the COOH–termini of their respective topo II isoforms and occur within 9–10 residues of an intron/exon boundary (43). Neither basic motif overlaps with previously identified functional NLS or NES sequences (21,22,24) and both are highly conserved in all vertebrate species for which complete topo II sequences are available (human, hamster, rat, mouse, chicken and pig).
DISCUSSION
Despite the established importance of the topo II isoforms in cell division and as clinically important drug targets, the cellular processes responsible for regulating their function and activity are still not fully defined. As the identification of protein–protein interactions can often provide insight into regulatory pathways, we undertook a search for novel topo II binding partners using yeast two-hybrid analysis. The sequence divergent topo II CTDs were used as bait because there is compelling evidence that many of their regulatory elements are contained in this region.
Since topo II α and β are normally localized to the nucleus, we initially found it surprising that our analysis identified PLSCR1, which is primarily localized to the endofacial plasma membrane in most cell types, as a topo II-binding protein. However, PLSCR1 can also be found in the nucleus and other organelles, albeit at much lower abundance (34,41). A non-classical functional NLS has been identified in PLSCR1 (35,44) and it has been demonstrated that PLSCR1 binds to a unique site in the nuclear import carrier protein importin α (44). Thus, there is strong evidence that under certain conditions, PLSCR1 is actively imported into the nucleus making its interaction with topo II in vivo feasible.
PLSCR1 at the plasma membrane is multiply palmitoylated which, like other forms of lipid modification, often induces or enhances membrane localization (34,45). The factors that regulate PLSCR1 palmitoylation are not known, nor is it known precisely how palmitoylation affects the non-plasma membrane localization or function of this protein. However, when palmitoylation of PLSCR1 is inhibited, either by mutation of the palmitoylated cysteines in the COOH-proximal end of the protein or by treatment of cells with 2-bromo-palmitate, PLSCR1 becomes primarily localized to the nucleus (34). Since the conditions used to prepare samples for the topo II pulldown and co-immunoprecipitation assays in the present study would be expected to yield predominantly palmitoylated and unpalmitoylated PLSCR1, respectively, our observations suggest that palmitoylation does not affect the interaction of PLSCR1 with topo II α or β, at least in vitro. However, because both topo II isoforms are almost exclusively found in the nucleus, it would seem likely that they interact primarily with unpalmitoylated PLSCR1 in vivo.
Protein–protein interactions involving both yeast and human topo II have been reported to result in several different functional consequences (26–33). In some cases, the topo II–protein interactions modulate the DNA decatenating activity of the enzyme which is critical for regulated chromosomal segregation. Thus, HDAC1 and the underphosphorylated form of the retinoblastoma protein inhibit the DNA decatenating activity of human topo IIα (27,29) while in contrast, the cyclic AMP-response element-binding protein (28), the mitotic cdc2 cyclin-dependent kinase (23) and CAD nuclease stimulate this activity (32). Our data indicates that PLSCR1 behaves like the latter proteins in its ability to stimulate DNA decatenation by topo IIα.
To better understand the molecular basis of the interaction between topo II and PLSCR1, we investigated the region of topo IIα responsible for its binding to PLSCR1. Using a series of nested truncated GST-fusion proteins in pull-down assays, a basic stretch of amino acids (residues 1432-1441) in topo IIα in a region of the enzyme that is predicted to be unstructured was shown to be critical. The importance of these amino acids was confirmed by demonstrating that a synthetic dodecapeptide corresponding to this sequence could inhibit binding of topo II and PLSCR1. However, while topo IIα residues1432-1441 are essential, it remains possible that they are not sufficient by themselves to mediate the interaction between the two proteins. Other proteins reported to bind to the topo II CTD include HDAC1 and HDAC2 (30) and p53 (26). It will be of interest to determine the relationships, if any, among the different topo II CTD binding proteins (which now includes PLSCR1) with respect to topo II functions (25).
Most studies of PLSCR1 have focused on its role in phospholipid mixing at the plasma membrane. More recently it has been suggested that PLSCR1 may have a role in suppressing tumorigenesis. For example, when human ovarian tumour cells stably transfected with PLSCR1 were implanted into athymic mice, the resulting tumours were much smaller and more differentiated than those arising from untransfected cells (46). Additionally, Huang et al. (47) showed that induction of PLSCR1 arrested proliferation of human myeloid leukemia cells and stimulated their differentiation. Elevated PLSCR1 mRNA levels have also been reported to be a positive prognostic factor in a clinical study of patients with acute myelogenous leukaemia (48). In contrast, when transplanted into syngeneic or athymic mice, an NH2-truncated form of murine Plscr1 (designated as TRA1) is leukemogenic (49). Ongoing studies are aimed at determining how the tumour-suppressor effects of PLSCR1 might be related to its effects on topo II activity.
The function(s) of nuclear PLSCR1 is presently not well understood. Recently, a domain within the putative NH2-proximal regulatory region of PLSCR1 was shown to stimulate expression of inositol 1,4,5-triphosphate receptor type 1 (IP3R1), a protein involved in regulating Ca2+ release from the endoplasmic reticulum, by binding directly to its promoter (50). It may be that PLSCR1 participates with other nuclear proteins to regulate expression of IP3R1 and possibly other genes by recruiting topo II or alternatively, topo II recruits PLSCR1 to specific regions of DNA in order to regulate topo II activity. For the topo II-nuclear protein interactions characterized thus far, it is the interacting protein partner that appears to be recruited by topo II (23,32,51). For example, it has been shown that chromatin remodelling occurs in response to topo II-mediated recruitment of the mitotic cdc2 cyclin-dependent kinase (51).
IP3R1 seems unlikely to be the only gene regulated by PLSCR1 and indeed, PLSCR1 has been reported to bind to promoter fragments from three other uncharacterized genes (50). PLSCR1 did not induce transcription from these promoter fragments in a luciferase reporter assay, an observation that could be explained if PLSCR1 is acting as a negative regulator of these genes or if a necessary adaptor protein was not present in the cells used. It has also been shown that nuclear levels of PLSCR1 can be enhanced substantially by treatment of at least some cell types with both type I and II interferons (46,52). Whether or not the ability of PLSCR1 to modulate topo II activity is involved in the cellular activities of interferons, however, remains to be determined.
In conclusion, our study shows for the first time a physical and functional interaction between topo II and PLSCR1. Further investigation of the interactions between these proteins is needed to better understand their possible role in tumour proliferation, as well as cellular differentiation and drug responsiveness.
ACKNOWLEDGEMENTS
The authors would like to thank Ms Kathy Sparks for expert technical assistance. This work was supported by a grant from Canadian Cancer Society/National Cancer Institute of Canada (NCIC) and a Terry Fox Foundation NCIC Postdoctoral Fellowship (J.P.W.). Funding to pay the Open Access publication charges for the article was provided by NCIC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 2.Woessner RD, Mattern MR, Mirabelli CK, Johnson RK, Drake FH. Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 1991;2:209–214. [PubMed] [Google Scholar]
- 3.Bauman ME, Holden JA, Brown KA, Harker WG, Perkins SL. Differential immunohistochemical staining for DNA topoisomerase II α and β in human tissues and for DNA topoisomerase IIβ in non-Hodgkin's lymphomas. Mod. Pathol. 1997;10:168–175. [PubMed] [Google Scholar]
- 4.Watanabe M, Tsutsui K, Inoue Y. Differential expressions of the topoisomerase IIα and IIβ mRNAs in developing rat brain. Neurosci. Res. 1994;19:51–57. doi: 10.1016/0168-0102(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 5.Tsutsui K, Sano K, Kikuchi A, Tokunaga A. Involvement of DNA topoisomerase IIβ in neuronal differentiation. J. Biol. Chem. 2001;276:5769–5778. doi: 10.1074/jbc.M008517200. [DOI] [PubMed] [Google Scholar]
- 6.Jensen S, Redwood CS, Jenkins JR, Andersen AH, Hickson ID. Human DNA topoisomerases IIα and IIβ can functionally substitute for yeast TOP2 in chromosome segregation and recombination. Mol. Gen. Genet. 1996;252:79–86. [PubMed] [Google Scholar]
- 7.Akimitsu N, Adachi N, Hirai H, Hossain MS, Hamamoto H, Kobayashi M, Aratani Y, Koyama H, Sekimizu K. Enforced cytokinesis without complete nuclear division in embryonic cells depleting the activity of DNA topoisomerase IIα. Genes Cells. 2003;8:393–402. doi: 10.1046/j.1365-2443.2003.00643.x. [DOI] [PubMed] [Google Scholar]
- 8.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIβ and neural development. Science. 2000;287:131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 9.Lyu YL, Wang JC. Aberrant lamination in the cerebral cortex of mouse embryos lacking DNA topoisomerase IIβ. Proc. Natl Acad. Sci. USA. 2001;100:7123–7128. doi: 10.1073/pnas.1232376100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaufmann SH, Earnshaw WC. Induction of apoptosis by cancer chemotherapy. Exp. Cell Res. 2000;256:42–49. doi: 10.1006/excr.2000.4838. [DOI] [PubMed] [Google Scholar]
- 11.Larsen AK, Escargueil AE, Skladanowski A. Catalytic topoisomerase II inhibitors in cancer therapy. Pharmacol. Ther. 2003;99:167–181. doi: 10.1016/s0163-7258(03)00058-5. [DOI] [PubMed] [Google Scholar]
- 12.Li TK, Liu LF. Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol. Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- 13.Harker WG, Slade DL, Parr RL, Feldhoff PW, Sullivan DM, Holguin MH. Alterations in the topoisomerase IIα gene, messenger RNA, and subcellular protein distribution as well as reduced expression of the DNA topoisomerase IIβ enzyme in a mitoxantrone-resistant HL-60 human leukemia cell line. Cancer Res. 1995;55:1707–1716. [PubMed] [Google Scholar]
- 14.Mirski SEL, Cole SPC. Cytoplasmic localization of a mutant M(r) 160,000 topoisomerase IIα is associated with the loss of putative bipartite nuclear localization signals in a drug-resistant human lung cancer cell line. Cancer Res. 1995;55:2129–2134. [PubMed] [Google Scholar]
- 15.Dereuddre S, Delaporte C, Jacquemin-Sablon A. Role of topoisomerase IIβ in the resistance of 9-OH-ellipticine-resistant Chinese hamster fibroblasts to topoisomerase II inhibitors. Cancer Res. 1997;57:4301–4308. [PubMed] [Google Scholar]
- 16.Wessel I, Jensen PB, Falck J, Mirski SEL, Cole SPC, Sehested M. Loss of amino acids 1490Lys-Ser-Lys1492 in the COOH-terminal region of topoisomerase IIα in human small cell lung cancer cells selected for resistance to etoposide results in an extranuclear enzyme localization. Cancer Res. 1997;57:4451–4454. [PubMed] [Google Scholar]
- 17.Mirski SEL, Sparks KE, Yu Q, Lang AJ, Jain N, Campling BG, Cole SPC. A truncated cytoplasmic topoisomerase IIα in a drug-resistant lung cancer cell line is encoded by a TOP2A allele with a partial deletion of exon 34. Int. J. Cancer. 2000;85:534–539. doi: 10.1002/(sici)1097-0215(20000215)85:4<534::aid-ijc15>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 18.Watt PM, Hickson ID. Structure and function of type II DNA topoisomerases. Biochem. J. 1994;303:681–695. doi: 10.1042/bj3030681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen S, Andersen AH, Kjeldsen E, Biersack H, Olsen EH, Andersen TB, Westergaard O, Jakobsen BK. Analysis of functional domain organization in DNA topoisomerase II from humans and Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:3866–3877. doi: 10.1128/mcb.16.7.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickey JS, Osheroff N. Impact of the C-terminal domain of topoisomerase II on the DNA cleavage activity of the human enzyme. Biochemistry. 2005;44:11546–11554. doi: 10.1021/bi050811l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirski SEL, Bielawski JC, Cole SPC. Identification of functional nuclear export sequences in human topoisomerase II α and β. Biochem. Biophys. Res. Commun. 2003;306:905–911. doi: 10.1016/s0006-291x(03)01077-5. [DOI] [PubMed] [Google Scholar]
- 22.Turner JG, Engel R, Derderian JA, Jove R, Sullivan DM. Human topoisomerase IIα nuclear export is mediated by two CRM-1-dependent nuclear export signals. J. Cell Sci. 2004;117:3061–3071. doi: 10.1242/jcs.01147. [DOI] [PubMed] [Google Scholar]
- 23.Escargueil AE, Plisov SY, Filhol O, Cochet C, Larsen AK. Mitotic phosphorylation of DNA topoisomerase IIα by protein kinase CK2 creates the MPM-2 phosphoepitope on Ser-1469. J. Biol. Chem. 2000;275:34710–34718. doi: 10.1074/jbc.M005179200. [DOI] [PubMed] [Google Scholar]
- 24.Mirski SEL, Gerlach JH, Cole SPC. Sequence determinants of nuclear localization in the α and β isoforms of human topoisomerase II. Exp. Cell Res. 1999;251:329–339. doi: 10.1006/excr.1999.4587. [DOI] [PubMed] [Google Scholar]
- 25.Linka RM, Porter ACG, Volkov A, Mielke C, Boege F, Christensen MO. C-terminal regions of topoisomerase IIα and IIβ determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm102. doi:10.1093/nar/gkm102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowell IG, Okorokov AL, Cutts SA, Padget K, Bell M, Milner J, Austin CA. Human topoisomerase IIα and IIβ interact with the C-terminal region of p53. Exp. Cell Res. 2000;255:86–94. doi: 10.1006/excr.1999.4772. [DOI] [PubMed] [Google Scholar]
- 27.Bhat UG, Raychaudhuri P, Beck WT. Functional interaction between human topoisomerase IIα and retinoblastoma protein. Proc. Natl Acad. Sci. USA. 1999;96:7859–7864. doi: 10.1073/pnas.96.14.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroll DJ, Sullivan DM, Gutierrez-Hartmann A, Hoeffler JP. Modification of DNA topoisomerase II activity via direct interactions with the cyclic adenosine-3',5'-monophosphate response element-binding protein and related transcription factors. Mol. Endocrinol. 1993;7:305–318. doi: 10.1210/mend.7.3.8387155. [DOI] [PubMed] [Google Scholar]
- 29.Tsai SC, Valkov N, Yang WM, Gump J, Sullivan D, Seto E. Histone deacetylase interacts directly with DNA topoisomerase II. Nat. Genet. 2000;26:349–353. doi: 10.1038/81671. [DOI] [PubMed] [Google Scholar]
- 30.Johnson CA, Padget K, Austin CA, Turner BM. Deacetylase activity associates with topoisomerase II and is necessary for etoposide-induced apoptosis. J. Biol. Chem. 2001;276:4539–4542. doi: 10.1074/jbc.C000824200. [DOI] [PubMed] [Google Scholar]
- 31.Kurz EU, Leader KB, Kroll DJ, Clark M, Gieseler F. Modulation of human DNA topoisomerase IIα by interaction with 14-3-3ε. J. Biol. Chem. 2000;275:13948–13954. doi: 10.1074/jbc.275.18.13948. [DOI] [PubMed] [Google Scholar]
- 32.Durrieu F, Samejima K, Fortune JM, Kandels-Lewis S, Osheroff N, Earnshaw WC. DNA topoisomerase IIα interacts with CAD nuclease and is involved in chromatin condensation during apoptotic execution. Curr. Biol. 2000;10:923–926. doi: 10.1016/s0960-9822(00)00620-5. [DOI] [PubMed] [Google Scholar]
- 33.Barker CR, Hamlett J, Pennington SR, Burrows F, Lundgren K, Lough R, Watson AJM, Jenkins JR. The topoisomerase II-Hsp90 complex: A new chemotherapeutic target? Int. J. Cancer. 2006;118:2685–2693. doi: 10.1002/ijc.21717. [DOI] [PubMed] [Google Scholar]
- 34.Wiedmer T, Zhao J, Nanjundan M, Sims PJ. Palmitoylation of phospholipid scramblase 1 controls its distribution between nucleus and plasma membrane. Biochemistry. 2003;42:1227–1233. doi: 10.1021/bi026679w. [DOI] [PubMed] [Google Scholar]
- 35.Ben-Efraim I, Zhou Q, Wiedmer T, Gerace L, Sims PJ. Phospholipid scramblase 1 is imported into the nucleus by a receptor-mediated pathway and interacts with DNA. Biochemistry. 2004;43:3518–3526. doi: 10.1021/bi0356911. [DOI] [PubMed] [Google Scholar]
- 36.Sakaguchi A, Miyaike M, Kuroda K, Nozaki N, Tanaka M, Hibino M, Fujii Y, Kato S, Kikuchi A. Epitope distribution of randomly established monoclonal antibodies against human type II DNA topoisomerases. J. Biochem. (Tokyo) 2002;132:409–416. doi: 10.1093/oxfordjournals.jbchem.a003237. [DOI] [PubMed] [Google Scholar]
- 37.Frasch SC, Henson PM, Nagaosa K, Fessler MB, Borregaard N, Bratton DL. Phospholipid flip-flop and phospholipid scramblase 1 (PLSCR1) co-localize to uropod rafts in formylated Met-Leu-Phe-stimulated neutrophils. J. Biol. Chem. 2004;279:17625–17633. doi: 10.1074/jbc.M313414200. [DOI] [PubMed] [Google Scholar]
- 38.Yamane K, Kawabata M, Tsuruo T. A DNA-topoisomerase-II-binding protein with eight repeating regions similar to DNA-repair enzymes and to a cell-cycle regulator. Eur. J. Biochem. 0000;250:794–799. doi: 10.1111/j.1432-1033.1997.00794.x. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Watt PM, Borts RH, Louis EJ, Hickson ID. The topoisomerase II-associated protein, Pat1p, is required for maintenance of rDNA locus stability in Saccharomyces cerevisiae. Mol. Gen. Genet. 1999;261:831–840. doi: 10.1007/s004380050027. [DOI] [PubMed] [Google Scholar]
- 40.Watt PM, Louis EJ, Borts RH, Hickson ID. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 41.Kametaka S, Shibata M, Moroe K, Kanamori S, Ohsawa Y, Waguri S, Sims PJ, Emoto K, Umeda M, Uchiyama Y. Identification of phospholipid scramblase 1 as a novel interacting molecule with β-secretase (β-site amyloid precursor protein (APP) cleaving enzyme (BACE)) J. Biol. Chem. 2003;278:15239–15245. doi: 10.1074/jbc.M208611200. [DOI] [PubMed] [Google Scholar]
- 42.Christensen MO, Larsen MK, Barthelmes HU, Hock R, Andersen CL, Kjeldsen E, Knudsen BR, Westergaard O, Boege F, Mielke C. Dynamics of human DNA topoisomerases IIα and IIβ in living cells. J. Cell. Biol. 2002;157:31–44. doi: 10.1083/jcb.200112023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lang AJ, Mirski SEL, Cummings HJ, Yu Q, Gerlach JH, Cole SPC. Structural organization of the human TOP2A and TOP2B genes. Gene. 1998;221:255–266. doi: 10.1016/s0378-1119(98)00468-5. [DOI] [PubMed] [Google Scholar]
- 44.Chen MH, Ben-Efraim I, Mitrousis G, Walker-Kopp N, Sims PJ, Cingolani G. Phospholipid scramblase 1 contains a nonclassical nuclear localization signal with unique binding site in importin α. J. Biol. Chem. 2005;280:10599–10606. doi: 10.1074/jbc.M413194200. [DOI] [PubMed] [Google Scholar]
- 45.Smotrys JE, Linder ME. Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 2004;73:559–587. doi: 10.1146/annurev.biochem.73.011303.073954. [DOI] [PubMed] [Google Scholar]
- 46.Silverman RH, Halloum A, Zhou A, Dong B, Al-Zoghaibi F, Kushner D, Zhou Q, Zhao J, Wiedmer T, Sims PJ. Suppression of ovarian carcinoma cell growth in vivo by the interferon-inducible plasma membrane protein, phospholipid scramblase 1. Cancer Res. 2002;62:397–402. [PubMed] [Google Scholar]
- 47.Huang Y, Zhao Q, Zhou CX, Gu ZM, Li D, Xu HZ, Sims PJ, Zhao KW, Chen GQ. Antileukemic roles of human phospholipid scramblase 1 gene, evidence from inducible PLSCR1-expressing leukemic cells. Oncogene. 2006;25:6618–6627. doi: 10.1038/sj.onc.1209677. [DOI] [PubMed] [Google Scholar]
- 48.Yokoyama A, Yamashita T, Shiozawa E, Nagasawa A, Okabe-Kado J, Nakamaki T, Tomoyasu S, Kimura F, Motoyoshi K, Honma Y, Kasukabe T. MmTRA1b/phospholipid scramblase 1 gene expression is a new prognostic factor for acute myelogenous leukemia. Leuk. Res. 2004;28:149–157. doi: 10.1016/s0145-2126(03)00189-9. [DOI] [PubMed] [Google Scholar]
- 49.Kasukabe T, Okabe-Kado J, Honma Y. TRA1, a novel mRNA highly expressed in leukemogenic mouse monocytic sublines but not in nonleukemogenic sublines. Blood. 1997;89:2975–2985. [PubMed] [Google Scholar]
- 50.Zhou Q, Ben-Efraim I, Bigcas JL, Junqueira D, Wiedmer T, Sims PJ. Phospholipid scramblase 1 binds to the promoter region of the inositol 1,4,5-triphosphate receptor type 1 gene to enhance its expression. J. Biol. Chem. 2005;280:35062–35068. doi: 10.1074/jbc.M504821200. [DOI] [PubMed] [Google Scholar]
- 51.Escargueil AE, Plisov SY, Skladanowski A, Borgne A, Meijer L, Gorbsky GJ, Larsen AK. Recruitment of cdc2 kinase by DNA topoisomerase II is coupled to chromatin remodeling. FASEB J. 2001;15:2288–2290. doi: 10.1096/fj.00-0726fje. [DOI] [PubMed] [Google Scholar]
- 52.Zhou Q, Zhao J, Al-Zoghaibi F, Zhou A, Wiedmer T, Silverman RH, Sims PJ. Transcriptional control of the human plasma membrane phospholipid scramblase 1 gene is mediated by interferon-α. Blood. 2000;95:2593–2599. [PubMed] [Google Scholar]