Abstract
Shaker-type potassium channels play important roles in determining the electrical excitability of cells. The native channel complex is thought to be formed by four pore-forming α subunits that provide four interaction sites for auxiliary modulatory Kvβ subunits. Because Kvβ subunits possess diverse modulatory activities including either up-regulation or down-regulation of potassium currents, differential assembly of the α–β complex could give rise to diverse current properties. However, the detailed physical and functional stoichiometry of the α–β complex remains unknown. Kvβ1 subunits reduce potassium currents through inactivation, whereas Kvβ2 subunits enhance potassium currents by inhibiting the Kvβ1-mediated inactivation and at the same time by promoting the surface expression of certain potassium channels. In this report we show that Kvβ1 and Kvβ2 of the Shaker-type potassium channels display distinct functional stoichiometry to interact with the Kv1 α subunits, a subfamily of Shaker-type potassium channels. The interaction of Kvβ1 subunits with α subunits is consistent with the α4βn model, where n equals 0, 1, 2, 3, or 4, depending upon the relative concentration of α and β subunits. The α4βn stoichiometry allows for gradual changes of the Kvβ1-mediated inactivation. In contrast, Kvβ2 subunits self-associate to form oligomers and interact with the α subunits via α4β4 stoichiometry, which permits effective multivalent associations with α subunits. Such distinct functional stoichiometry of Kvβ1 and Kvβ2 provides a molecular mechanism that is well suited to their contrasting activities of up-regulation or down-regulation of potassium currents.
In the nervous system, potassium currents are essential for regulating membrane potentials, cardiac pacemaking, and neurotransmitter release (1–4). Investigation of the mechanisms involved in regulating potassium ion conduction is, therefore, essential for the full understanding of potassium channels in electrical signaling. One key process that leads to reduction or elimination of potassium currents during depolarization is known as inactivation, which includes fast and slow inactivation. In the Shaker-type potassium channels, fast inactivation is primarily mediated by a cytoplasmic inactivation gate that is thought to close the ion conduction pathway by “plugging” into the opened conduction pore (5–7). Because the inactivation gates can be donated by either the pore-forming α subunits or auxiliary Kvβ subunits (6, 8), the degree of inactivation is determined by the presence and number of inactivation gate(s) in a given channel complex. Thus, the composition and stoichiometry of α–β complexes could ultimately determine channel inactivation properties.
A total of nine genes encoding various Kvβ subunits have been reported (J.X. and M.L., unpublished work). On the basis of amino acid sequence, each member of this class of hydrophilic subunits can be divided into two parts, a conserved core region and a variable N-terminal region. Evidence from binding and electrophysiological studies in heterologous expression systems has shown that Kvβ subunits interact selectively with α subunits. Kvβ1, for example, inactivates only a subset of Kv1 α subunits through specific interaction with the NAB domain, a conserved region within the N-terminal hydrophilic domain that is also involved in α–α association (10, 11). The assembly and inactivation of Kvβ1 subunits are thought to be mediated by two separable interactions: (i) NAB of α subunits to the core region of β subunits and (ii) interaction gates of β subunits with their receptors on α subunits. In contrast, Kvβ2, although it also interacts with the Kv1 subfamily of α subunits, displays no modulatory activity on fast inactivation of α subunits by itself. Instead, Kvβ2 inhibits the Kvβ1-mediated inactivation in transfected cells (12), which could result from either the formation of heteromultimers with Kvβ1 or effective competition for the binding sites of α subunits.
The functional stoichiometry of the β subunit of Shaker-type potassium channels in fast inactivation is unknown. The current view of the stoichiometry of the α–β complex is based primarily on two lines of evidence. (i) Hydrodynamic studies show experimentally that the native Kv1.2–Kvβ2 complex is consistent with an α/β molar ratio of 1:1 (13), suggestive of α4β4 stoichiometry because a functional channel formed by α subunits is a tetramer (14, 15). (ii) Because Shaker-type α subunits and β subunits are two groups of homologous proteins (8, 16–22), the α4β4 model is assumed to apply to all α–β complexes in Shaker-type potassium channels.
If the above α4β4 model were correct, a cell that contained an excess amount of a noninactivating Kv1 α subunit and an inactivating Kvβ1 subunit should possess two populations of assembled channels: α4 and α4β4. Electrophysiological analysis should, therefore, show a summation of inactivating current of α4β4 and noninactivating of α4. At the molecular level, the model would predict that the β–β interaction is necessary to determine the α4β4 stoichiometry because α subunits can form functional channels in the absence of β subunits. In this study, we have directly tested the β–β interactions and mapped regions that mediate the interaction by using the yeast two-hybrid system. The functional stoichiometry of α–β complexes was examined by electrophysiological analyses of various wild-type and chimeric β subunits. Our results showed that Kvβ1 and Kvβ2 interact with α subunits via two distinct functional stoichiometries: α4βn for Kvβ1 and α4β4 for Kvβ2. These two modes of subunit interaction may play a role in their contrasting physiological functions in vivo.
MATERIALS AND METHODS
Methods for the Yeast Two-Hybrid System.
The procedures were performed according to a published protocol (23) by using the HF7c yeast strain (MATα ura3–52 his-200 ade2–101 lys2–801 trp1–901 leu2–3,112 gal4–542 gal80–538 LYS2∷GAL1UAS-GAL1TATA-HIS3 URA3∷GAL417 mer(x3)-CYCLTATA-lacZ) as host cells; this strain was provided by the laboratory of David Beach (24).
Vector Construction.
Plasmid constructions were performed according to standard recombinant DNA techniques that we have previously described (12). The expression of cDNAs encoding partial or entire α subunits in COS cells was driven by the cytomegalovirus promoter using the pCMV vector (Invitrogen) or its derivatives. Kvβ1 and deletions of Kvβ2 were tagged with the 12CA5 monoclonal epitope as described (11, 12). Construction of the ShBΔ(6–46)Δ(59–95) was carried out as follows: First, two HindIII sites were introduced into the ShBΔ(6–46) coding sequence at amino acid positions 59 and 227 as described (25). The coding sequence between amino acids 95 and 227 was amplified by high-fidelity PCR and ligated in the frame between amino acid positions 59 and 227, which resulted in ShBΔ(6–46)Δ(59–95). Chimeric Kvβ subunits were constructed by first cloning the sequence for amino acids 1–72 of Kvβ1 into the vector and then inserting either the C-terminal region or the full-length Kvβ2 after it. Sequences of oligonucleotides used for constructing the deletion and chimeric constructs are available upon request from the authors. All constructs used in the experiments have been confirmed by DNA sequencing.
Transient Expression of Potassium Channel Subunits.
COS cells were used for our current studies. Culture of COS cells, plasmid purification, and transfection were carried out as described (11). Cotransfection of Shaker α subunits and Kvβ1, unless otherwise indicated, was done at a ratio of 1:6 (3 μg of ShB and 18 μg of Kvβ1). A plasmid containing CD4 (2 μg) marker gene was cotransfected to aid the identification of the transfected cells. Whole-cell recordings were carried out 36–72 h after transfection.
Whole-Cell Patch-Clamp Recording.
Whole-cell voltage-clamp recordings were carried out according to the published protocol (26). Electrodes (Kimble Glass, Vineland, NJ) were pulled from a two-stage vertical puller (Narishige, Tokyo). When filled with intracellular solution, their resistance varied from 2 to 8 MΩ. The intracellular solution contained 110 mM KF, 30 mM KCl, 5 mM NaCl, 2 mM MgCl2, 10 mM EDTA, and 10 mM Hepes; the pH was adjusted to 7.2 with KOH. During recording, the cells were constantly superfused with the extracellular solution containing 145 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 33 mM glucose, and 25 mM HEPES; the pH was adjusted to 7.4 with NaOH. The liquid junction potential was calculated to be 7.13 mV by using jpcalc software (27) and corrected for the holding potential. The current response to a voltage step from a holding potential of −77 mV to 13 mV is illustrated unless otherwise mentioned.
An Axopatch 200A (Axon Instruments, Foster City, CA) amplifier was used in the experiments. Whole-cell capacitance and series resistance were compensated. Voltage protocols were generated by pclamp6 software (Axon Instruments). Typically, the cell was held at −77 mV and the holding voltage was then jumped up from this potential to +73 mV in 10-mV increments for 300 ms. Current data were filtered at 1 KHz, digitized at 100-μs intervals and stored in a computer (Dell, 486/33) for later analysis. Data analysis was done by using clampfit (pclamp6, Axon Instruments). Basal leak current was subtracted. Data were then transferred into a sigma plot (Jandel, San Rafael, CA) for final analysis.
Mathematical Treatment.
Prediction of the time constant–steady-state current (Iss)/peak current (Ipk) curve was done according to the following simplified model previously proposed by MacKinnon and coworkers (15, 28):
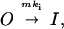 |
where m is the number of Kvβ1 inactivation particles and mki is the inactivation rate constant for channels with m inactivation particles. We have performed outside-out patch recording and found that inactivation constant did not change in as long as 30 min. The recovery rate, which is in the range of one to several minutes (data not shown), is negligible. Denoting A(m) as the fraction of channels with m inactivation particles, then the time constant of any cell is a weighted average of channels with all possible time constants:
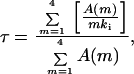 |
where τ4 denotes the inactivation time constant for channels with four inactivation gates; we used τ4 = 12.5 ms in our calculations, which reflects an averaged inactivation time constant from traces with Iss/Ipk less than 0.4. Iss/Ipk indicates the ratio of steady-state current over peak current, or the proportion of noninactivating current can be given by A(0) + f (f is between 0.2 and 0.3 under our recording conditions), where f is a correction factor for systematic noninactivating current. For a model where each individual inactivation gate acts independently, a simple binomial distribution (see the formula below) was used to assess A(m), where P is the probability of finding one β subunit associating with a given α subunit.
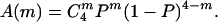 |
For the cooperative α–β association model (i.e., α4β4) to demonstrate the degree of cooperativity, a binomial distribution was weighted against a linear input of tetramer portion, i.e., A(4) was allowed to be changed until the best fit was obtained. Multiple fitting trials have been tested. Four multiple fitting trials are shown in Fig. 4: the α4β4(2500) is derived from an A(4) input with 2,500 times that of the binomial distribution; the α4β4(250) is derived from an A(4) input with 250 times that of the binomial distribution; the α4β2(2) is derived from an A(2) input with half of the oligomers in the α4β2 stoichiometry. A moderate change of A(2) caused significant deviation from our experimental data, suggesting that the tetrameric contribution is dominant.
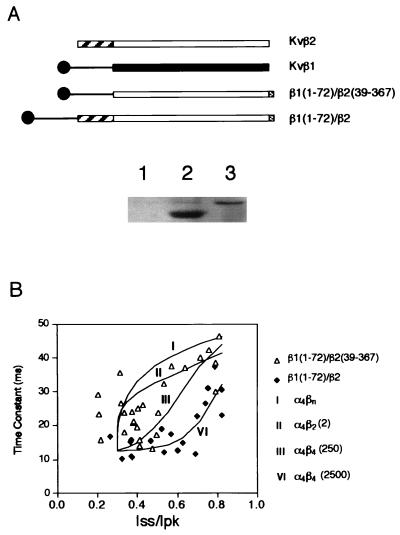
Figure 4.
Inactivation mediated by the chimeric Kvβ subunits. (A Upper) Schematic diagram showing the coding sequence of Kvβ1, Kvβ2, and two chimeric constructs. The open or solid boxes represent the core regions of either Kvβ2 or Kvβ1. The C-terminal shaded boxes show 12CA5 epitope tag. (Lower) Immunoblot showing the monoclonal antibody 12CA5 detection of chimeric β subunit protein from mock-transfected cells (lane 1), cells transfected with β1(1–72)/β2(39–367) (lane 2), and cells transfected with β1(1–72)/β2 (lane 3). (B) Inactivation time constants in milliseconds obtained from cells transfected with ShBΔ(6–46) and with chimeric Kvβ constructs were plotted against Iss/Ipk (steady-state current/peak current). ▵, Cotransfection with β1(1–72)/β2(39–367) (n = 26); ⧫, cells cotransfected with β1(1–72)/β2 (n = 19). Curves show four fitting trials representing the theoretical prediction of stoichiometry for α4βn (I), α4β2 (2) (II), and α4β4 under two conditions: α4β4 is favored by either 250-fold [α4β4(250)] (III) or 2,500-fold [α4β4(2,500)] (VI) (see Materials and Methods for details).
RESULTS
Heterogeneity of α–Kvβ1 Stoichiometry.
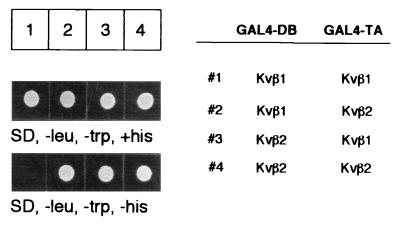
Kvβ1 and Kvβ2 appear to show contrasting modulatory activity on potassium currents (see Introduction). To test the β–β interaction, we cloned the full-length cDNA of Kvβ1 and Kvβ2 into a yeast expression vector and tested their interaction in different pairwise combinations by the yeast two-hybrid system (23, 29, 30). In this experiment, Kvβ subunits were expressed as fusion proteins with either the DNA binding domain or transactivation domain of GAL4. If two Kvβ subunits could associate, the resultant interaction confers the ability of the yeast transformants to grow on synthetic medium lacking histidine. Fig. 1, columns 1–4, respectively, shows the results of four combinations: Kvβ1/Kvβ1, Kvβ1/Kvβ2, Kvβ2/Kvβ1, and Kvβ2/Kvβ2. In the absence of α subunits, Kvβ2 could interact either homomerically with Kvβ2 or heteromerically with Kvβ1. In contrast, no growth was observed for the Kvβ1/Kvβ1 combination, which is consistent with no interaction (Fig. 1). The failure to observe the Kvβ1–Kvβ1 interaction raises the question of whether Kvβ1 interacts with α subunits in a stoichiometry different from the proposed α4β4 model of the α–Kvβ2 complex.
Figure 1.
Differential interactions of Kvβ1 and Kvβ2. HF7C yeast cells were transformed by pairwise combinations of the yeast two-hybrid constructs that express fusion proteins of either the DNA binding domain of GAL4 (DB) or the transcription activation domain of GAL4 (TA) (Right). Yeast transformants carrying the two fusion proteins were first selected by dextrose synthetic drop-out medium lacking leucine and tryptophan (Left) (SD, −leu, −trp, +his) to confirm that the transformants had taken both plasmids. An identical number of cells from each transformation were inoculated on the same medium lacking histidine (Left) (SD, −leu, −trp, −his). The cells were allowed to grow at 30°C for 48 h.
There are at least two possible modes by which Kvβ1 may interact with the α subunit. One is through α4β4 stoichiometry, and the other is through α4βn (n = 0, 1, 2, 3, 4) stoichiometry. In the first case, when the noninactivating α subunit is present in excess, one should see a fixed inactivation rate regardless of the α4/α4β4 ratio. The variation of the α4/α4β4 ratio would only change the level of steady-state current. Under the same conditions, the latter case would give rise to a different inactivation constant depending upon the expression level of Kvβ1 in reference to the α subunits. We cotransfected COS cells with two plasmids encoding either Kvβ1 or ShBΔ(6–46), a mutated Shaker potassium channel lacking the intrinsic inactivation gate (6, 31). Because individual transfected cells may have different ratios of uptaken plasmids, the analyses of inactivation kinetics of randomly selected cells may allow one to distinguish the two modes of interaction. When different transfected cells were recorded by whole-cell voltage clamp, we observed inactivation time constants that varied from 8 ms to more than 36 ms (data not shown). Because the ShBΔ(6–46) normally does not display fast inactivation, the observed variation of inactivation time constant is in agreement with the heterogeneous assembly of α4 with various numbers of β subunits.
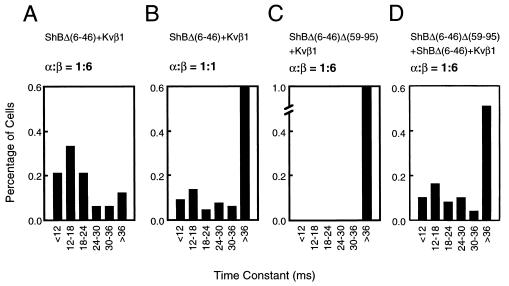
The α4βn model would predict dosage-dependent inactivation by Kvβ1. Thus, within a given cell, higher Kvβ1 expression would result in a higher percentage of α4β4 and α4β3 and thereby the cell would exhibit faster inactivation. If, however, there were fewer Kvβ1 present, the inactivation should be slower because more complexes would be in the forms of α4β1 and α4β0. To test this, we performed experiments by transfecting COS cell with ShBΔ(6–46) and Kvβ1 in two plasmid input ratios, 3 μg/18 μg (1:6) and 3 μg/3 μg (1:1), respectively. The transfected cells were first examined by immunoblot analysis using antibodies specific for the Kvβ1 polypeptide (11, 12), and the higher expression of Kvβ1 in the 1:6 transfection was confirmed (data not shown). We recorded 33 transfected cells for the 1:6 input and 66 cells for the 1:1 Kvβ1 input. The recorded traces at +13 mV for each cell were analyzed and fit with two exponential functions to obtain inactivation constants. When ShBΔ(6–46) and Kvβ1 were transfected in a 1:6 ratio, we observed that more than 50% of transfected cells had a fast inactivation time constant of less than 18 ms (Fig. 2A). When ShBΔ(6–46) and Kvβ1 are in a 1:1 ratio, less than 25% of cells showed an inactivation time constant of less than 18 ms (Fig. 2B). As the fraction of the inactivating component became smaller, the rate of inactivation became slower, consistent with the α4βn model, which predicts that decreasing the availability of the β subunits gradually reduces the inactivation rate.
Figure 2.
Distribution of inactivation time constant of the Kvβ1-mediated inactivation of ShBΔ(6–46). The inactivation constants (horizontal axis) were plotted against cell number (in percentage of recorded cells that express Shaker current; vertical axis). The plasmid combinations and α/β ratio of individual transfections are indicated on the top of each histogram. (A) ShBΔ(6–46) (3 μg) + Kvβ1 (18 μg) (n = 33.). (B) ShBΔ(6–46) (3 μg) + Kvβ1 (3 μg) + vector (14 μg) (n = 66). (C) ShBΔ(6–46)Δ(59–95) (3 μg) + Kvβ1 (18 μg) (n = 39). (D) ShBΔ(6–46) (1 μg) + ShBΔ(6–46)Δ(59–95) (2 μg) + Kvβ1 (18 μg) (n = 49).
To further test the α4βn hypothesis, we constructed a Shaker deletion mutant, ShBΔ(6–46)Δ(59–95), that is insensitive to the Kvβ1-mediated fast inactivation as a result of a reduced ability to interact with Kvβ1 (Fig. 2C). This mutant is functional and coassembles with ShBΔ(6–46). We measured the Kvβ1-mediated inactivation rates from cells cotransfected with ShBΔ(6–46)Δ(59–95), ShBΔ(6–46), and Kvβ1 with a plasmid input of 2 μg, 1 μg, and 18 μg, respectively. Although the ratio of the total α subunits and Kvβ1 was maintained at 1:6, in contrast to the ShBΔ(6–46)/Kvβ1 transfection in Fig. 2A, a large portion of recordings from the triple transfection showed a slow inactivation rate similar to the results of the 1:1 plasmid input (Fig. 2D, n = 49). Thus, these results argue in favor of the α4βn model.
Molecular Determinants in β–β Interaction.
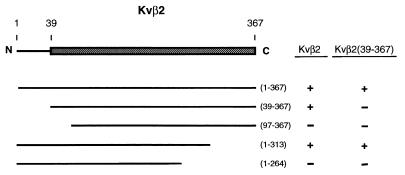
The failure to detect Kvβ1–Kvβ1 interaction suggests that they appear to act independently to bind α subunits. Because Kvβ2 can form homomultimers (ref. 12 and Fig. 1) and the amino acid sequences of Kvβ1 and Kvβ2 show about 85% amino acid identity at their C-terminal core regions, it would be valuable to know the specific region in Kvβ2 that mediates the β–β interaction. Previously, we have shown that the C-terminal core region of Kvβ2 is the minimal region required for interacting with full-length Kvβ2 (12). What is the minimal region of Kvβ2 that is sufficient to interact with the core region of Kvβ2? To answer this, we constructed a series of deletions of Kvβ2 and tested for their ability to interact with either Kvβ2 or the core region of Kvβ2 [amino acids 39–367, denoted as Kvβ2(39–367)] by using the yeast two-hybrid system. The results of growth selection on plates lacking histidine are summarized in Fig. 3. On the basis of these results, we found that the core region of Kvβ2 failed to interact with itself. The interaction can be restored when the N-terminal domain is present. One interesting observation is the interaction between Kvβ2(1–313) and Kvβ2(39–367), which suggests that the N-terminal 313 amino acids of Kvβ2 interact with the C-terminal core region. This result implies that the interaction between Kvβ2 subunits is not a homophilic interaction between the conserved core regions. Instead, the Kvβ2–Kvβ2 interaction is polarized and the N-terminal portion of Kvβ2 is required to interact with the core region of Kvβ2 (Fig. 3). Thus, the failure to form the Kvβ1–Kvβ1 homomultimer is presumably due to the lack of an appropriate determinant within the N-terminal region that mediates β–β interaction in the presence of the conserved core region.
Figure 3.
Molecular determinants that mediate the Kvβ2–Kvβ2 interaction. (Left) A schematic diagram representing the coding sequence of Kvβ2 is shown with the C-terminal core region highlighted as a shaded box. Deletion constructs expressing different coding regions of Kvβ2 in GAL4–TA fusions are shown. The numbers in the parentheses indicate the beginning and ending amino acids of the individual deletion mutants. (Right) The ability of the deletion mutants to interact with full-length Kvβ2 or Kvβ2(39–367) was tested in the yeast two-hybrid system. Results from the growth assay are shown (+, growth; −, no growth).
The above binding studies indicate the potential molecular determinants responsible for the difference between Kvβ1 and Kvβ2 in forming homomultimers. However, one cannot rule out that the two-hybrid interaction results were tainted by the lack of efficient protein folding or sufficient stability for some deletion mutants. To demonstrate this functionally, we constructed two chimeric Kvβ subunits. One links amino acids 1–71 of Kvβ1 (corresponding to the inactivation particle and its putative “chain,” see ref. 8) to the core region of Kvβ2 [denoted as (β1(1–72)/β2(39–367)]; the other fuses Kvβ1(1–71) to the full-length Kvβ2 [denoted as β1(1–72)/β2] (Fig. 4A). Both chimeric constructs contain inactivation particles but differ in their ability to form homomultimers, i.e., only the β1(1–72)/β2 chimera can form oligomers and presumably trigger fast inactivation regardless of its expression level with respect to the interacting α subunits. The Kvβ2 express considerably higher than Kvβ1 even in an identical vector (12). Transient expression of these chimeric constructs has shown comparable protein level (Fig. 4A). We cotransfected COS cells with ShBΔ(6–46) plus either of these two constructs in various plasmid inputs to achieve various α/β ratios and performed recordings and analyses similar to those shown in Fig. 2. The β1(1–72)/β2(39–367) chimera inactivates in a mode quite similar to that of the native Kvβ1. In contrast, the β1(1–72)/β2 chimera showed a significant degree of cooperativity in α–β assembly (Fig. 4). Even at an Iss/Ipk value of 0.7 (where only a very low amount of β was present), the inactivation time constant is still as fast as that obtained at an Iss/Ipk value of 0.2 (see Fig. 4 and Materials and Methods for mathematical fitting). This result and the binding data support the idea that the ability to form a β–β multimer allows for cooperative assembly and accelerated inactivation. It also provides evidence that the poorly cooperative α4β4 assembly of wild-type Kvβ1 is due to the lack of Kvβ1–Kvβ1 interaction.
DISCUSSION
The composition and stoichiometry of the α–β complex of Shaker-type potassium channels ultimately determine the kinetic and gating properties of the resultant channel protein. In this report, we have presented several lines of evidence supporting the model that Kvβ1 and Kvβ2, two abundant and homologous β subunits that interact with the same subset of α subunits, coassemble with α subunits via two distinct modes. The Kvβ1 subunit binds to α subunits through a model consistent with α4βn stoichiometry, where n = 0, 1, 2, 3, 4, depending upon the relative concentration of α and β subunits. In contrast, the Kvβ2 subunits form oligomers (probably tetramers) and interact with α subunits via α4β4 stoichiometry.
Distinct Functional Stoichiometry of Kvβ1 and Kvβ2.
Three lines of evidence have been presented in support of the notion that Kvβ1 does not oligomerize efficiently. As a result, it acts on α subunits independently depending upon its concentration relative to the expression level of α subunits. (i) We failed to detect the Kvβ1–Kvβ1 interaction in the yeast two-hybrid system. Kvβ1 interacts with Kvβ2 and N-terminal domains of the Kv1 α subunits under the same conditions. It is therefore unlikely that the inability to detect the Kvβ1–Kvβ1 interaction is due to a technical error such as a failure of GAL4–Kvb1 fusion to enter the nucleus in yeast. (ii) The speed of inactivation correlates with the Kvβ1 plasmid input in the cotransfection. This argues against the α4β4 model, which predicts that inactivation constants should be similar regardless of the expression level of Kvβ1. (iii) A channel containing both Kvβ1-sensitive and -insensitive α subunits exhibits much reduced inactivation even under the condition of the high Kvβ1 plasmid input.
The inactivation properties of Kvβ1 in many ways are similar to that mediated by intrinsic inactivation gates of α subunits. However, it should be noted that there are several differences between these two types of inactivation gates. For example, the off-rate of the Kvβ1 inactivation gate is on the order of seconds (data not shown) in contrast to the 50 ms reported for ShB (28). In addition, a weak Kvβ1–Kvβ1 interaction was implicated because the rate of recovery from inactivation correlates with the number of Kvβ1 inactivation gates (data not shown).
In contrast, the Kvβ2 subunits self-associate and presumably form a tetramer. This was first suggested by hydrodynamic studies of the native preparation (13) and later Kvβ2 was found to interact in the yeast two-hybrid system in the absence of α subunits (12). In this report, the formation of Kvβ2 oligomers was further demonstrated by two sets of experiments. (i) The determinants that mediate the Kvβ2–Kvβ2 interaction were tested in the yeast two-hybrid system. Interestingly, the formation of Kvβ2 oligomers is not through homophilic interactions. Instead, the Kvβ2–Kvβ2 association is the result of “polarized” interactions. This result provides a molecular explanation as to why Kvβ1 does not interact with itself to form an oligomer. (ii) The Kvβ2–Kvβ2 interaction was implicated functionally by using a chimeric construct that transplants the inactivation gate of Kvβ1 to Kvβ2. The resultant chimera has shown strong cooperative α4β4 assembly regardless of the expression level of β subunits. In fact, our data can be fitted by a model that favors cooperative tetrameric assembly of the β chimera by a factor of 2,500-fold over an independent assembly model (see Materials and Methods).
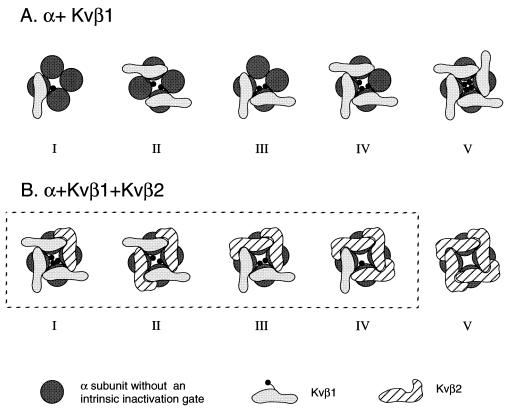
Thus, we propose a working model, which is illustrated in Fig. 5. In a cell that contains both α subunits and Kvβ1 subunits, depending upon the α/β ratio, the channels can be occupied by from zero up to four Kvβ1 subunits (Fig. 5A). If all α subunits, Kvβ1 and Kvβ2 subunits are present in a given cell or neuron, in principle, we should expect several states (Fig. 5), depending on the α/Kvβ1/Kvβ2 ratio. Under this condition, because Kvβ2 is capable of forming oligomers, it conceivably has higher avidity to interact with α subunits as a result of multivalent interactions. If this were the case, the intermediate states (I–IV) would not exist or would be present at a very low level. All α subunits would be occupied by tetrameric Kvβ2 subunits (Fig. 5B, state V). Indeed, coexpression of Kvβ1 and Kvβ2 in transfected cells showed that Kvβ2 completely inhibits the Kvβ1-mediated inactivation and that intermediate inactivation cannot be observed (12). It should be noted that the proposed model is simplified. For example, one can speculate that Kvβ2 could form linear polymers with more than four subunits. This hypothesis can be tested by a number of ways including biochemical analyses of purified Kvβ2 protein. In addition, the difference of Kvβ1 and Kvβ2 in β–β interaction may be further influenced by differential posttranslational modifications, such as phosphorylation, and by other channel-interacting proteins or subunits.
Figure 5.
Proposed stoichiometry of α–β complexes. (A) Diagram illustrating the α–Kvβ1 complexes with increasing availability of Kvβ1. (B) Diagram illustrating the possible α–Kvβ1–Kvβ2 complexes. The model may not represent the steady-state species of the α–β complexes (see Discussion).
Molecular Interplay of Kvβ1- and Kvβ2-Mediated Modulation.
Both Kvβ1 and Kvβ2 bind to the N-terminal domains of Kv1 α subunits (10–12). So far three Kvβ1 splice variants have been identified, known as Kvβ1.1, Kvβ1.2, and Kvβ1.3 (17, 18, 32). These three splice variants have identical C-terminal core regions but differ in the sequence and length of their N-terminal domains. Their shared C-terminal core region mediates the interaction with α subunits and Kvβ2, although the distinct N-terminal domains appear to be responsible for conferring the heterogeneity of their modulatory activity. In contrast to Kvβ1, the Kvβ2 subunit exhibits little modulation on fast inactivation. Instead, it seems to play a role in facilitating surface expression of certain α subunits (9). More importantly, Kvβ2 is capable of inhibiting the Kvβ1-mediated inactivation (12).
The heterogeneity of Kvβ1 inactivation, presumably as a result of α4βn stoichiometry, offers a mechanism to incrementally strengthen the ability of Kvβ1 to accelerate fast inactivation as a result of increasing expression of the Kvβ1 subunit. Because the off-rate of the Kvβ1 inactivation gates is in the range of seconds (data not shown), the inactivation of Kvβ1 subunits could provide a prolonged reduction of potassium currents. In contrast to Kvβ1, Kvβ2 appears to interact with the α subunits via α4β4 stoichiometry, which allows for effective competition for the Kvβ1 binding sites on α subunits, thereby inhibiting the Kvβ1-mediated inactivation. Combining the high avidity of the α4–Kvβ2 interaction with its ability to promote surface expression, the outcome of the Kvβ2-mediated modulation is to increase potassium currents. Thus, a difference in the functional stoichiometry of these two overlappingly expressed β subunits provides a molecular mechanism to either up-regulate or down-regulate potassium currents.
Acknowledgments
We thank Dr. Peter Gillespie and members of the Li lab for critical reading of the manuscript and Robyne Butzner for help with the manuscript preparation. This work is supported by grants (to M.L.) from the National Institutes of Health and the Council for Tobacco Research, Inc. M.L. is an American Heart Association–Pfizer Fellow.
References
- 1.Connor J A, Stevens C F. J Physiol. 1971;213:21–30. doi: 10.1113/jphysiol.1971.sp009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrne J H. J Neurophysiol. 1980;43:651–668. doi: 10.1152/jn.1980.43.3.651. [DOI] [PubMed] [Google Scholar]
- 3.Rogawski M A. Trends Neurosci. 1985;8:214–219. [Google Scholar]
- 4.Hille B. Ionic Channels of Excitable Membrane. Sunderland, MA: Sinauer; 1991. pp. 58–75. , 99–116. [Google Scholar]
- 5.Armstrong C M, Bezanilla F, Rojas E. J Gen Physiol. 1973;62:375–391. doi: 10.1085/jgp.62.4.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoshi T, Zagotta W N, Aldrich R W. Science. 1990;250:533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- 7.Jan L Y, Jan Y N. Annu Rev Physiol. 1992;54:537–555. doi: 10.1146/annurev.ph.54.030192.002541. [DOI] [PubMed] [Google Scholar]
- 8.Rettig J, Heinemann S H, Wunder F, Lorra C, Parcej D N, Dolly J O, Pongs O. Nature (London) 1994;369:289–294. doi: 10.1038/369289a0. [DOI] [PubMed] [Google Scholar]
- 9.Shi G, Nakahira K, Hammond S, Rhodes K, Schechter L, Trimmer J. Neuron. 1996;16:843–852. doi: 10.1016/s0896-6273(00)80104-x. [DOI] [PubMed] [Google Scholar]
- 10.Sewing S, Roeper J, Pongs O. Neuron. 1996;16:455–463. doi: 10.1016/s0896-6273(00)80063-x. [DOI] [PubMed] [Google Scholar]
- 11.Yu W F, Xu J, Li M. Neuron. 1996;16:441–453. doi: 10.1016/s0896-6273(00)80062-8. [DOI] [PubMed] [Google Scholar]
- 12.Xu J, Li M. J Biol Chem. 1997;272:11728–11735. doi: 10.1074/jbc.272.18.11728. [DOI] [PubMed] [Google Scholar]
- 13.Parcej D N, Scott V E, Dolly J O. Biochemistry. 1992;31:11084–11088. doi: 10.1021/bi00160a018. [DOI] [PubMed] [Google Scholar]
- 14.Li M, Unwin N, Stauffer K A, Jan Y N, Jan L Y. Curr Biol. 1994;4:110–115. doi: 10.1016/s0960-9822(94)00026-6. [DOI] [PubMed] [Google Scholar]
- 15.MacKinnon R. Nature (London) 1991;350:232–235. doi: 10.1038/350232a0. [DOI] [PubMed] [Google Scholar]
- 16.Scott V E, Rettig J, Parcej D N, Keen J N, Findlay J B, Pongs O, Dolly J O. Proc Natl Acad Sci USA. 1994;91:1637–1641. doi: 10.1073/pnas.91.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.England S K, Uebele V N, Shear H, Kodali J, Bennett P B, Tamkun M M. Proc Natl Acad Sci USA. 1995;92:6309–6313. doi: 10.1073/pnas.92.14.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.England S K, Uebele V N, Kodali J, Bennett P B, Tamkun M M. J Biol Chem. 1995;270:28531–28534. doi: 10.1074/jbc.270.48.28531. [DOI] [PubMed] [Google Scholar]
- 19.Chouinard S W, Wilson G F, Schlimgen A K, Ganetzky B. Proc Natl Acad Sci USA. 1995;92:6763–6767. doi: 10.1073/pnas.92.15.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majumder K, De Biasi M, Wang Z, Wible B A. FEBS Lett. 1995;361:13–16. doi: 10.1016/0014-5793(95)00120-x. [DOI] [PubMed] [Google Scholar]
- 21.Morales M J, Castellino R C, Crews A L, Rasmusson R L, Strauss H C. J Biol Chem. 1995;270:6272–6277. doi: 10.1074/jbc.270.11.6272. [DOI] [PubMed] [Google Scholar]
- 22.Fink M, Duprat F, Lesage F, Heurteaux C, Romey G, Barhanin J, Lazdunski M. J Biol Chem. 1996;271:26341–26348. doi: 10.1074/jbc.271.42.26341. [DOI] [PubMed] [Google Scholar]
- 23.Xu J, Yu W, Jan Y N, Jan L Y, Li M. J Biol Chem. 1995;270:24761–24768. doi: 10.1074/jbc.270.42.24761. [DOI] [PubMed] [Google Scholar]
- 24.Feilotter H E, Hannon G J, Ruddell C J, Beach D. Nucleic Acids Res. 1994;22:1502–1503. doi: 10.1093/nar/22.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Jan Y N, Jan L Y. Science. 1992;257:1225–1230. doi: 10.1126/science.1519059. [DOI] [PubMed] [Google Scholar]
- 26.Hamill O P, Marty A, Nehre E, Sakmann B, Sigworth F J. Pfluegers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 27.Barry P H. J Neurosci Methods. 1994;51:107–116. doi: 10.1016/0165-0270(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 28.MacKinnon R, Aldrich R W, Lee A W. Science. 1993;262:757–759. doi: 10.1126/science.7694359. [DOI] [PubMed] [Google Scholar]
- 29.Chevray P M, Nathans D. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fields S, Song S-K. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 31.Zagotta W N, Hoshi T, Aldrich R W. Science. 1990;250:568–571. doi: 10.1126/science.2122520. [DOI] [PubMed] [Google Scholar]
- 32.McCormack K, McCormack T, Tanouye M, Rudy B, Stuhmer W. FEBS Lett. 1995;370:32–36. doi: 10.1016/0014-5793(95)00785-8. [DOI] [PubMed] [Google Scholar]