Abstract
The rapidly growing understanding of human genetic pathways, including those that mediate cancer biology and drug response, leads to an increasing need for extensive and reliable mutation screening on a population or on a single patient basis. Here we describe s-RT-MELT, a novel technology that enables highly expanded enzymatic mutation scanning in human samples for germline or low-level somatic mutations, or for SNP discovery. GC-clamp-containing PCR products from interrogated and wild-type samples are hybridized to generate mismatches at the positions of mutations over one or multiple sequences in-parallel. Mismatches are converted to double-strand breaks using a DNA endonuclease (Surveyor™) and oligonucleotide tails are enzymatically attached at the position of mutations. A novel application of PCR enables selective amplification of mutation-containing DNA fragments. Subsequently, melting curve analysis, on conventional or nano-technology real-time PCR platforms, detects the samples that contain mutations in a high-throughput and closed-tube manner. We apply s-RT-MELT in the screening of p53 and EGFR mutations in cell lines and clinical samples and demonstrate its advantages for rapid, multiplexed mutation scanning in cancer and for genetic variation screening in biology and medicine.
INTRODUCTION
Screening for genetic changes to unveil molecular attributes of human specimens is important for a variety of medical applications, including genotyping for inherited disorders, prediction of the pathologic behavior of malignancies, identification of cancer biomarkers and can affect treatment decisions for individual patients (1–3). For example, mutations in genes like EGFR can profoundly influence chemotherapeutic response in lung cancer (2–5) and the response is modulated by mutations in other genes of the same signaling pathway [e.g. K-ras, HER2, ErbB-3 (1,6)]. Therefore there is a need for efficient and high-throughput mutation screening of multiple genes along identified signal transduction pathways in tumor samples. Because a large portion of cancer-causing genetic changes remains unknown and can occur in numerous positions along tumor suppressor genes (e.g. p53, ATM, PTEN) mutation scanning rather than detection of specific mutations is frequently required for molecular cancer profiling.
Sequencing is often considered the gold standard for comprehensive mutation analysis. Multi-capillary electrophoresis, re-sequencing arrays or pyrosequencing provide platforms for highly parallel genetic analysis (7–13). However, the expense associated with these techniques is currently high both for instrumentation and for running-costs. Since somatic mutations for most genes are relatively rare events it can be inefficient to scan for mutations using expensive approaches that in several cases provide unnecessary data (14,15). Another issue with direct sequencing or re-sequencing arrays is the difficulty in detecting a small fraction of mutated alleles in the presence of a high excess normal alleles, which is frequently the case with clinical cancer samples (16). As a less expensive alternative, rapid pre-screening methods such as SSCP, DGGE, dHPLC, CCM, CDCE or HR-melting are widely utilized to identify DNA fragments that contain mutations prior to performing full sequencing (14,16–20). Enzymatic mutation detection based on mismatch scanning enzymes like MutY, TDG or T4 endonuclease VII for mutation pre-screening has also been employed (21–25), albeit with modest success since these enzymes cannot detect all possible mutations and deletions (22) and some of them have substantial activity on homoduplex DNA (16). Recently an enzymatic mutation scanning method based on the Surveyor™ (CELI/II) nuclease (26,27) combined with dHPLC or gel electrophoresis detection was introduced that shows satisfactory selectivity and reliability (1% mutant to wild-type alleles is detectable) while it also identifies all base substitutions and small deletions that are important to cancer (17,28) or to biotechnology and plant genetic applications [TILLING method (29–34)]. While reliable, the use of dHPLC for examining Surveyor™-generated DNA fragments is a slow endpoint detection method restricted to examining a single DNA fragment at a time and the resulting DNA fragments cannot be sequenced. This limits analysis of cancer specimens when numerous samples or genetic regions need to be screened.
We introduce a new approach that enables Surveyor™ to scan for mutations over one or several PCR products simultaneously and selectively amplify and isolate the mutation-containing DNA fragment(s) via linker-mediated PCR. By selectively amplifying mutation-containing DNA from wild-type fragments, the present approach de-couples enzymatic mutation scanning from the endpoint detection step. As a result, following enzymatic action on mismatches any chosen DNA detection method (real-time PCR, gel/capillary electrophoresis, microarray-based detection) can potentially be used to identify the mutated DNA fragments in a simplex or multiplex fashion. Here we utilize real-time PCR coupled with melting curve analysis (Surveyor™-mediated Real Time Melting, s-RT-MELT) to validate the new technology. We demonstrate that this approach increases the mutation scanning throughput by 1–2 orders of magnitude when several (>100) samples are to be pre-scanned for mutations, enables mutation scanning over several PCR fragments simultaneously and mutation-positive samples can be directly sequenced when somatic mutations are at a low-level (∼1–10% mutant-to-wild-type ratio) in surgical cancer specimens.
METHODS
Samples and controls
Genomic DNA from cell lines with defined mutations in p53 exons, DU145 (exon 6), SW480 (exons 8 and 9), DLD1 (exon 7) and BT483 (exon 7) was extracted from cell lines purchased from the American Type Culture Collection (ATCC), or purchased as purified DNA when available. Surgical colon and lung cancer tumor samples were obtained from the Massachusetts General Hospital Tumor Bank following Internal Review Board approval. DNA from the EGFR mutation-positive cell lines A549, HCC827, H1975 and LU011 and from formalin-fixed-paraffin-embedded lung cancer samples were obtained from the Lowe Center for Thoracic Oncology, Dana Farber Cancer Institute following Internal Review Board approval. We isolated genomic DNA using DNeasy™ Tissue Kit (Qiagen).
PCR with primers containing 5′-GC-clamp and 5′-M13
Sequences for the 5′M13 and GC-clamp portion of the primers, as well as the gene-specific portion of the primers used in this investigation are listed in Supplementary Table 1. The M13f and GC-clamp sequence was added to the 5′ end of forward and reverse gene-specific primers respectively, or vice versa. Twenty microliter PCR reactions were performed from genomic DNA with final concentrations of reagents as follows: 1X JumpStart™ buffer (Sigma), 0.2 mM each dNTP, 0.2 μM forward and reverse primer, 1X JumpStart™ Taq polymerase (Sigma). PCR cycling was done on a Perkin Elmer 9600 PCR machine. The cycling conditions were: 94°C, 90 s; (94°C, 20 s/65°C, 20 s/68°C, 1 min) × 10 cycles, with annealing temperature decreasing 1°C/cycle, touch-down PCR; (94°C, 20 s/55°C, 20 s/68°C, 1 min) × 30 cycles; 68°C, 5 min. This PCR program was linked to a program for denaturation and re-annealing of the PCR product over 10 min.
Treatment of cross-hybridized sequences with the Surveyor™ endonuclease
Five-microliter PCR product (300–500 ng) was mixed with 0.5 μl Enhancer™ and 0.5 μl Surveyor™ (Transgenomic) and incubated at 42°C for 20 min followed by adding 0.5 μl Stop-solution, as per manufacturer's protocol. The inactivated Surveyor™-digested product was purified with PCR QiaQuick™ purification kit (Qiagen) and eluted in 35 μl water. In some experiments, the PCR product was mixed with an approximately equal amount of PCR product from wild-type DNA prior to forming cross-hybridized sequences, to facilitate detection of homozygous mutations.
Addition of polyA-tail on the 3′-end
Following purification of the Surveyor™-treated sample, Poly-adenine ‘tail’ was added to the 3′-ends of DNA fragments. For each reaction, we added 5 μl purified surveyor-digested PCR product to a final volume of 20 μl with final concentration of 1X reaction buffer-4, 1X CoCL2, 0.2 mM dATP, 4 U Terminal Transferase (New England Biolabs). The reaction was incubated at 37°C for 10 min and inactivated by heating at 75°C for 10 min.
Real-time PCR, melting curve analysis and dHPLC
The real-time PCR amplification was performed using Titanium-Taq™ polymerase (BD-Biosciences - Clontech) in a Smart Cycler (Cepheid) real-time PCR machine. For each reaction, we added 0.5 μl polyA-tailed DNA to a final volume of 20 μl with final concentration of 1X Titanium buffer, 0.2 mM each dNTP, 0.1 × LCGreen (Idaho Technologies), 0.2 μM m13f primer, 0.2 μM oligodT-anchor mix GACCACGCGTATCGATGTCGACTTTTTTTTTTTTTTTTV
[V represents A, C and G each oligodT-anchor concentration is 0.067 μM, as per RACE protocol (35)], 1 × Titanium™ polymerase (Clontech— BD Biosciences). The thermocycling program was as following: 1 cycle of 94°C for 2 min, 25 cycles of 94°C for 15 s, 55°C for 20 s and 68°C for 30 s for reading fluorescence. Temperature titration was performed using different denaturation temperatures, 94–82°C to experimentally determine conditions that selectively enable mutation-containing fragments to amplify.
The real-time PCR step was immediately followed by real-time differential melting curve analysis using the SmartCycler™ machine. DNA melting was performed immediately following PCR on the Smart Cycler I machine. Samples were heated from 70°C to 95°C at 0.1°C/s. Differential fluorescent intensity curves (−dF/dT) were smoothed and used for identification of melting peak (s).
Altenatively, real-time PCR products were examined via dHPLC chromatography on a WAVE™ system (Transgenomic). Mutation-positive PCR products were purified via PCR purification kit (Qiagen) and sequenced using the M13f primer. All experiments were repeated at least three times in independent runs from genomic DNA.
Real-time PCR and melting using the OpenArray™ platform
The OpenArray™ high-throughput, massively parallel real-time PCR platform (36) (BioTrove) was tested for compatibility with s-RT-MELT. Forty-eight samples of p53 exon 8 PCR products were generated from 48 different lung adenocarcinoma samples and mutation-containing cell lines and processed via the hybridization and enzymatic steps of s-RT-MELT. Real-time PCR in the OpenArray™ platform was performed with the LightCycler FastStart™ DNA Master SYBR Green™ I (Roche) using 0.2 μM M13f and 0.2 μM oligodT-anchor-mix as primers pre-positioned on the array through-holes (36) and polyA-tailed DNA as template. The cycling conditions were as follows: 1 cycle at 94°C for 2 min, 25 cycles of 90°C for 15 s, 55°C for 20 s and 68°C for 30 s for reading fluorescence using a high sensitivity imaging camera (36). The real-time PCR step was immediately followed by real-time differential melting curve analysis. Raw data were exported in Excel software for further analysis. The OpenArray™ experiment was repeated twice at the company's headquarters.
Prediction of melting temperatures
To estimate Tm,min, the PCR denaturation temperature below which PCR is not efficient it was assumed, as an initial approximation, that >95% hypochromicity must be present for PCR to work (i.e. any given sequence must be completely denatured, otherwise it re-forms immediately when temperature is lowered in the reaction and inhibits primer binding). The percent melting (hypochromicity)-versus-temperature relations for GC-clamp-containing PCR products and Surveyor™ activity-generated products were estimated using the POLAND algorithm (37), and the thermodynamic parameters determined by Blake and Delcourt for 75 mM NaCl in the solution (38) were used. In order to force agreement at a single point, predicted and observed values for a p53 exon 8 sequence containing a short GC-clamp were normalized at 88°C. This shift accounts for the influence on Tm,min of NaCl and Mg++ content in the reaction, the presence of the SYBR-GREEN/LC-GREEN dyes and the proprietary composition of PCR buffers. The Tm,min of all other PCR products was then estimated using these semi-empirically determined parameters.
RFLP-based verification of low-level mutation detected in codon 273, p53 exon 8
The ‘enriched PCR’ method by Behn et al. (39) was used to sequence codon 273 mutation of p53 exon 8 from sample CT20 and wild-type samples. In addition, a second method [Amplification via Primer-Ligation At The Mutation (40,41)] was used to distinguish mutant and wild-type samples by virtue of the de novo Nla-III site generated in the mutant sample by the p53 codon 273 G > A mutation.
RESULTS
Overview of the s-RT-MELT assay
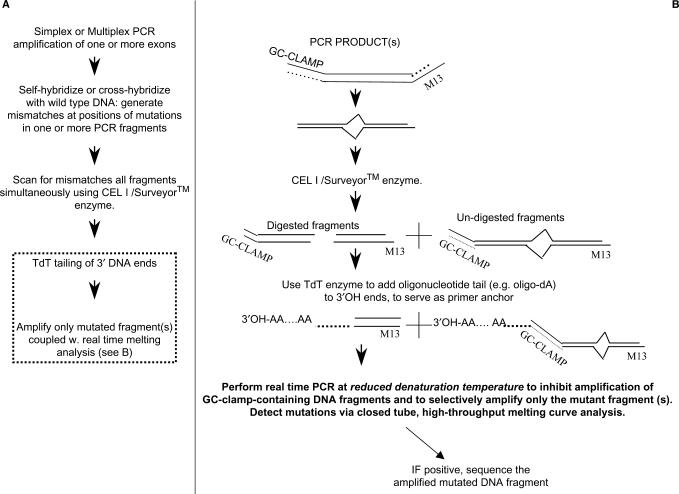
The s-RT-MELT assay converts PCR fragments generated at positions of mutations by the Surveyor™ enzyme to fully amplifiable sequences that enable selective PCR amplification in a subsequent quantitative PCR detection method. Following denaturation and re-annealing of PCR products that leads to formation of cross-hybridized sequences at the positions of mutations (Figure 1A) the sample is exposed to Surveyor™ endonuclease that recognizes base pair mismatches or small loops with high specificity (28) and generates a break on both DNA strands 3′ to the mismatch. The resulting DNA fragments participate in a terminal transferase (TdT) reaction that leads to polynucleotide ‘tailing’ (sequential addition of adenine, poly-A-tail) at the 3′-ends. A real-time PCR reaction is subsequently performed using adjusted conditions that enable selective amplification of the mutant-only fragments, followed by real-time melting curve analysis for identification of mutations in the presence of SYBR-GREEN™ or LC-GREEN™ DNA dye.
Figure 1.
s-RT-MELT for rapid mutation scanning using enzymatic selection and real-time DNA-melting. (A) General outline of the approach. The dotted line contains the new steps involved in s-RT-MELT relative to previous approaches, i.e. the addition of a 3′-polynucleotide tail followed by real-time PCR that enables selective amplification of the Surveyor™-cut sequences and real time melting curve analysis. (B) Detailed outline of the procedure used to selectively amplify the mutation-containing fragments in s-RT-MELT.
To enable selective amplification of the mutation-containing fragments in the real-time PCR step, modified primers are employed for the original amplification from genomic DNA (Figure 1B). The forward primer contains a region specific to the target gene and a high melting domain (GC-clamp), while the reverse primer contains a region specific to the target gene and an M13 tail (or vice versa). Following the TdT tailing reaction, the M13 primer is used for real-time PCR in conjunction with a primer that binds to the poly-A tail. The denaturation temperature of the real-time PCR reaction is lowered to enable PCR amplification only for fragments that do not contain GC-clamps. Because the PCR products that escape digestion by Surveyor™ contain GC-clamps (Figure 1B), these fragments do not amplify efficiently during PCR, thereby enabling selective amplification of Surveyor™-selected fragments, i.e. an effective ‘purification’ of mutation-containing fragments. The subsequent closed-tube melting curve analysis enables clear separation of true mutant sequences from PCR dimers or other artifacts.
Because s-RT-MELT does not require size-separation for identification of enzymatically generated fragments, more than one sequence can be scanned in parallel for unknown mutations in a single-tube reaction of Surveyor™. This simple procedure enables the specificity of the Surveyor™ enzyme to be combined with the throughput and convenience of real-time PCR for rapid mutation scanning. Finally, because the amplified mutated sequences contain defined primers at their ends, direct sequencing of enzymatically selected PCR products is readily possible following the real-time melting step, enabling sequencing of low-level mutations identified by Surveyor™.
Detection of p53 exon 8 mutations using s-RT-MELT
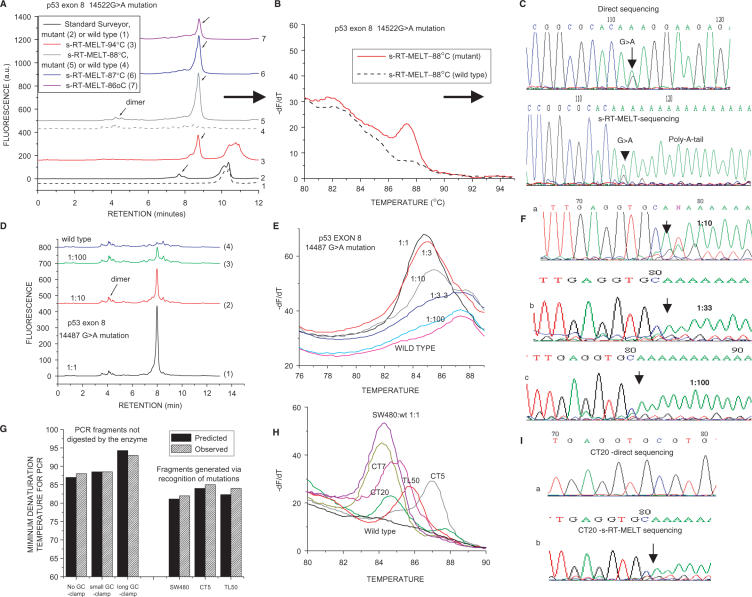
To provide initial proof of principle for unknown mutation scanning using s-RT-MELT we utilized cell lines and tumor samples containing sequencing-identified mutations at several positions of p53 exon 8. Figure 2A depicts dHPLC chromatograms of the products obtained using a sample containing a p53 exon 8 G > A mutation or a wild-type sample. The standard Surveyor™-dHPLC approach (28) was first employed to identify the mutation following PCR amplification of exon 8 from genomic DNA. The resulting dHPLC traces contain a single product for the wild-type and two products for the mutation-containing sequences (Figure 2A, curves 1 and 2, respectively). Next, s-RT-MELT was used to screen the same p53 exon 8 sequence. Following PCR amplification with GC/M13-modified primers we cross-hybridized PCR products and exposed them to Surveyor™ and TdT tailing. The subsequent real-time PCR was run at different denaturation temperatures and the products were examined either via dHPLC or via real-time melting-curve analysis. At the standard denaturation temperature of 94°C the mutation-containing sample contains two peaks, corresponding to the anticipated amplification of both Surveyor™-digested and un-digested fragments (Figure 2A, curve 3). However, when the PCR denaturation temperature is lowered (e.g. 86–88°C) a single PCR product is generated for the mutant sample, while the wild-type sample demonstrates no product (Figure 2A, curves 4–7). In Figure 2B, real-time differential melting curves for the PCR reaction run at 88°C are depicted. A peak corresponding to the PCR product from the mutant sample is again clearly evident, which is absent in the wild-type sample. Finally, Figure 2C depicts sequencing of the s-RT-MELT-generated PCR fragment, as well as the direct sequencing from genomic DNA. The G > A mutation is evident in both samples. In the s-RT-MELT product the anticipated addition of the poly-A tail at the 3′-position next to the mutation is illustrated.
Figure 2.
Detection of p53 exon 8 mutations using s-RT-MELT. (A) dHPLC chromatograms of the products obtained using the standard Surveyor™-dHPLC approach (28), versus the new technology on a sample containing a p53 exon 8 14522G > A mutation or a wild-type sample. Curves 1 and 2: Standard Surveyor™-dHPLC on wild- type and mutant samples, respectively. Curves 3–7: s-RT-MELT products when real-time PCR is performed at different denaturation temperatures. (B) Real-time differential melting curves for a PCR denaturation temperature of 88°C. (C) Sequencing of the s-RT-MELT-generated PCR fragment for a PCR denaturation temperature of 88°C. Direct sequencing of the same PCR product from genomic DNA is also depicted. D. dHPLC chromatograms of s-RT-MELT products obtained using serial dilution of DNA from SW480 cells in wild-type DNA. Real-time PCR was performed at 88°C denaturation temperature. (E) Melting curve analysis of the s-RT-MELT products obtained using serial dilution of SW480 in wild-type DNA at 88°C denaturation temperature. (F) s-RT-MELT-sequencing of s-RT-MELT products obtained using serial dilution of SW480 in wild-type DNA. (G) Predicted-versus-observed minimum denaturation temperatures for generation of s-RT-MELT products following real-time PCR. The influence of the GC-clamp length (no GC-clamp; 26-nt GC-clamp; or 117-nt GC-clamp), and the position of the mutation along the sequence are depicted. (H) s-RT-MELT screening of unknown p53 exon 8 mutations of 48 colon and lung tumor DNA: representative results from mutation-positive samples and wild-type sample are depicted. (I) Sequencing of low-level p53 exon 8 mutation (colon tumor sample CT20) by direct sequencing and by s-RT-MELT sequencing.
To examine the selectivity of s-RT-MELT, dilutions of mutant to wild-type DNA were performed using DNA from SW-480 cells that harbor a p53 exon 8 14487G > A homozygous mutation. The real-time PCR reaction was again performed at 88°C and mutant-to wild-type ratios of ∼1–10% were distinguished from the wild-type using either dHPLC (Figure 2D) or melting curve analysis (Figure 2E). In these samples, direct di-deoxy-sequencing could not identify a mutation if the ratio of mutant-to-wild-type was <∼30–40% (data not shown). On the other hand, sequencing of s-RT-MELT products was possible including the lower dilutions (Figure 2F). sRT-MELT sequencing generated traces with poly-A tails depicting the presence and the position of the mutation, although the exact nucleotide change was less clear than the one in exon 5 (i.e. the position ±1 base from the mutation might also be confused to be a mutation). The reason for this ±1 base ambiguity of the exact position of the mutation can be probably understood. The PCR performed following poly-A tail addition contains an equimolar mixture of three reverse primers (3′ ending in V = G, A or C). Depending on the exact nucleotide at the mutation, the correct primer should in theory be preferred, while the other two primers should not allow efficient polymerase extension due to the mismatched 3′-end. However, in practice this ‘allele-specific PCR’ step occasionally allows 3′-mismatched primer extension, enabling more than one version of the primer to amplify over the position of the mutation, or alternatively the incorporation of the poly-A tail may occur ±1 base from the exact position of the mutation. We conclude that in certain cases sRT-MELT indicates the position of the mutation to within 1 base, while in others (e.g. p53 exon 5) it indicates the position ‘and’ the actual nucleotide change.
Next, p53 exon 8 was amplified using DNA from a group of 48 surgical lung adenocarcinoma samples and s-RT-MELT was used for the screening of unknown mutations via melting curve analysis. Mutations at different positions along exon 8 were present in several of these clinical samples, as indicated by the shift in melting profiles obtained (Figure 2H) and subsequently verified via sequencing. In this set of samples, sRT-MELT-sequencing detected a low-level mutation on a colon cancer specimen (CT20) that direct sequencing failed to identify (Figure 2I). As with Figure 2F, sequencing of sample CT20 indicated the position of poly-A tail addition to within one base, but the actual nucleotide change was difficult to identify. To exclude the possibility for a false positive, two independent RFLP-based methods were used to verify the presence of the mutation. Thus, since the position of poly-A tail addition was known (Figure 2I, codon 273 of p53 exon 8) the mismatched primer approach by Behn et al. (42) was used to introduce an MluI restriction site for the wild-type p53 sample but not for the codon 273 mutants. Subsequent restriction with MluI enzyme followed by PCR generated a product with a 14487G > A mutation for the CT20 sample but not for the wild-type sample (Supplementary Figure 1, Frame A). As an additional verification for the low-level CT20 mutation, we observed that G > A mutation generates a de-novo Nla-III site at the position of the mutation. Accordingly, we applied ‘Amplification via Primer-Ligation At The Mutation’, a method that we described previously (40,41) to ligate a primer at the Nla-III-digested site, and preferentially amplified the mutant fragment in a second PCR. The sequenced PCR product identified again the 14487G > A mutation (Supplementary Figure 1, Frame B). In conclusion, sRT-MELT identified correctly a p53 codon 273 low-level mutation on CT20 that was missed by regular sequencing. This is very significant as p53 exon 8 mutations at codon 273 have been associated with bad prognosis in cancer (43,44).
Table 2 of Supplementary Data depicts a good agreement between standard Surveyor screening, s-RT-MELT screening and di-deoxy-sequencing, except for the low-level mutation discovered on sample CT20 via s-RT-MELT. s-RT-MELT-sequencing traces for two samples with p53 exons 6 and 7 mutations are also depicted.
The data in Figures 2A–D and H indicate a lack of substantial PCR amplification at denaturation temperatures ⩽88°C for fragments containing the GC-clamp and a selective amplification of the mutation-containing fragments for several different mutation positions on p53 exon 8. To estimate the influence of the GC-clamp length on PCR efficiency versus temperature and the PCR amplification of fragments generated for mutations lying at different positions along the sequence, a calculation based on the POLAND algorithm (37) was performed. The predicted minimum temperatures for substantial PCR amplification were then plotted versus the experimentally observed values. Three possibilities were simulated, no GC-clamp, 26 nucleotides (nt) GC-clamp and 117-nt GC-clamp. DNA fragments corresponding to mutations at several positions along exon 8 were also simulated and compared to the experimentally observed minimum temperatures for generating a PCR product for three samples that contained mutations at different positions along p53 exon 8 (SW480, CT5 and TL50). The results (Figure 2G) indicate agreement to within ∼1.0°C between theoretical prediction and experimental observation. For denaturation temperatures in the region 85–88°C in combination with a 26-nt GC-clamp all the available mutations on p53 exon 8 are predicted to result in selective amplification of the mutation-containing fragment and inhibition of the GC-clamp-containing fragment. This prediction is consistent with the experimental results obtained from PCR temperature-titration experiments (Figure 2G). The developed calculation algorithm can thus be used to predict the appropriate PCR denaturation temperature for additional PCR fragment/GC-clamp combinations.
Detection of p53 exons 5–9 mutations and EGFR mutations in clinical samples
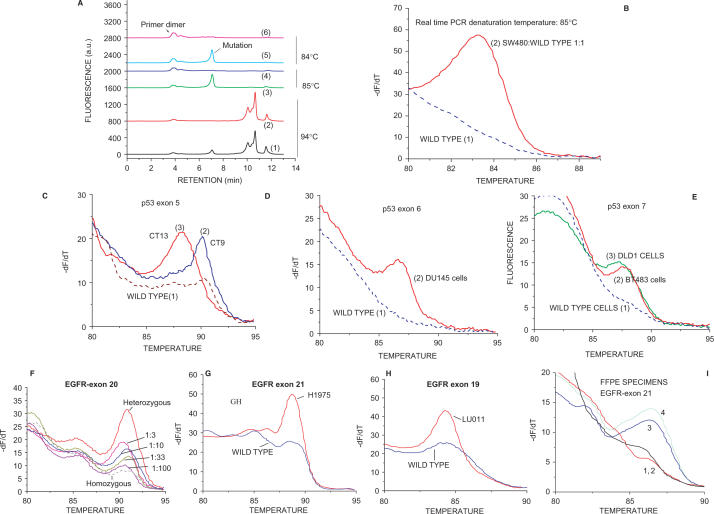
As a further validation for s-RT-MELT, we utilized the method to identify mutations in additional p53 exons. Figure 3A depicts the chromatographs obtained when a 1:1 mixture of DNA from SW-480 cells (homozygous mutation at p53 14686 C > T exon 9) and from wild-type cells was screened. The real-time PCR reaction was performed at different denaturation temperatures and the products were examined both via dHPLC and via melting curve analysis for comparison. As also observed for p53 exon 8, at 94°C denaturation temperature both the Surveyor™-digested and the undigested PCR products are amplified during real-time PCR (Figure 3A, curves 1 and 2, mutant and wild-type, respectively). By lowering denaturation temperature to 85°C or 84°C, a single PCR product is obtained from the mutant while no product, other than primer dimer, is obtained by the wild-type sample (Figure 3A, curves 3–6). Figure 3B depicts the melting curves obtained following real-time PCR at 85°C denaturation temperature for the mutant and wild-type samples. s-RT-MELT was subsequently applied in the same manner to screen for p53 mutations in exons 5–7 from cell lines and surgical colon samples harboring sequencing-identified mutations including a single-base frameshift mutation in exon 7 (listed in Supplementary Table 2). The melting curves from mutant and wild-type samples in p53 exons 5–7 are depicted in Figure 3C–E. The data indicate that results similar to those obtained for p53 exon 8 are also obtained for p53 exons 5, 6, 7 and 9.
Figure 3.
Detection of p53 exons 5–9 mutations and EGFR mutations in clinical samples and cell lines. (A) dHPLC chromatograms of the s-RT-MELT p53 exon 9 products obtained using DNA from wild type cells, or a 1:1 mixture of SW480 and wild-type cells, at various real-time PCR denaturation temperatures. Curves 1 and 2: mutant and wild- type s-RT-MELT products, respectively, at 94°C; Curves 3 and 4: mutant and wild-type s-RT-MELT products, respectively, at 85°C; Curves 5 and 6: mutant and wild-type s-RT-MELT products, respectively, at 84°C. (B) Melting curves obtained following s-RT-MELT of p53 exon 9 at 85°C denaturation temperature for wild-type DNA, or a 1:1 mixture of SW480 and wild-type DNA. (C) Melting curves obtained following s-RT-MELT of p53 exon 5 for colon cancer surgical samples CT9, CT13 and wild-type samples, curves 2, 3 and 1, respectively. (D) Melting curves obtained following s-RT-MELT of p53 exon 6 using DNA from cell line DU145 and wild-type samples. (E) Melting curves obtained following s-RT-MELT of p53 exon 7 using DNA from mutant cell lines DLD1, BT483 and wild-type samples. (F) Melting curves obtained following s-RT-MELT of EGFR exon 20 for serial dilution of a DNA sample containing a heterozygous single nucleotide polymorphism (SNP) into a homozygous DNA sample. (G) Melting curves obtained following s-RT-MELT of EGFR exon 21 for lung tumor cell line H1975 (L858R mutation) and wild-type samples. (H) Melting curves obtained following s-RT-MELT of EGFR exon 19 for lung tumor cell line LU011 (L747-E749 deletion) and from wild-type samples. (I) Melting curves obtained following s-RT-MELT of EGFR exon 21 for FFPE lung tumor samples #2, #21, #6 and #10, curves 3, 1, 2 and 4, respectively.
Detection of mutations in EGFR exons 18–21 is of particular clinical interest as these alterations can modulate response to EGFR inhibitors in lung adenocarcinoma patients (2,3). Figures 3F, G and H depict the application of s-RT-MELT for screening DNA from lung cancer cell lines that harbor dHPLC-identified alterations in EGFR exons 19–21, including a two-codon deletion (del L747-E749, exon 19). The ability of s-RT-MELT for detecting low-level EGFR mutations was evaluated by performing DNA dilutions of a heterozygous EGFR exon 20 into a homozygous sample. A 1–10% mutant-to-wild-type ratio was detectable in this dilution experiment (Figure 3F). Finally, the application of s-RT-MELT in detecting mutations in DNA from formalin-fixed paraffin-embedded (FFPE) samples was examined by screening four clinical FFPE lung adenocarcinoma specimens. Two of these samples were known to harbor EGFR exon 21 mutations (L858R), while the other two were negative for mutations when independently evaluated via dHPLC (28). Figure 3I demonstrates the identification of the mutational status of these samples via s-PCR-MELT.
Multiplex s-RT-MELT or OpenArray™-based s-RT-MELT increases the throughput of mutation scanning
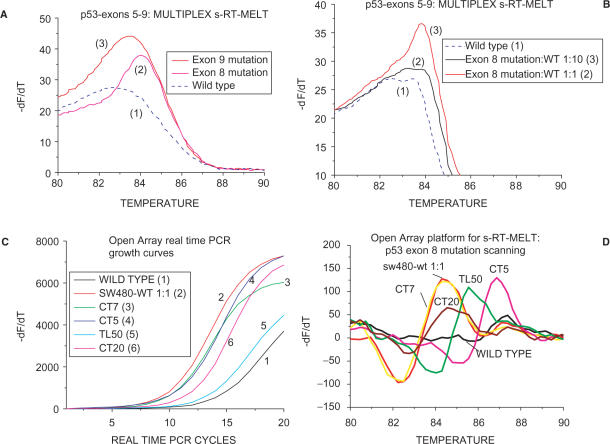
A significant potential advantage of enzymatic mutation scanning is the ability to screen several sequences simultaneously for mutations. To demonstrate that s-RT-MELT can be used for parallel scanning of mutations in several PCR products, we mixed equimolar amounts of PCR products from p53 exons 5–9 containing mutations either in exon 8 or in exon 9. We then formed ‘cross-hybridized sequences’ and screened the mixture for mutations in p53 exons 5–9 in a single tube using s-RT-MELT, as depicted in Figure 1A. Following real-time PCR and melting curve analysis, the exon 8 or exon 9 mutants were clearly distinguished from the wild-type sample (Figure 4A, curves 1–3). Next, the mutant exon 8 DNA sample was first diluted 10-fold into wild-type exon 8 and the equimolar mixture of p53 exons 5–9 was prepared and screened again in a single tube via s-RT-MELT. The exon 8 mutation was again distinguished from the wild-type mixture of exons (Figure 4B, curves 1–3). Since >80% of p53 mutations in human tumors are encountered in exons 5–9 (45), the multiplex single-tube s-RT-MELT reaction could be used to identify most p53 mutations encountered in clinical tumor samples. Combined with multiplex PCR directly from genomic DNA, this approach could result to a convenient, high-throughput method for mutation scanning.
Figure 4.
Multiplex s-RT-MELT or OpenArray™-based s-RT-MELT. (A) Melting curves obtained following multiplex s-RT-MELT for mixture of p53 exons 5–9 (exon 8 mutation, curve 2) or exon 9 mutation (curve 3) or wild-type (curve 1). (B) Melting curves obtained following multiplex s-RT-MELT for mixture of p53 exons 5–9 (exon 8 mutation, curve 3) or 10-fold diluted into wild-type exon 8 mutation (curve 2) and wild-type (curve 1). (C) OpenArray™ based s-RT-MELT PCR growth curves for p53 exon 8 using DNA from lung and colon surgical specimens and cell lines. (D). Melting curves obtained following OpenArray™ based s-RT-MELT of p53 exon 8 using DNA from lung and colon surgical specimens and cell lines.
By adopting a real-time PCR platform as endpoint detection for s-RT-MELT, the throughput for mutation scanning increases drastically over other mutation pre-screening approaches that utilize dHPLC, or capillary and gel electrophoresis. To demonstrate better this point, a highly parallel nano-technology platform was adopted for the real-time PCR step of s-RT-MELT that enables an array of 3072 nl volume real-time PCR reactions (OpenArray™ system) to be carried-out simultaneously followed by differential melting curve analysis (36). As a proof of principle of the compatibility of s-RT-MELT with OpenArray™, p53 exon 8 PCR products were generated from 48 different lung adenocarcinoma samples and mutation-containing cell lines and processed via the hybridization and enzymatic steps of s-RT-MELT. The 48 samples were each dispensed in 10 replicate nano-liter volume reactions on OpenArray™ plates pre-fabricated to contain the appropriate primers and amplified in 3072 real-time PCR reactions using a denaturation temperature of 90°C in the presence of SYBR-GREEN I dye. Melting curves were subsequently obtained using the OpenArray™ melting curve analysis mode. The PCR growth curves and smoothed differential melting curves obtained distinguish clearly the mutation-containing samples from wild-type samples (Figure 4C and D, representative results from 3072 reactions). Furthermore, identification of mutation-containing samples is in good agreement between the conventional and the nano-technology platforms (Figure 4D versus Figure 2H). These data indicate that s-RT-MELT is compatible with high-throughput nano-technology detection formats and reiterates the advantage of de-coupling enzymatic selection from the detection step. Comparison of the throughput using conventional pre-screening method (dHPLC or dHPLC/Surveyor™) to s-RT-MELT (Table 1) indicates that s-RT-MELT is 1–2 orders of magnitude faster when a large number of samples (>100) are screened for mutations. If the multiplex s-RT-MELT format is adopted, the throughput can increase further.
Table 1.
Comparison of throughput in mutation scanninga. Plus the ability to sequence and identify low-level somatic mutations
| One sample | 16 samples (e.g. Cepheid QRT-PCR machine) | 96 samples (96-well QRT-PCR machine) | 384 samples (ABI QRT-PCR machine) | 3,072 samples (OpenArray™) | Detects low-level mutations (1–10% mutant-to-wild type) | Nucleotide change and/or position of the mutation | |
|---|---|---|---|---|---|---|---|
| dHPLC/Surveyorb | 0.5 h | 3-4 h | 24 h | 96 h | 768 h | Yes | No |
| s-RT-MELTc | 1.5 h | 2.0 h | 3.0 h | 4.0 h | 17 h | Yes | Yes |
aPost-PCR treatment time to accomplish pre-screening for unknown mutations.
bSurveyor™ treatment requires ∼15–20 minutes. The dHPLC screens one sample at a time (12-15 minutes/sample including wash).
cAn additional half hour for every batch of 96 samples purified via 96-sample Qiagen purification kit, following Surveyor™ treatment, was accounted for s-RT-MELT.
DISCUSSION
The intrinsic potential of enzymatic mutation scanning for parallel identification of mutations can, in principle, be very high since the enzyme operates on numerous distinct mismatch-containing sequences on a molecule-to-molecule basis thus providing highly parallel mutation scanning. However, in the past the selectivity of the enzymes used and the endpoint detection method has limited the realization of this potential. Here we enabled Surveyor™, an endonuclease that recognizes selectively mismatches formed by mutations and small deletions following ‘cross-hybridized sequence’ formation, to generate mutation-specific DNA fragments that are amplified and screened via differential melting curve analysis. The replacement of size-separation methods (capillary/gel electrophoresis, dHPLC) by real-time PCR technology as the endpoint detection platforms and the ability to scan more than one sequences in parallel result in a highly increased throughput for s-RT-MELT while retaining the ability to detect diverse mutations at low-levels.
Cel I/II endonucleases have also been known to have exonuclease activity on 5′ DNA-ends (26,27). For this reason, s-RT-MELT was designed to attach an oligonucleotide linker to the 3′-DNA ends via terminal transferase (TdT) instead of using the 5′-DNA ends. The exonuclease activity also tends to degrade the attached 5′-GC-clamps in s-RT-MELT, thereby eliminating their influence in reducing amplification of un-digested fragments. We found that if exposure of DNA ‘cross-hybridized sequences’ to Surveyor™ is limited to 15–20 min, the substantial degradation of 5′-GC-clamps is avoided. For multiplexing mutation detection using several PCR products simultaneously, the size of the GC-clamp on each PCR amplicon may need to be individually adjusted to ensure that mutations along all sequence positions of the PCR products included in the mixture can be screened at a single real-time PCR temperature and that undigested fragments do not amplify. The calculational tools developed in this work can be used to guide the individual design of GC-clamps. s-RT-MELT detects heterozygous SNPs as well as mutations. As with other mutation pre-screening techniques, the presence of a SNP concurrently with a mutation might be difficult to identify without performing sequencing. Because SNPs occur at fixed positions, melting peaks originating from SNPs have a reproducible pattern and melting temperatures (46,47) thus in many cases they should be distinguishable from mutations. Finally, it is noteworthy that s-RT-MELT is a general methodology that may also be applied to isolate mutations using mismatch-cutting enzymes other than Surveyor™ when enzymes with satisfactory properties for mutation detection become available. Detection platforms other than real-time PCR/melting (e.g. DNA microarray-based) may also be envisioned following enzymatic mutation selection.
In summary, we developed a new method for rapid mutation scanning, s-RT-MELT that utilizes the Cel I/II (Surveyor™) and terminal deoxy-nucleotide transferase (TdT) enzymes to isolate and amplify mutation-containing DNA fragments without the requirement of DNA size-dependent techniques. Besides enabling highly increased throughput, multiplexed mutation screening and direct sequencing of the identified mutant DNA fragments, s-RT-MELT also retains the advantages of the Surveyor endonuclease over alternative pre-screening methods, such as reliability and identification of genetic alterations present at low (1–10%) fractions in the sample. s-RT-MELT provides a significant advancement in unknown mutation scanning in cancer research and diagnostics as well as for general medical, biological and biotechnology applications.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The assistance of Mohamet Miri and Frank Haluska, MD in obtaining tissue specimens from the Massachusetts General Hospital Tumor Bank is gratefully acknowledged. This work was supported by NCI grants 1 R21 CA111994-01 and CA 115439-01, by training grant 5 T32 CA09078 (JL) and by the Joint Center for Radiation Therapy Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Baselga J. Targeting tyrosine kinases in cancer: the second wave. Science. 2006;312:1175–1178. doi: 10.1126/science.1125951. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, et al. EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 4.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 5.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl Acad. Sci. USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int. J. Cancer. 2006;118:257–262. doi: 10.1002/ijc.21496. [DOI] [PubMed] [Google Scholar]
- 7.Kheterpal I, Mathies RA. Capillary array electrophoresis DNA sequencing. Anal. Chem. 1999;71:31A–37A. doi: 10.1021/ac990099w. [DOI] [PubMed] [Google Scholar]
- 8.Mitchelson KR. The application of capillary electrophoresis for DNA polymorphism analysis. Methods Mol. Biol. 2001;162:3–26. doi: 10.1385/1-59259-055-1:3. [DOI] [PubMed] [Google Scholar]
- 9.Hacia JG. Resequencing and mutational analysis using oligonucleotide microarrays. Nat. Genet. 1999;21:42–47. doi: 10.1038/4469. [DOI] [PubMed] [Google Scholar]
- 10.Wong CW, Albert TJ, Vega VB, Norton JE, Cutler DJ, Richmond TA, Stanton LW, Liu ET, Miller LD. Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays. Genome Res. 2004;14:398–405. doi: 10.1101/gr.2141004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmadian A, Ehn M, Hober S. Pyrosequencing: history, biochemistry and future. Clin. Chim. Acta. 2006;363:83–94. doi: 10.1016/j.cccn.2005.04.038. [DOI] [PubMed] [Google Scholar]
- 12.Thomas RK, Nickerson E, Simons JF, Janne PA, Tengs T, Yuza Y, Garraway LA, Laframboise T, Lee JC, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat. Med. 2006;12:852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 13.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen LA, Christiansen M, Vuust J, Andersen PS. Recent developments in high-throughput mutation screening. Pharmacogenomics. 2001;2:387–399. doi: 10.1517/14622416.2.4.387. [DOI] [PubMed] [Google Scholar]
- 15.Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, Teague J, Butler A, Edkins S, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 16.Nollau P, Wagener C. Methods for detection of point mutations: performance and quality assessment. IFCC Scientific Division, Committee on Molecular Biology Techniques. Clin. Chem. 1997;43:1114–1128. [PubMed] [Google Scholar]
- 17.Yeung AT, Hattangadi D, Blakesley L, Nicolas E. Enzymatic mutation detection technologies. Biotechniques. 2005;38:749–758. doi: 10.2144/05385RV01. [DOI] [PubMed] [Google Scholar]
- 18.Cotton RG, Campbell RD. Reactivity of cytosine and thymine in single base pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc. Natl Acad. Sci. USA. 1988;83:4397–4401. doi: 10.1073/pnas.85.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khrapko K, Coller HA, Li-Sucholeiki XC, Andre PC, Thilly WG. High resolution analysis of point mutations by constant denaturant capillary electrophoresis (CDCE) Methods Mol. Biol. 2001;163:57–72. doi: 10.1385/1-59259-116-7:57. [DOI] [PubMed] [Google Scholar]
- 20.Chou LS, Lyon E, Wittwer CT. A comparison of high-resolution melting analysis with denaturing high-performance liquid chromatography for mutation scanning: cystic fibrosis transmembrane conductance regulator gene as a model. Am. J. Clin. Pathol. 2005;124:330–338. doi: 10.1309/BF3M-LJN8-J527-MWQY. [DOI] [PubMed] [Google Scholar]
- 21.Maulik G, Botchway S, Chakrabarti S, Tetradis S, Price B, Makrigiorgos GM. Novel non-isotopic detection of MutY enzyme-recognized mismatches in DNA via ultrasensitive detection of aldehydes. Nucleic Acids Res. 1999;27:1316–1322. doi: 10.1093/nar/27.5.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Kaur M, Price BD, Tetradis S, Makrigiorgos GM. An amplification and ligation-based method to scan for unknown mutations in DNA. Hum. Mutat. 2002;20:139–147. doi: 10.1002/humu.10106. [DOI] [PubMed] [Google Scholar]
- 23.Makrigiorgos GM. PCR-based detection of minority point mutations. Hum. Mutat. 2004;23:406–412. doi: 10.1002/humu.20024. [DOI] [PubMed] [Google Scholar]
- 24.Youil R, Kemper BW, Cotton RG. Screening for mutations by enzyme mismatch cleavage with T4 endonuclease VII. Proc. Natl Acad. Sci. USA. 1995;92:87–91. doi: 10.1073/pnas.92.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Gregorio L, Gallinari P, Gariboldi M, Manenti G, Pierotti MA, Jiricny J, Dragani TA. Genetic mapping of thymine DNA glycosylase (Tdg) gene and of one pseudogene in the mouse. Mamm. Genome. 1996;7:909–910. doi: 10.1007/s003359900267. [DOI] [PubMed] [Google Scholar]
- 26.Yang B, Wen X, Kodali NS, Oleykowski CA, Miller CG, Kulinski J, Besack D, Yeung JA, Kowalski D, et al. Purification, cloning, and characterization of the CEL I nuclease. Biochemistry. 2000;39:3533–3541. doi: 10.1021/bi992376z. [DOI] [PubMed] [Google Scholar]
- 27.Oleykowski CA, Bronson Mullins CR, Godwin AK, Yeung AT. Mutation detection using a novel plant endonuclease. Nucleic Acids Res. 1998;26:4597–4602. doi: 10.1093/nar/26.20.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janne PA, Borras AM, Kuang Y, Rogers AM, Joshi VA, Liyanage H, Lindeman N, Lee JC, Halmos B, et al. A rapid and sensitive enzymatic method for epidermal growth factor receptor mutation screening. Clin. Cancer Res. 2006;12:751–758. doi: 10.1158/1078-0432.CCR-05-2047. [DOI] [PubMed] [Google Scholar]
- 29.Till BJ, Reynolds SH, Greene EA, Codomo CA, Enns LC, Johnson JE, Burtner C, Odden AR, Young K, et al. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 2003;13:524–530. doi: 10.1101/gr.977903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Till BJ, Burtner C, Comai L, Henikoff S. Mismatch cleavage by single-strand specific nucleases. Nucleic Acids Res. 2004;32:2632–2641. doi: 10.1093/nar/gkh599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perry JA, Wang TL, Welham TJ, Gardner S, Pike JM, Yoshida S, Parniske M. A TILLING reverse genetics tool and a web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiol. 2003;131:866–871. doi: 10.1104/pp.102.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurlstone AF, Haramis AP, Wienholds E, Begthel H, Korving J, Van Eeden F, Cuppen E, Zivkovic D, Plasterk RH, et al. The Wnt/beta-catenin pathway regulates cardiac valve formation. Nature. 2003;425:633–637. doi: 10.1038/nature02028. [DOI] [PubMed] [Google Scholar]
- 33.Till BJ, Zerr T, Bowers E, Greene EA, Comai L, Henikoff S. High-throughput discovery of rare human nucleotide polymorphisms by Ecotilling. Nucleic Acids Res. 2006;34:e99. doi: 10.1093/nar/gkl479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilchrist EJ, O’Neil NJ, Rose AM, Zetka MC, Haughn GW. TILLING is an effective reverse genetics technique for Caenorhabditis elegans. BMC Genomics. 2006;7:262. doi: 10.1186/1471-2164-7-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rapid amplification of 5′ complementary DNA ends (5′ RACE) Nat. Methods. 2005;2:629–630. doi: 10.1038/nmeth0805-629. [DOI] [PubMed] [Google Scholar]
- 36.Morrison T, Hurley J, Garcia J, Yoder K, Katz A, Roberts D, Cho J, Kanigan T, Ilyin SED, et al. Nanoliter high throughput quantitative PCR. Nucleic Acids Res. 2006;34:e123. doi: 10.1093/nar/gkl639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steger G. Thermal denaturation of double-stranded nucleic acids: prediction of temperatures critical for gradient gel electrophoresis and polymerase chain reaction. Nucleic Acids Res. 1994;22:2760–2768. doi: 10.1093/nar/22.14.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blake RD, Delcourt SG. Thermal stability of DNA. Nucleic Acids Res. 1998;26:3323–3332. doi: 10.1093/nar/26.14.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Behn M, Qun S, Pankow W, Havemann K, Schuermann M. Frequent detection of ras and p53 mutations in brush cytology samples from lung cancer patients by a restriction fragment length polymorphism- based “enriched PCR” technique. Clin. Cancer Res. 1998;4:361–371. [PubMed] [Google Scholar]
- 40.Kaur M, Zhang Y, Liu WH, Tetradis S, Price BD, Makrigiorgos GM. Ligation of a primer at a mutation: a method to detect low level mutations in DNA. Mutagenesis. 2002;17:365–374. doi: 10.1093/mutage/17.5.365. [DOI] [PubMed] [Google Scholar]
- 41.Liu WH, Kaur M, Makrigiorgos GM. Detection of hotspot mutations and polymorphisms using an enhanced PCR-RFLP approach. Hum. Mutat. 2003;21:535–541. doi: 10.1002/humu.10184. [DOI] [PubMed] [Google Scholar]
- 42.Behn M, Qun S, Pankow W, Havemann K, Schuermann M. Frequent detection of ras and p53 mutations in brush cytology samples from lung cancer patients by a restriction fragment length polymorphism-based “enriched PCR” technique. Clin. Cancer Res. 1998;4:361–371. [PubMed] [Google Scholar]
- 43.Huang C, Taki T, Adachi M, Konishi T, Higashiyama M, Miyake M. Mutations in exon 7 and 8 of p53 as poor prognostic factors in patients with non-small cell lung cancer. Oncogene. 1998;16:2469–2477. doi: 10.1038/sj.onc.1201776. [DOI] [PubMed] [Google Scholar]
- 44.Huang CL, Taki T, Adachi M, Konishi T, Higashiyama M, Kinoshita M, Hadama T, Miyake M. Mutations of p53 and K-ras genes as prognostic factors for non-small cell lung cancer. Int. J. Oncol. 1998;12:553–563. doi: 10.3892/ijo.12.3.553. [DOI] [PubMed] [Google Scholar]
- 45.Steger G. Thermal denaturation of double-stranded nucleic acids: prediction of temperatures critical for gradient gel electrophoresis and polymerase chain reaction. Nucleic Acids Res. 1994;22:2760–2768. doi: 10.1093/nar/22.14.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hernandez-Boussard T, Rodriguez-Tome P, Montesano R, Hainaut P. IARC p53 mutation database: a relational database to compile and analyze p53 mutations in human tumors and cell lines. International Agency for Research on Cancer. Hum. Mutat. 1999;14:1–8. doi: 10.1002/(SICI)1098-1004(1999)14:1<1::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 47.Lipsky RH, Mazzanti CM, Rudolph JG, Xu K, Vyas G, Bozak D, Radel MQ, Goldman D. DNA melting analysis for detection of single nucleotide polymorphisms. Clin. Chem. 2001;47:635–644. [PubMed] [Google Scholar]
- 48.Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin. Chem. 2004;50:1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.