Abstract
Carbon catabolite repression (CCR) is the prototype of a signal transduction mechanism. In enteric bacteria, cAMP was considered to be the second messenger in CCR by playing a role reminiscent of its actions in eukaryotic cells. However, recent results suggest that CCR in Escherichia coli is mediated mainly by an inducer exclusion mechanism. In many Gram-positive bacteria, CCR is triggered by fructose-1,6-bisphosphate, which activates HPr kinase, presumed to be one of the most ancient serine protein kinases. We here report cloning of the Bacillus subtilis hprK and hprP genes and characterization of the encoded HPr kinase and P-Ser-HPr phosphatase. P-Ser-HPr phosphatase forms a new family of phosphatases together with bacterial phosphoglycolate phosphatase, yeast glycerol-3-phosphatase, and 2-deoxyglucose-6-phosphate phosphatase whereas HPr kinase represents a new family of protein kinases on its own. It does not contain the domain structure typical for eukaryotic protein kinases. Although up to now the HPr modifying/demodifying enzymes were thought to exist only in Gram-positive bacteria, a sequence comparison revealed that they also are present in several Gram-negative pathogenic bacteria.
Keywords: histidine-containing protein, protein phosphorylation, carbon catabolite repression
Carbon catabolite repression (CCR) is the paradigm of signal transduction. It allows bacteria to alter catabolic gene expression in response to the availability of rapidly metabolizable carbon sources. Discovered in the early 1940s in Bacillus subtilis and termed the “diauxic phenomenon” (1), one type of molecular mechanism was deciphered in the 1960s in Escherichia coli; in enteric bacteria, changes in the level of cAMP were thought to provide the signal for CCR (2). However, recent results on lacZ expression in E. coli suggest that an increase in the cAMP level reduces only the lag phase of diauxic growth but that the major CCR mechanism is based on inducer exclusion mediated by EIIAGlc of the phosphoenolpyruvate:sugar phosphotransferase system (PTS) (3). It was only in the last decade that the molecular mechanisms underlying CCR in bacilli and other Gram-positive bacteria were partly elucidated (refs. 4–7; for a review, see ref. 8). In these organisms, the complex regulatory cascade is triggered by the ATP-dependent, fructose-1,6-bisphosphate (FBP)-stimulated phosphorylation of Ser-46 in histidine-containing protein (HPr) (9–11), a phosphocarrier protein implicated in carbohydrate transport effected via PTS (12). Signal transduction in CCR continues with a phosphorylation-controlled protein–protein interaction between HPr and the transcriptional repressor/activator catabolite control protein A (CcpA) (13, 14). ATP-dependent phosphorylation at Ser-46 is a prerequisite for the interaction of HPr with CcpA whereas phosphoenolpyruvate-dependent phosphorylation of HPr at His-15 prevents the complex formation, thus linking PTS-mediated sugar transport to CCR (13). The protein complex formed between CcpA and P-Ser-HPr interacts specifically with an operator site called catabolite responsive element (cre) (15, 16). A recently discovered HPr-like protein of B. subtilis, catabolite repression HPr (Crh) (11), is also phosphorylated by the HPr kinase at Ser-46 and is involved in CCR of certain operons. The only poorly characterized components in this signal transduction pathway were the kinase that modifies HPr, and the phosphatase acting on P-Ser-HPr. The genes encoding these two enzymes previously had not been identified. In this communication, we report the identification of the B. subtilis hprK and hprP genes and the characterization of the encoded HPr kinase and P-Ser-HPr phosphatase.
MATERIALS AND METHODS
Plasmid Construction and Protein Purification.
By using the two oligonucleotides 5′-CAGGAGGAAGGATCCGTGGCAAAGGTTCGC-3′ and 5′-ATTTAACGCAAGCTTCTATTCTTCTTGTTC-3′, a 945-bp DNA fragment containing the B. subtilis hprK gene (previously called yvoB) with a BamHI site at the 5′ end and a HindIII site at the 3′ end, was amplified by PCR and cloned into plasmid pQE30 (Qiagen, Chatsworth, CA) cut with the same restriction enzymes to give plasmid pAG4. E. coli strain M15 containing plasmid pREP4 (Qiagen) was transformed with pAG4. Using a similar approach, hprP (previously called yvoE) of B. subtilis was amplified by using oligonucleotides 5′-GACGACGACAAGATGAGTGACAAACAAGTAACG-3′ and 5′-GAGGAGAAGCCCGGTCTACTTCACTCCAACGATTTGTAATAG-3′ and cloned into vector pET-32LIC (Novagen) by single strand annealing. The resulting plasmid was transformed into E. coli strain BL21(DE3), allowing the expression of a thioredoxin-P-Ser-HPr phosphatase fusion protein with a His-tag located between the fused proteins. Induction of hprK and hprP and purification of HPr kinase(His)6, thioredoxin-P-Ser-HPr phosphatase(His)6, Crh(His)6, and HPr(His)6 on Ni-NTA-agarose columns were carried out as described (11). When the thioredoxin domain and the His-tag of thioredoxin-P-Ser-HPr phosphatase(His)6 were cut off with enterokinase, P-Ser-HPr phosphatase was repurified on a Ni-NTA-column.
Protein Phosphorylation and Dephosphorylation Assays.
Phosphorylation of HPr(His)6 or Crh(His)6 by the HPr kinase was carried out as described by Galinier et al. (11). A typical assay contained, in a total volume of 20 μl, 1 μg of Crh(His)6 or HPr(His)6, which was incubated for 10 min at 37°C with 0.5 μg of purified HPr kinase in 20 mM Tris⋅HCl (pH 7.4), 1 mM DTT, 10 mM FBP, 10 mM MgCl2, and 50 μM γ-[32P]ATP (4 μCi). The phosphorylation reaction was stopped by adding an equal volume of sample buffer to the assay mixtures before loading them onto a SDS/polyacrylamide gel (17). After electrophoresis, gels were treated for 5 min with boiling 16% trichloroacetic acid before they were dried for 2 h and exposed to autoradiography.
Dephosphorylation of P-Ser-HPr or P-Ser-Crh by P-Ser-HPr phosphatase was measured on nondenaturing 12.5% polyacrylamide gels. On nondenaturing polyacrylamide gels, seryl-phosphorylated HPr or Crh are well separated from the corresponding unphosphorylated protein, thus allowing detection of P-Ser-HPr phosphatase activity (18). Either 2 μg of P-Ser-HPr or P-Ser-Crh was incubated with 10 mM MgCl2 and with or without 1 μg of P-Ser-HPr phosphatase for 60 min at 37°C, or 4 μg of P-Ser-HPr or P-Ser-Crh was incubated with 10 mM MgCl2 and with 1 μg of thioredoxin-P-Ser-HPr phosphatase(His)6 for 15 min. The reaction was carried out in the absence or presence of phosphate, which stimulates P-Ser-HPr phosphatase activity. When indicated, the reaction mixture contained 1 μg of native or heat-denatured (10 min at 80°C) HPr kinase(His)6. The reaction was stopped by keeping the samples for 10 min at 60°C.
Bacterial Strains, Growth Conditions, and Enzyme Assays.
A 391-bp fragment ranging from nucleotide 189–580 in the hprK gene was amplified by PCR with the two oligonucleotides 5′-GCTTGGCTGTGTCGGAGAACAGCTTCCTGAAGAGGAGA-3′ and 5′-TCCGTGCCCTCCTGGAGAGCGTTTCCCACAAGAGTATC-3′ and cloned into the integration vector pMutin-Fse (V. Vagner, E. Dewyn, and S. D. Ehrlich, unpublished data, and F. Denizot, unpublished data) by single strand annealing resulting in a fusion of the hprK fragment with the lacZ gene present in this plasmid. The annealing site in pMutin-Fse is preceded by an erm gene and the spac promoter and is followed by the lacZ gene. The erythromycin-resistant hprK mutant OBKO13 was constructed by Campbell type integration of pMutin-Fse carrying the hprK fragment into the genome of SG82, a trpC2 lacA mutant strain derived from B. subtilis 168 (19). As a consequence of this integration, the lacZ gene in OBKO13 is expressed under control of the yvo promoter whereas the genes following hprK in the yvo operon are expressed under control of the spac promoter. The ptsH1 crh∷aphA3 double mutant strain QB7102 is identical to the previously described QB7103 (11) but does not contain the levD′–′lacZ fusion integrated at the amyE site.
β-Xylosidase activities were measured after growth of B. subtilis strains in minimal medium supplemented with succinate and glutamate. Induction was carried out by diluting an overnight culture one- to threefold in minimal medium supplemented with the appropriate sugars. The minimal medium had the following composition (40 mM Mops, pH 7.4/11.4 mM K2SO4/3.4 mM NaCitrate/0.8 mM MgSO4/2 mM K2HPO4/50 μg/ml tryptophane/2 mg/ml glutamine/6 g/liter potassium succinate/8 g/liter sodium glutamate/7 mg/ml FeCl3(H2O)6/0.2 mg/ml MnSO4). When indicated, sugars were added at the following concentrations: 0.27% xylose, 0.27% glucose, and 0.67% glycerol. After growth for 1 h at 37°C, β-xylosidase activity was measured in 100-μl samples of the culture medium by using 4-methylumbelliferyl β-xyloside as substrate. Ten microliters of 4-methylumbelliferyl β-xyloside (10 mg/ml dissolved in dimethyl sulfoxide) was added to 100 μl of the culture medium and incubated at 25°C. The reaction was stopped by adding 100 μl of 0.5 M sodium carbonate. Fluorescence was measured by using a Millipore cytofluor. β-Xylosidase activity is expressed as the increase of fluorescence intensity per min (excitation at 390 nm and emission at 460 nm when using a sensitivity level of 4). Induction of β-xylosidase was always ≈1.5-fold higher in mutant strains compared with induction in the wild-type strain SG82.
RESULTS
The yvoB Gene of B. subtilis Encodes HPr Kinase.
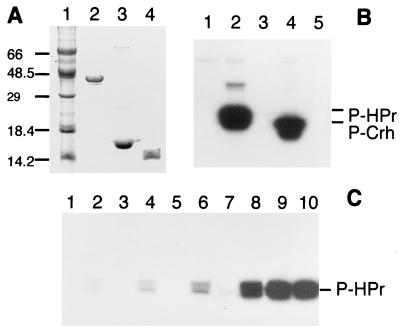
To identify the B. subtilis hprK gene within the completed genome sequence, we first purified the HPr kinase from Enterococcus faecalis to homogeneity. Using degenerate oligonucleotides based on the N-terminal sequence and on the sequence of a tryptic peptide of purified enterococcal HPr kinase, we were able to amplify by PCR a fragment of the hprK gene encoding the first 99 residues of HPr kinase (European Molecular Biology Laboratory accession no. Y14027). The complete sequence of the E. faecalis hprK gene and characterization of the enterococcal HPr kinase will be published elsewhere. Comparison of the enterococcal hprK gene fragment with the genome sequence of B. subtilis revealed a putative ORF (yvoB, GenBank accession no. Z99121) encoding a protein with sequence similarity to the N-terminal sequence of the enterococcal HPr kinase. yvoB is part of an operon comprised of six ORFs [yvoB to yvoF; see B. subtilis genome sequence (20)]. After amplification of the yvoB gene by PCR, the putative protein encoded by this gene was synthesized in E. coli and purified (Fig. 1A, lane 2). The purified protein was able to phosphorylate both HPr and Crh (Fig. 1B). Replacement of Ser-46 by Ala in HPr or Crh completely prevented their ATP-dependent phosphorylation by YvoB (data not shown). As previously described for the HPr kinase, FBP was found to be an allosteric activator of YvoB-catalyzed HPr phosphorylation (Fig. 1C) whereas inorganic phosphate (50 mM) acted as an inhibitor (data not shown). In summary, these results clearly establish that YvoB is identical with the B. subtilis HPr kinase previously reported to phosphorylate HPr and Crh (11). In accordance with the E. faecalis gene designation, yvoB was renamed “hprK.”
Figure 1.
(A) SDS gel of purified proteins; lanes: 1, protein marker; 2, HPr kinase(His)6; 3, HPr(His)6; and 4, Crh(His)6. (B) Phosphorylation of HPr or Crh by purified HPr kinase; lanes: 1, 1 μg of HPr kinase(His)6; 2, 1 μg of HPr kinase(His)6 with HPr(His)6; 3, HPr(His)6; 4, 1 μg of HPr kinase(His)6 with Crh(His)6; and 5, Crh(His)6. (C) Effect of 20 mM FBP on the phosphorylation of 1 μg HPr(His)6; only samples loaded on lanes with even numbers contained FBP; lanes 1 and 2 contained 0.5 ng; lanes 3 and 4, 5 ng; lanes 5 and 6, 50 ng; lanes 7 and 8, 250 ng; and lanes 9 and 10, 750 ng of HPr kinase(His)6.
HPr Kinase Is Present in Both Gram-Positive and Gram-Negative Bacteria.
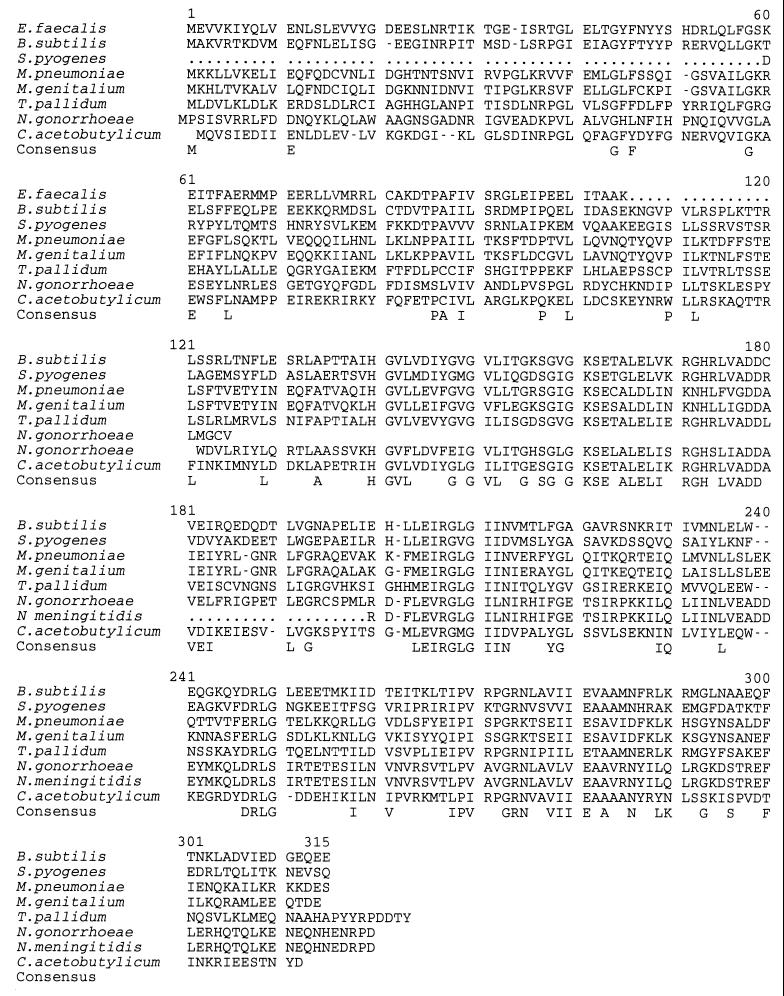
A sequence comparison indicated that HPr kinase was present also in Streptococcus pyogenes, Mycoplasma pneumoniae, Mycoplasma genitalium, Treponema pallidum, Neisseria gonorrhoeae, Neisseria meningitidis, and Clostridium acetobutylicum (Fig. 2). Using clustal alignment, sequence identity ranged from 32.3% (B. subtilis HPr kinase sequence compared with that of N. gonorrhoeae) to 44.5% (B. subtilis HPr kinase sequence compared with that of T. pallidum or C. acetobutylicum) whereas sequence similarity in these alignments ranged from 56.1 to 68.4%. No gene exhibiting homology to B. subtilis hprK could be detected in the completely sequenced genomes of the Gram-negative bacteria E. coli, Haemophilus influenzae, and Borrelia burgdorferi. Up to now, ATP-dependent HPr phosphorylation was thought to exist only in Gram-positive bacteria with low GC content. The presence of a HPr kinase-like protein in the Gram-negative bacteria Treponema pallidum, a spirochete with a GC content of 52.4–53.7% (21), and in neisseriae, which belong to the β subdivision of eubacteria, suggests that HPr of these bacteria also might be phosphorylated at Ser-46.
Figure 2.
Multiple alignment of HPr kinase sequences. A consensus sequence was indicated when, for a given position, an identical amino acid was found in at least five HPr kinase sequences. A sequence corresponding to the putative A motif or P-loop (24) of nucleotide binding proteins (GXXXXGK[TS]) is located in positions 153–160 of the B. subtilis HPr kinase. The different sequences can be found on the World Wide Web at the following Internet addresses: E. faecalis (http://expasy.hcuge.ch/sprot/sprot_top.html, Swiss-Prot accession no. 007664); S. pyogenes (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-ouacgtbl (contig183)); M. pneumoniae (http://nbrfa.georgetown.edu/nbrf/get.html, Protein Identification Resource, accession no. S73934); M. genitalium (http://expasy.hcuge.ch/sprot/sprot_top.html, Swiss-Prot accession no. P47331); T. pallidum [http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-tigrbl (tp_5580)]; N. gonorrhoeae [http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-ouacgtbl (contig268)]; N. meningitidis (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-tigrbl); and C. acetobutylicum (http://www.genomecorp.com/htdocs/sequences/clostridium/clospage.html). The genome sequences of E. faecalis, S. pyogenes, N. gonorrhoeae, N. meningitidis, and C. acetobutylicum are not yet completed. The hprK sequences of E. faecalis, S. pyogenes, and N. meningitidis are incomplete, and semicolons in the deduced HPr kinase sequences represent unknown amino acids. We used the taxonomy browser from the National Center for Biotechnology Information to characterize the position of each bacterium.
The yvoE Gene of B. subtilis Encodes P-Ser-HPr Phosphatase.
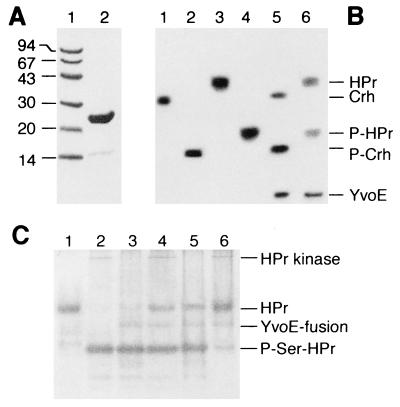
Comparison of the different ORFs of the putative yvo operon of B. subtilis with the complete genome sequence of T. pallidum revealed that only the yvoE gene exhibited similarity to a putative gene in T. pallidum. In addition, the yvoE gene shows similarity to phosphoglycolate phosphatase of several bacteria [Lactobacillus casei (22), Alcaligenes eutrophus (23), and the cyanobacterium Synechocystis sp. (24); see also genome sequences of E. coli, H. influenzae, and B. burgdorferi] and to the yeast enzymes glycerol-3-phosphatase 1, glycerol-3-phosphatase 2, and 2-deoxyglucose-6-phosphate phosphatase 1. Because YovE exhibited a molecular mass (23 kDa) identical to the molecular mass of one of three proteins that were present in a partially purified P-Ser-HPr phosphatase preparation (18), we assumed that yvoE might encode P-Ser-HPr phosphatase. The yvoE gene was integrated into the expression plasmid pET-32LIC, the encoded fusion protein was purified, and part of it was cleaved with enterokinase (Fig. 3A). Slow dephosphorylation of P-Ser-HPr and P-Ser-Crh by YvoE (Fig. 3B) and of P-Ser-HPr by the thioredoxin fusion protein could be demonstrated (Fig. 3C, lane 3). Of interest, the presence of HPr kinase in the assay mixture stimulated dephosphorylation of P-Ser-HPr by YvoE fused to thioredoxin (Fig. 3C, lane 4). No P-Ser-HPr phosphatase activity was present in the HPr kinase preparation (Fig. 3, lane 2), and heat-inactivated HPr kinase had no stimulatory effect on P-Ser-HPr dephosphorylation (Fig. 3C, lane 3). As described for P-Ser-HPr phosphatase, YvoE-catalyzed dephosphorylation of P-Ser-HPr was found to have a stringent requirement for magnesium ions (data not shown) and to be stimulated by inorganic phosphate (Fig. 3C, lanes 5 and 6). A similar effect of Mg2+, phosphate and HPr kinase was observed on the thioredoxin-YvoE(His)6-catalyzed dephosphorylation of P-Ser-Crh and on YvoE-catalyzed dephosphorylation of P-Ser-HPr and P-Ser-Crh (data not shown). These results strongly suggest that yvoE encodes P-Ser-HPr phosphatase, and the yvoE gene was renamed “hprP.” A homologue of the hprP gene was detected in most bacteria that contained the hprK gene. Of interest, only in B. subtilis were hprK and hprP found to be located in the same operon. Compared with HPr kinase, sequence identity between the different P-Ser-HPr phosphatases was less pronounced (data not shown). A conserved sequence (F,L)DLDGTL, located very close to the N terminus, was present not only in the various P-Ser-HPr phosphatases but also in bacterial P-glycolate phosphatases and the yeast enzymes glycerol-3-phosphatase 1, glycerol-3-phosphatase 2, and 2-deoxyglucose-6-phosphate phosphatase 1, suggesting that this consensus sequence is necessary for the phosphatase activity of these enzymes. Sequence identity between P-Ser-HPr phosphatase and the yeast phosphatases ranges from 24.6 to 26.8%.
Figure 3.
(A) SDS/polyacrylamide gel of protein markers (lane 1) and purified phosphatase (lane 2). (B) Nondenaturing polyacrylamide Pharmacia Phast-gel stained with Coomassie blue. P-Ser-HPr phosphatase-catalyzed dephosphorylation of P-Ser-HPr and P-Ser-Crh during 60-min incubation at 37°C. Lanes: 1, 2 μg of Crh(His)6; 2, 2 μg of P-Ser-Crh(His)6; 3, 4 μg of HPr(His)6; 4, 4 μg of P-Ser-HPr(His)6; 5, 2 μg of P-Ser-Crh(His)6 incubated with 1 μg of P-Ser-HPr phosphatase; 6, 2 μg of P-Ser-HPr(His)6 incubated with 1 μg P-Ser-HPr phosphatase. (C) Nondenaturing 12.5% polyacrylamide gel stained with Coomassie blue. The effects of HPr kinase (native or denatured by keeping it for 10 min at 80°C) and 50 mM potassium phosphate (pH 7.4) on the activity of thioredoxin-P-Ser-HPr phosphatase(His)6 were measured. Samples were incubated for 15 min at 37°C. Lanes: 1, 4 μg of HPr; 2, 4 μg of P-Ser-HPr, 1 μg of HPr kinase, and 50 mM phosphate; 3, 4 μg of P-Ser-HPr, 1 μg of P-Ser-HPr phosphatase, and 1 μg of denatured HPr kinase; 4, 4 μg of P-Ser-HPr, 1 μg of P-Ser-HPr phosphatase, and 1 μg of native HPr kinase; 5, 4 μg of P-Ser-HPr, 1 μg of P-Ser-HPr phosphatase, 1 μg of denatured HPr kinase, and 50 mM phosphate; and 6, 4 μg of P-Ser-HPr, 1 μg of P-Ser-HPr phosphatase, 1 μg of native HPr kinase, and 50 mM phosphate. YvoE is identical with P-Ser-HPr phosphatase. YvoE-fusion, the thioredoxin-P-Ser-HPr(His)6 fusion protein.
Effect of a hprK Gene Disruption on CCR.
To demonstrate the role of HPr kinase in CCR, we inactivated hprK of B. subtilis by inserting a plasmid containing a lacZ reporter gene (pMutin-Fse plasmid). The effect of the inactivation of hprK on β-xylosidase activity in cells grown in minimal medium in the presence of xylose, xylose plus glucose, or xylose plus glycerol was studied. Whereas in the wild-type strain β-xylosidase activity was repressed strongly by the presence of glucose, no repressive effect of glucose could be observed in the hprK mutant strain OBKO13 (Table 1). By contrast, the repressive effect of glycerol was only slightly relieved with 99% inhibition in the wild-type strain and 78% in the hprK mutant. The hprK mutant strain exhibited β-xylosidase activities similar to those observed in a ptsH1 crh∷aphA3 double mutant.
Table 1.
Effect of the B. subtilis hprK gene disruption on CCR of β-xylosidase
| Sugar added | SG82 (crh+ ptsH+) | OBKO13 (hprK−) | QB7102 (crh::aphA3 ptsH1) |
|---|---|---|---|
| Xylose | 24.3 ± 1.3* | 42.3 ± 4.5 | 32.8 ± 0.7 |
| Xylose + glucose | 2.5 ± 0.1 (90%) | 48.2 ± 2.5 (−14%) | 32.3 ± 1.5 (1%) |
| Xylose + glycerol | 0.2 ± 0.1 (99%) | 9.3 ± 2.4 (78%) | 24.2 ± 4 (26%) |
Enzyme activities are expressed in arbitrary units and are the mean of three independent experiments. SDs and the percentage of CCR (in parantheses) also are indicated.
DISCUSSION
It appears that all major components participating in a physiological phenomenon described extensively for B. subtilis more than 50 years ago by Jacques Monod and termed “diauxie” (1) (later on better known as CCR) now have been identified. Compared with enteric bacteria, in which PTS-mediated inducer exclusion and cAMP/catabolite activator protein-dependent activation of catabolic gene expression seem to regulate CCR and diauxic growth, respectively (2, 3), a much more complex, negative mechanism seems to be operative in Gram-positive bacteria. In these bacteria, a great number of catabolic genes or operons sensitive to CCR were found to contain, in the promoter region or at the 5′ end of the first gene, an operator site (cre) (25) to which a repressor, CcpA, can bind (15, 16). In contrast to most other repressors, the major corepressor regulating the binding of CcpA to the various cre sequences was not a low molecular mass metabolite but the seryl-phosphorylated form of the protein HPr. Nevertheless, two low molecular mass compounds, FBP and inorganic phosphate, were found to regulate the activities of HPr kinase and P-Ser-HPr phosphatase, which determine the intracellular concentration of P-Ser-HPr.
We here report the identification of the B. subtilis hprK and hprP genes that encode HPr kinase and P-Ser-HPr phosphatase, respectively. Several bacterial kinases with similarity to eukaryotic protein kinases have been discovered (26–28). However, HPr kinase does not exhibit similarity to eukaryotic serine/threonine kinases (29). The domain structure typical of eukaryotic serine/threonine kinases is completely missing in HPr kinase (30). In addition, the ATP binding motif of HPr kinase resembles the A-motif (P-loop) of nucleotide binding proteins (31, 32) rather than the ATP binding site of eukaryotic protein kinases. Because the PTS is thought to have developed early in evolution (33), HPr kinase is considered one of the most ancient protein kinases. HPr kinase does not exhibit similarity to B. subtilis serine kinases, such as SpoIIAB (34) and PrkA (35), or to other known bacterial protein kinases (36). Bacteria that have been shown to possess HPr kinase contain a consensus sequence around Ser-46 of HPr: (V/G)(N/D)XKS(L/I)(M/I)(G/N)(V/L). This motif, which differs from the sequence around Ser-46 in HPr of Gram-negative bacteria not possessing the hprK gene, such as E. coli, H. influenzae, and B. burgdorferi, probably is recognized by the HPr kinase. Of interest, HPr of Alcaligenes eutrophus also contains this consensus sequence. It is therefore tempting to assume that this Gram-negative bacterium, which belongs to a branch of the β subdivision of eubacteria different from the one containing neisseriae, also possesses the HPr kinase.
In addition to hprK, the putative B. subtilis yvo operon also contains the hprP (former yvoE) gene encoding P-Ser-HPr phosphatase. As described previously for the E. faecalis P-Ser-HPr phosphatase, dephosphorylation of P-Ser-HPr or P-Ser-Crh by the B. subtilis P-Ser-HPr phosphatase required Mg ions and was stimulated by inorganic phosphate. Most interesting, dephosphorylation of P-Ser-HPr or P-Ser-Crh also was found to be stimulated by the presence of HPr kinase. This activation probably occurs by allosteric interaction. However, because the P-Ser-HPr and P-Ser-Crh preparations contained some remaining ATP, we cannot exclude the possibility that HPr kinase stimulates P-Ser-HPr phosphatase via phosphorylation. P-Ser-HPr phosphatase shows sequence similarity to phosphoglycolate phosphatase of several bacteria (E. coli, A. eutrophus, L. casei, and H. influenzae). The hprP gene was found in most bacteria possessing hprK. Because some of the genome sequences are not yet completed, hprP is awaited to be discovered in those bacteria that contain hprK but for which hprP has not yet been detected. P-Ser-HPr phosphatase exhibits no significant homology to known eukaryotic or prokaryotic P-protein phosphatases, including SpoIIE of B. subtilis (37). Together with bacterial phosphoglycolate phosphatase and the yeast enzymes glycerol-3-phosphatase 1, glycerol-3-phosphatase 2, and 2-deoxyglucose-6-phosphate phosphatase 1, it forms a new family of phosphatases.
We could demonstrate that HPr kinase participates in CCR. β-Xylosidase activity in the hprK mutant strain OBKO13 was not repressed by glucose, and activities similar to those observed in a ptsH1 crh∷aphA3 double mutant were measured. It is most likely that HPr kinase also is involved in CCR of all operons, the expression of which is sensitive to ptsH1 or crh1 mutations (11). CCR mediated by the non-PTS carbohydrate glycerol was relieved only partially by the hprK mutation, suggesting an additional CCR mechanism for glycerol and possibly other non-PTS substrates.
In summary, our results clearly establish that the main mechanism of CCR in B. subtilis is mediated by protein phosphorylation involving HPr (and for certain operons Crh), HPr kinase, and P-Ser-HPr phosphatase. The discovery of HPr kinase- and P-Ser-HPr phosphatase-like proteins in T. pallidum and neisseriae suggests that HPr phosphorylation also exists in certain Gram-negative bacteria. It will be interesting to find out whether the presumed ATP-dependent HPr phosphorylation in these Gram-negative bacteria also plays a role in CCR, which would make the HPr kinase-mediated CCR mechanism more common than the CCR mechanism operative in Gram-negative enteric bacteria or whether it serves another function. Although the HPr kinase-mediated CCR mechanism is reminiscent of signal transduction pathways in eukaryotic cells, neither HPr kinase nor P-Ser-HPr phosphatase shows significant homology to eukaryotic protein kinases or protein phosphatases, respectively.
Acknowledgments
We are grateful to M. Blüggel and H. E. Meyer for N-terminal and peptide microsequencing, to I. Martin-Verstraete, V. Vagner, and F. Denizot for providing us with strain QB7102, pMutin1, and modified pMutin-Fse, respectively, to J. M. Jault for stimulating discussions, and to E. Hanssen, S. Roche, C. van Herrewege, and A. Bosch for their help with iconography. This research was supported by the European Community Biotech Programme Contract No. BIO4-CT96–0380, the Actions Concertées Coordonnées, the Deutsche Forschungsgemeinschaft, and the Centre National de la Recherche Scientifique.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: CCR, carbon catabolite repression; FBP, fructose 1,6 bisphosphate; HPr, histidine containing HPr; PTS, phosphoenolpyruvate:sugar phosphotransferase system; CcpA, catabolite control protein A; cre, catabolite responsive element; Crh, catabolite repression HPr.
References
- 1.Monod J. Ph.D. thesis. Paris, France: Université Paris 7; 1942. [Google Scholar]
- 2.Botsford J L, Harman J G. Microbiol Rev. 1992;56:100–122. doi: 10.1128/mr.56.1.100-122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inada T, Kimata K, Aiba H. Genes Cells. 1996;1:293–301. doi: 10.1046/j.1365-2443.1996.24025.x. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson W L, Park Y-K, Henkin T M, Won M, Weickert M J, Gaskell J A, Chambliss G H. J Mol Biol. 1987;198:609–618. doi: 10.1016/0022-2836(87)90204-x. [DOI] [PubMed] [Google Scholar]
- 5.Weickert M J, Chambliss G H. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 7.Deutscher J, Reizer J, Fischer C, Galinier A, Saier M H, Jr, Steinmetz M. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saier M H, Jr, Chauvaux S, Deutscher J, Reizer J, Ye J J. Trends Biochem Sci. 1995;20:267–271. doi: 10.1016/s0968-0004(00)89041-6. [DOI] [PubMed] [Google Scholar]
- 9.Deutscher J, Engelmann R. FEMS Microbiol Lett. 1984;23:157–162. [Google Scholar]
- 10.Reizer J, Novotny M J, Hengstenberg W, Saier M H., Jr J Bacteriol. 1984;160:333–340. doi: 10.1128/jb.160.1.333-340.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galinier A, Haiech J, Kilhoffer M-C, Jaquinod M, Stülke J, Deutscher J, Martin-Verstraete I. Proc Natl Acad Sci USA. 1997;94:8439–8444. doi: 10.1073/pnas.94.16.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kundig W, Ghosh S, Roseman S. Proc Natl Acad Sci USA. 1964;52:1067–1074. doi: 10.1073/pnas.52.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 14.Jones E J, Dossonnet V, Küster E, Hillen W, Deutscher J, Klevit R. J Biol Chem. 1997;272:26530–26535. doi: 10.1074/jbc.272.42.26530. [DOI] [PubMed] [Google Scholar]
- 15.Fujita Y, Miwa Y, Galinier A, Deutscher J. Mol Microbiol. 1995;17:953–960. doi: 10.1111/j.1365-2958.1995.mmi_17050953.x. [DOI] [PubMed] [Google Scholar]
- 16.Gösseringer R, Küster E, Galinier A, Deutscher J, Hillen W. J Mol Biol. 1997;266:665–676. doi: 10.1006/jmbi.1996.0820. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Deutscher J, Kessler U, Hengstenberg W. J Bacteriol. 1985;163:1203–1209. doi: 10.1128/jb.163.3.1203-1209.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniel R A, Haiech J, Denizot F, Errington J. J Bacteriol. 1997;179:5636–5638. doi: 10.1128/jb.179.17.5636-5638.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, et al. Nature (London) 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 21.Miao R, Fieldsteel A H. J Bacteriol. 1978;133:101–107. doi: 10.1128/jb.133.1.101-107.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toy J, Bognar A L. J Biol Chem. 1990;265:2492–2499. [PubMed] [Google Scholar]
- 23.Schäferjohann J, Je-Geun Y, Kusian B, Bowien B. J Bacteriol. 1993;175:7329–7340. doi: 10.1128/jb.175.22.7329-7340.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, et al. DNA Res. 1996;3:185–209. doi: 10.1093/dnares/3.3.185. [DOI] [PubMed] [Google Scholar]
- 25.Hueck C J, Hillen W. Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 26.Munoz-Dorado J, Inouye S, Inouye M. Cell. 1991;67:995–1006. doi: 10.1016/0092-8674(91)90372-6. [DOI] [PubMed] [Google Scholar]
- 27.Smith R F, King K Y. Prot Sci. 1995;4:126–129. doi: 10.1002/pro.5560040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janda L, Tichy P, Spizek J, Petricek M. J Bacteriol. 1996;178:1487–1489. doi: 10.1128/jb.178.5.1487-1489.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunter T. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 30.Taylor S S, Buechler J A, Yonemoto W. Annu Rev Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- 31.Saraste M, Sibbald P R, Wittinghofer Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 32.Walker J E, Saraste M, Runswick M J, Gay N J. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reizer J, Saier M H., Jr Curr Opin Struct Biol. 1997;7:407–415. doi: 10.1016/s0959-440x(97)80059-0. [DOI] [PubMed] [Google Scholar]
- 34.Min K-T, Hildtich C M, Diederich B, Errington J, Yudkin M D. Cell. 1993;74:735–742. doi: 10.1016/0092-8674(93)90520-z. [DOI] [PubMed] [Google Scholar]
- 35.Fischer C, Geourjon C, Bourson C, Deutscher J. Gene. 1996;168:55–60. doi: 10.1016/0378-1119(95)00758-x. [DOI] [PubMed] [Google Scholar]
- 36.Cozzone A J. J Cell Biochem. 1993;51:7–13. doi: 10.1002/jcb.240510103. [DOI] [PubMed] [Google Scholar]
- 37.Arigoni F, Duncan L, Aler S, Losick R, Stragier P. Proc Natl Acad Sci USA. 1996;93:3238–3242. doi: 10.1073/pnas.93.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]