Abstract
Stomata are innovations of land plants that allow regulated gas exchange. Stomatal precursor cells are produced by asymmetric cell division, and once formed, signal their neighbors to inhibit the formation of stomatal precursors in direct contact. We report a gene of Arabidopsis thaliana, EPIDERMAL PATTERNING FACTOR 1 (EPF1) that encodes a small secretory peptide expressed in stomatal cells and precursors and that controls stomatal patterning through regulation of asymmetric cell division. EPF1 activity is dependent on the TOO MANY MOUTHS receptor-like protein and ERECTA family receptor kinases, suggesting that EPF1 may provide a positional cue interpreted by these receptors.
Keywords: Stomata, mediator, asymmetric cell division, meristemoid, secretory peptide, Arabidopsis thaliana
Although multicellularity evolved independently in animals and plants (Baldauf 2003), both utilize asymmetric cell division for creating new cell lineages during development (Scheres and Benfey 1999; Betschinger and Knoblich 2004). In plants, stomatal development offers an excellent, tractable system to study asymmetric cell division. Stomata consist of two guard cells and a pore between them for gas exchange. The guard cells are produced through a series of asymmetric and symmetric cell divisions that not only produce the differentiated cells, but control the overall density and pattern of stomata on the organ surface (Bergmann 2003). In most dicot leaves, stomata follow a “one-cell-spacing” rule in which two stomata are separated by at least one intervening nonstomatal epidermal cell (Sachs 1991). This spacing is hypothesized to be important for efficient gas exchange (Nadeau and Sack 2002b). The first morphologically discernible stomatal precursor cell is the meristemoid, which is the smaller, triangular-shaped daughter cell produced by asymmetric cell division. The larger daughter cell may differentiate into a nonstomatal epidermal cell (pavement cell) or it may undergo its own asymmetric cell division, creating a satellite meristemoid. A meristemoid has a self-renewing capability and can continue asymmetric divisions, but eventually converts into a guard mother cell (GMC), which then divides symmetrically to form two guard cells that constitute a stoma. The major enforcer of the one-cell-spacing pattern appears to be a signal sent from stomata and precursors (guard cells, GMCs, and mature meristemoids) to undifferentiated neighbor cells that influences the plane of cell division (Geisler et al. 2000).
A number of key regulators of the asymmetric cell divisions that ensure the one-cell-spacing rule and control stomatal densities are known. Mutations in TOO MANY MOUTHS (TMM) (Yang and Sack 1995; Geisler et al. 2000; Nadeau and Sack 2002a), STOMATAL DENSITY AND DISTRIBUTION1 (SDD1) (Berger and Altmann 2000; von Groll et al. 2002), YODA (YDA) (Bergmann et al. 2004), and a triple mutant of ERECTA (ER); ERECTA-LIKE1 (ERL1); ERL2 (Shpak et al. 2005) result in plants with excess and clustered stomata. TMM is a membrane-anchored protein with an extracellular leucine-rich repeat (LRR) domain (LRR receptor-like protein; LRR-RLP), but no cytoplasmic signaling domain (Nadeau and Sack 2002a). ER, ERL1, and ERL2 are LRR receptor kinases (LRR kinases) (Shpak et al. 2005). TMM and ER family proteins have been proposed to function together in the stomatal lineage to perceive positional information in relation to pre-existing stomata and precursors (Ingram 2005; Shpak et al. 2005). Because TMM mutations appear to affect only stomatal development, whereas the ER family is important for a broader range of biological processes (Shpak et al. 2004), it may be that a presumed TMM/ER family complex would rely on TMM for specificity of the positional information and the kinase-containing ER family proteins to transmit signals to downstream elements. One potential downstream target is YDA, a mitogen-activated protein kinase kinase kinase (MAPKKK) that has been shown to act as a switch between stomatal and pavement cell fate (Bergmann et al. 2004). The extracellular-localized subtilisin protease SDD1 was proposed to process a precursor of an unknown mobile signal that controls stomatal density, and to a lesser extent, stomatal pattern (Berger and Altmann 2000). These cell–cell signal(s) are thought to ultimately affect the transcription factors that direct cell stomatal cell division and cell-type differentiation (Lai et al. 2005; Ohashi-Ito and Bergmann 2006; Macalister et al. 2007; Pillitteri et al. 2007).
The cellular mechanism of stomatal pattern and the molecular identities of the previously identified mediators strongly suggest that local activation of a signaling pathway in cells neighboring stomata or precursors regulates placement of meristemoid-forming division planes. Missing from the previous models was the identity of the (presumed proteinaceous) activating signal. In this manuscript we present evidence that the EPIDERMAL PATTERNING FACTOR 1 (EPF1) gene encodes a protein capable of being such a signal.
Results and Discussion
Identification of the EPF1 gene
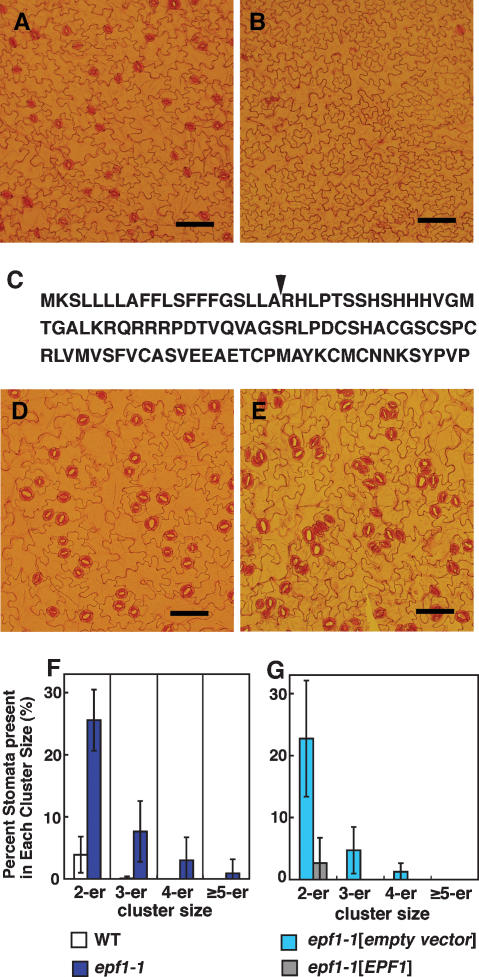
In a genome-scale project to identify novel secreted peptide mediators that regulate plant development, we assayed the effect on Arabidopsis development of individually overexpressing 153 genes predicted to encode small (<150-amino-acid) secreted peptides. Through this screen, we identified a gene (At2g20875) that decreased stomatal density when overexpressed using a modified Cauliflower Mosaic Virus 35S promoter (Promoter2x35S-Ω) (Fig. 1A,B). We designated the gene EPF1. The extent to which stomatal density decreased was correlated with the expression levels of EPF1, with the most severely affected plants producing no stomata (Supplementary Fig. S1). Such plants were significantly smaller than wild type, and were infertile. Transgenic lines with lower EPF1 overexpression levels and only 10%–20% of the wild-type numbers of stomata did not exhibit any other discernable developmental phenotypes, suggesting that EPF1 overexpression specifically affects stomatal development, and that the poor growth and infertility are secondary physiological consequences of a lack of stomata.
Figure 1.
The EPF1 gene, encoding a secretory protein, regulates stomatal patterning. (A,B) Overexpression of EPF1 results in decreased number of stomata. An abaxial side of a cotyledon of Arabidopsis transformed with the pTK016 vector (A) and Promoter2x35S-Ω-EPF1 (B) are shown. After 7 d selection on BASTA (phosphinotricin) plate, resistant plants were further grown on nutrient plates for 7 d. (C) Deduced amino acid sequence of EPF1 product. The predicted cleavage site flanking the N-terminal signal sequence is shown with an arrowhead. (D,E) epf1-1 has clustered stomata. Abaxial sides of 20-d-old wild-type (D) and epf1-1 (E) cotyledons are shown. (F,G) Percent stomata (mean ± SD) present in stomatal clusters. (F) White column shows wild type and blue column shows epf1-1. (G) Light-blue column shows epf1-1 transformed with the control vector, and gray column shows epf1-1 transformed with the EPF1 gene. Bars, 100 μm.
The deduced coding region of the EPF1 product is 104 amino acids long, and is predicted to have an N-terminal secretory signal peptide, followed by a cleavage site be-tween the 20th and 21st amino acid residues (Fig. 1B; SignalP 3.0 program, http://www.cbs.dtu.dk/services/SignalP; Nielsen et al. 1997). Outside of the signal peptide, EPF1 contains no significant similarity to proteins (or protein domains) of known function.
Loss-of-function epf1 mutant has a defect in one-cell-spacing rule of stomatal pattern
The overexpression phenotypes of EPF1 suggest that this protein is capable of influencing stomatal pattern. We next sought to determine whether EPF1 was required for normal stomatal development. For this purpose, a T-DNA insertion allele of EPF1 (epf1-1) was characterized. In the homozygous mutant, both an increase in stomatal density and violations of the one-cell-spacing rule (clustering of stomata) were observed (Fig. 1D–F; Supplementary Table 1; Supplementary Fig. S3). The epf1-1 mutant was complemented with a 3.3-kb genomic region spanning At2g20875 (Supplemental Material), indicating that the phenotype is due to disruption of this locus (Fig. 1G). epf1-1 had no appreciable phenotypes unrelated to stomatal development, again indicating the specificity of EPF1 in stomatal patterning. Stomatal clusters were most evident in cotyledon and rosette leaves, but clustering occurred in all stomata-producing organs examined (Supplementary Table 1). The clustered phenotype suggests that, in the absence of wild-type EPF1, the orientation of cell divisions in the stomatal lineage is incorrect. Indeed, we observed formation of new meristemoids adjacent to guard cells or precursors (Supplementary Fig. S2).
EPF1 is expressed in the stomatal precursor cells
Stomatal development occurs only in the aerial portions of the plant and predominantly in young leaves. EPF1 transcript was detected in the shoot, but not the root, by RT–PCR (Fig. 2A). RNA gel blot analysis using RNA isolated from different developmental stages revealed that the EPF1 transcript was highly expressed in developing leaves, contemporaneous with TMM and SDD1 expression (Fig. 2B). We examined the cell-type specificity of EPF1 expression by in situ RNA hybridization. In the epidermis of immature leaves, EPF1 signal was detected in a punctuate pattern (Fig. 2C). In the 2- to 3-mm leaves, stomatal development is completed in the distal region. In such leaves, the punctuate EPF1 signal was detected only in the proximal region, where stomatal precursor cells exist (data not shown). The signal pattern was similar to that of SDD1, which was reported to be expressed in stomatal precursor cells (von Groll et al. 2002), although EPF1 signal was weaker than that of SDD1 (Fig. 2C,E). Because outlines of cells in the whole-mount in situ hybridization samples were unclear, to better understand the expression pattern we created a β-glucuronidase (GUS) fusion to the EPF1 promoter (PromoterEPF1-GUS) and green fluorescent protein (GFP) fusion to the EPF1 promoter (PromoterEPF1-erGFP) and analyzed reporter expression in developing plants. The reporter signals are absent from the shoot meristem (data not shown), are first detectable in young leaves when the stomatal lineage initiates (Supplementary Fig. S4), and are restricted to meristemoids, GMCs, and young guard cells (Fig. 2F,G). EPF1 signal in the in situ RNA hybridization experiment was undetectable in young guard cells; however, the GUS and GFP reporter signals were positive. This discrepancy in expression may be due to stability of the reporters or, alternatively, to limited accessibility of RNA probes in the whole-mount in situ RNA hybridization. Interestingly, in contrast to genes for putative receptors (TMM and ER family) that are also expressed in the stomatal lineage neighbor cells (that respond to positional cues) (8,13), all measures of EPF1 expression indicate that EPF1 is only found in cells implicated as signaling sources.
Figure 2.
Expression of EPF1. (A) RT–PCR analysis for EPF1 using RNA from roots (R), rosette leaves (RL), cauline leaves (CL), stems (St), and floral buds (FB). 18S rRNA was used as a control target. (B) RNA gel blot analysis using RNA from cotyledons (Co), rosette leaves of different developmental stages (1–12), and shoot apex of 16-d-old wild-type plants. EPF1, TMM, SDD1, and 18S rRNA were used as probes. (C–E) In situ hybridization for abaxial side of rosette leaves, probed with antisense EPF1 (C), sense EPF1 (D), and antisense SDD1 (E). Arrows indicate mature stomata and arrowheads indicate hybridization signal. (F) GUS expression in 14-d-old Arabidopsis transformed with PromoterEPF1-GUS. (G) Expression of PromoterEPF1∷GFP (green) in abaxial rosette leaf. To highlight the outline of cells, leaves were stained with FM4-64, which accumulates in membranes (red color). The arrow and arrowhead indicate a meristemoid and a GMC, respectively. Bars: B,F, 5 mm; C–E,G, 50 μm.
EPF1 is genetically upstream of genes for TMM receptor-like protein, ER family receptor-like kinases, and the YDA MAPKKK
A simple unifying model for stomatal pattern generation would be that SDD1 cleaves and activates the EPF1 peptide, EPF1 serves as a ligand for TMM and the ER family (Ingram 2005; Shpak et al. 2005), and this positional cue is relayed through a YDA-dependent signaling cascade. This model was tested in a series of genetic experiments. Promoter2x35S-Ω-EPF1 decreases stomatal density (Fig. 3); this decrease in stomatal density is dependent on functional TMM, ER family genes and YDA because both the increased stomatal density and stomatal clustering phenotypes characteristic of tmm-1, yda, and er;erl1;erl2 are epistatic to Promoter2x35S-Ω-EPF1 (Fig. 3). Somewhat surprisingly, the EPF1 overexpression phenotype did not depend on SDD1 (Fig. 3). These results suggest that EPF1 functions upstream of YDA, TMM, and ER family genes, but acts independently from SDD1.
Figure 3.
Effect of EPF1 overexpression on stomatal density in different genetic backgrounds. Spots represent stomatal densities of independent T1 plants that had been transformed with the control vector (open spots) and Promoter2x35S-Ω-EPF1 (closed spots). (Top) Abaxial side of cotyledons of 15-d-old plants. (Bottom) Abaxial side of primary leaves of 20-d-old plants.
To further verify the epistatic relationships, we made epf1;tmm and epf1;sdd1 double mutants. The degree of stomatal clustering and increase in stomatal density in epf1;tmm was similar to that in the tmm single mutant (Fig. 4A; Supplmentary Fig. S3); in agreement with the idea that TMM is a receptor for EPF1. TMM is a negative regulator of stomatal formation in cotyledons and leaves, but is a positive regulator of stomatal formation in the stem (Yang and Sack 1995). Therefore, we also examined genetic interactions between epf1 and tmm in the stem. The epf1 mutation alone moderately increased the number of stomata in the stem (Supplementary Table 2; Supplementary Fig. S5), a phenotype opposite of tmm (Supplementary Fig. S5). The double tmm;epf1 mutant lacked stomata in the stem, again indicating that tmm is epistatic to epf1 (Supplementary Fig. S5). In contrast, the effects of epf1 and sdd1 mutations on clustering and density of stomata were additive (Fig. 4B; Supplementary Fig. S3), strengthening the conclusion that EPF1 and SDD1 function independently (Fig. 3). While these results suggest that SDD1 does not process the EPF1 ligand for use in stomatal patterning (von Groll et al. 2002), it is possible that SDD1 processes an yet unknown signaling molecule, or that SDD1 processes a different class of protein, such as TMM.
Figure 4.
Size distribution of stomatal clusters. (A) epf1, tmm, and epf1;tmm. (B) epf1, sdd1, and epf1;sdd1. Single stomata are not shown.
Mutations in known stomatal patterning genes affect both stomatal density and clustering. However, the effects of mutations on stomatal density and the one-cell-spacing rule are different between affected genes: TMM is crucial for limiting density of stomata and for orienting spacing divisions. SDD1 appears to have a primary role in controlling density and a secondary role in spacing (Nadeau and Sack 2003). epf1-1 has a consistent defect in enforcing the one-cell-spacing pattern, yet its effect on stomatal density is moderate compared with sdd1 or tmm (Supplementary Fig. S3; Supplementary Tables 1, 2), suggesting that EPF1 has a primary role in orienting spacing divisions.
A model of stomatal patterning
We have shown that EPF1 is expressed in the signaling cells of the stomatal lineage and influences stomatal density and pattern. The EPF1 overexpression phenotype is dependent on functional TMM and ER family genes, and tmm is epistatic to epf1. This, together with the predicted secretory nature of the EPF1 product and receptor-like structure of TMM and ER family proteins is consistent with the hypothesis that EPF1 diffuses from stomatal precursors, is perceived by TMM and ER family receptors in neighbor cells and provides the positional information to regulate asymmetric cell division, which in turn, results in proper cell fate determination. Our results do not exclude the possibility that EPF1 has additional functions through TMM and ER family proteins to enforce pavement cell fate after the asymmetric cell division occurs, or to decrease cell division frequency of cells that contact stomata or precursors. The use of positional information to bias the orientation of asymmetric divisions is a universal theme in development (Horvitz and Herskowitz 1992; Scheres and Benfey 1999; Betschinger and Knoblich 2004). In animals ranging from Caenorhabditis elegans to Drosophila and vertebrates, positional information—well-known positional cues for asymmetries during cell division are the Wnt-Frizzled family of signals and receptors—is translated through a set of conserved proteins into establishment of cell polarity (Goldstein et al. 2006). Plants achieved multicellularity independent of the animal lineages, and while both accomplish cell division and cell-type differentiation, the plant cells face constraints by the cell walls during development. Recruitment of a unique set of signaling molecules and receptors may have reflected the innovation by plants to orient asymmetric cell divisions and coordinate pattern formation.
Materials and methods
Plant materials and growth conditions
Arabidopsis ecotype Columbia was used in all experiments unless noted. Plants were grown in plates with GM medium (MS salts, 1% sucrose, 1/100 volume of 2.5% MES-KOH at pH 5.7, 0.3% Phytagel) under continuous light at 22°C.
Mutants used in this study are as follows: yda-Y295 (Bergmann et al. 2004), er, erl1-2, erl2-1 (Shpak et al. 2004), tmm (SALK_011959), sdd1 (GABI, 627-D04), and epf1-1 (SALK_137549). In epf1-1 (SALK_137549), the T-DNA is inserted 10 base pairs (bp) upstream of the translational initiation codon. The sequence around the T-DNA insert is as follows: 5′-cgtccgcaatgtgttattatcgttgtctaagttgtttctatttccttgttttcATAATA TATCATGAAGTCTCTTCTTCTCCTTGCCTTTTTCC-3′. In this sequence, the T-DNA region is shown with lowercase letters, the Arabidopsis genome sequence is shown with capital letters, and the translation initiation codon of EPF1 is underlined. In tmm (SALK_011959), a T-DNA is present immediately upstream of the 518th nucleotide of the coding region of the single-exon gene TMM, with its LB flanking the 518th nucleotide. In sdd1 (GABI, 627-D04), a T-DNA is present immediately upstream of the 137th nucleotide of the coding region of the single-exon gene SDD1, with its LB flanking the 137th nucleotide. Double homozygous mutants were selected from offspring of plants generated by crossing.
Screen for bioactive secretory peptides
A collection of all genes of Arabidopsis coding for small proteins (<150 amino acids in length) was analyzed using the PSORT program to predict the destination of the gene products. A set of genes coding for products predicted to enter the secretory pathway was selected for further study. Entire coding regions of a set of genes were PCR-amplified and cloned in the SmaI site of a plant expression vector, pTK014. pTK014 was made by replacing the GUS gene and the NOS terminator of binary vector pGPTV-Kan (Becker et al. 1992) with duplicated CaMV35S RNA gene enhancers, the CaMV 35S RNA gene minimum promoter, and the Ω leader sequence from pE2113 (Mitsuhara et al. 1996), a multicloning site including the SmaI site and CaMV 35S RNA gene terminator. One-hundred-fifty-three genes were transformed into Arabidopsis Columbia ecotype for overexpression. For most clones, >10 independent T1 transformants were examined for phenotypes. Through this screening, we found At2g20875 (EPF1), which caused a decrease in stomatal density when overexpressed. Subsequently, At2g20875 was also identified in the stomatal-enriched population in microarray data sets (Bergmann et al. 2004), and as a gene coexpressed with regulatory genes of stomatal development using Microarray data set (AtGenExpress Tissue Set analyzed by Expression Angler; http://bbc.botany.utoronto.ca/ntools/cgi-bin/ntools_expression_angler.cgi) (K.U. Torii, unpubl., data not shown).
In situ RNA hybridization
Whole-mount in situ RNA hybridization was performed using digoxigenin-labeled cRNA probes and alkaline phosphatase-labeled anti-digoxigenin, following the procedures outlined in Hejatko et al. (2006).
Plasmid constructs used in this study
For overexpression of EPF1, genomic sequence was PCR-amplified with the primers K148 (5′-AAAATGAAGTCTCTTCTTCTCCTTGCCTT-3′) and K149 (5′-AAGGAAAACAAAACGGTTGAATGCATAGA-3′). The underlined AAA sequence preceding the translation initiation codon ATG in K148 was added to improve translational efficiency because it matches the most frequent sequence preceding translational initiation codons in Arabidopsis. The amplified fragment was cloned into pTK014 (giving kanamycin resistance to plants) or a similar vector, pTK016 (BASTA resistance in plants). The resulting constructs—pTK014-EPF1 and pTK016-EPF1—were transformed into Arabidopsis using the flower-dipping method (Clough and Bent 1998).
For complementation of epf1-1, a 3308-bp genomic sequence consisting of a 2390-bp promoter, the entire coding region, and 517 bp of 3′ region was PCR-amplified from the genome of wild-type Columbia using primers no. 2731 (5′-CCGCTCGAGTGGTTATGAAATATTTTCCCCA CTT-3′) and no. 2786 (5′-CCCGAGCTCTAATTTTATTTTATTTTGTT GTAGAGGAGA-3′). Underlined regions introduce XhoI and SacI sites. The amplified fragment was digested with XhoI and SacI, and cloned between the XhoI and SacI sites of pGWB1, which is a binary vector with Hygromycin-selectable marker in plants. The resulting plasmid was transformed into plants homozygous for epf1-1.
For construction of PromoterEPF1-erGFP, a 2457-bp upstream region of EPF1 was PCR-amplified with the primers K176 (5′-ACGACGATGT CCTCTTTTGTCTTTGAGAA-3′) and K197 (5′-CCGCTCGAGGATAT ATTATCGCAAGTGGTAAAAGTAA-3′; the underlined region indicates an added XhoI site), and cloned between the SmaI and XhoI sites of pRK2, which carries erGFP (GFP with ER targeting signal). For construction of PromoterEPF1-erGFP, a 2457-bp upstream region of EPF1 was PCR-amplified with the use of the primers K176 (5′-ACGACGATGT CCTCTTTTGTCTTTGAGAA-3′) and K177 (5′-TGATATATTATCGC AAGTGGTAAAAGTAA-3′) and blunt-end-cloned in the SmaI site of pGPTV-Kan (Becker et al. 1992).
Phenotypic analysis
For analysis of the stomatal pattern, plants were fixed in 90% ethanol/10% acetic acid, washed in water, stained with 1 μg/mL Safranin O, washed with water, then examined with a confocal microscope (FV300, Olympus) using an excitation wavelength of 543-nm and emission spectrum of 570-nm light.
For determining the number of stomata in each cluster size and stomatal density, four square areas of 0.49 mm2 were counted per organ, and the numbers were averaged to yield an estimated cluster distribution per organ. Organs from at least six plants were used for each genotype.
For quantitative examination of the stem, pedicel, and silique, and for images in Figure 1, the epidermis of fixed samples was peeled and stained with 1 mg/mL Safranin O, and then examined under a transmission light microscope. Images of EPF1 expression were obtained by examining plants expressing PromoterEPF1-erGFP (488-nm Argon laser excitation, 510- to 530-nm emission) and counterstaining with a membrane marker FM4-64 (10 μg/mL) (488-nm Argon laser excitation, >570-nm emission).
Acknowledgments
We thank T. Nakagawa for pGWB1, D. Becker for pGPTV-Kan, and Y. Ohashi for pE2113. This work was supported by Grants-in-Aid for Scientific Research 15107001 (Japan Society for the Promotion of Science) and 17027017 (Ministry of Education, Sports, Science, and Technology), NSF grant IOB-0544895 and DOE grant DE-FG02-06ER15810 to D.C.B., and NSF grant IOB-0520548 to K.U.T. K.U.T. was an investigator of CREST, Japan Science and Technology Agency, and is an affiliate faculty at the University of Washington, Institute for Stem Cell and Regenerative Medicine.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1550707
References
- Baldauf S.L. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- Becker D., Kemper E., Schell J., Masterson R., Kemper E., Schell J., Masterson R., Schell J., Masterson R., Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Berger D., Altmann T., Altmann T. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes & Dev. 2000;14:1119–1131. [PMC free article] [PubMed] [Google Scholar]
- Bergmann D.C. Integrating signals in stomatal development. Curr. Opin. Plant Biol. 2003;7:26–32. doi: 10.1016/j.pbi.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Bergmann D.C., Lukowitz W., Somerville C.R., Lukowitz W., Somerville C.R., Somerville C.R. Stomatal development and pattern controlled by a MAPKK kinase. Science. 2004;304:1494–1497. doi: 10.1126/science.1096014. [DOI] [PubMed] [Google Scholar]
- Betschinger J., Knoblich J.A., Knoblich J.A. Dare to be different: Asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr. Biol. 2004;14:R674–R685. doi: 10.1016/j.cub.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Geisler M., Nadeau J., Sack F.D., Nadeau J., Sack F.D., Sack F.D. Oriented asymmetric divisions that generate the stomatal spacing pattern in arabidopsis are disrupted by the too many mouths mutation. Plant Cell. 2000;12:2075–2086. doi: 10.1105/tpc.12.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B., Takeshita H., Mizumoto K., Sawa H., Takeshita H., Mizumoto K., Sawa H., Mizumoto K., Sawa H., Sawa H. Wnt signals can function as positional cues in establishing cell polarity. Dev. Cell. 2006;10:391–396. doi: 10.1016/j.devcel.2005.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejatko J., Blilou I., Brewer P.B., Friml J., Scheres B., Benkova E., Blilou I., Brewer P.B., Friml J., Scheres B., Benkova E., Brewer P.B., Friml J., Scheres B., Benkova E., Friml J., Scheres B., Benkova E., Scheres B., Benkova E., Benkova E. In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat. Protocols. 2006;1:1939–1946. doi: 10.1038/nprot.2006.333. [DOI] [PubMed] [Google Scholar]
- Horvitz H.R., Herskowitz I., Herskowitz I. Mechanisms of asymmetric cell division: Two Bs or not two Bs, that is the question. Cell. 1992;68:237–255. doi: 10.1016/0092-8674(92)90468-r. [DOI] [PubMed] [Google Scholar]
- Ingram G.C. Plant development: Spacing out stomatal pores. Curr. Biol. 2005;15:R663–R665. doi: 10.1016/j.cub.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Lai L.B., Nadeau J.A., Lucas J., Lee E.K., Nakagawa T., Zhao L., Sack F.D., Nadeau J.A., Lucas J., Lee E.K., Nakagawa T., Zhao L., Sack F.D., Lucas J., Lee E.K., Nakagawa T., Zhao L., Sack F.D., Lee E.K., Nakagawa T., Zhao L., Sack F.D., Nakagawa T., Zhao L., Sack F.D., Zhao L., Sack F.D., Sack F.D. The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell. 2005;17:2754–2767. doi: 10.1105/tpc.105.034116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macalister C.A., Ohashi-Ito K., Bergmann D.C., Ohashi-Ito K., Bergmann D.C., Bergmann D.C. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature. 2007;445:537–540. doi: 10.1038/nature05491. [DOI] [PubMed] [Google Scholar]
- Mitsuhara I., Ugaki M., Hirochika H., Ohshima M., Murakami T., Gotoh Y., Katayose Y., Nakamura S., Honkura R., Nishimiya S., Ugaki M., Hirochika H., Ohshima M., Murakami T., Gotoh Y., Katayose Y., Nakamura S., Honkura R., Nishimiya S., Hirochika H., Ohshima M., Murakami T., Gotoh Y., Katayose Y., Nakamura S., Honkura R., Nishimiya S., Ohshima M., Murakami T., Gotoh Y., Katayose Y., Nakamura S., Honkura R., Nishimiya S., Murakami T., Gotoh Y., Katayose Y., Nakamura S., Honkura R., Nishimiya S., Gotoh Y., Katayose Y., Nakamura S., Honkura R., Nishimiya S., Katayose Y., Nakamura S., Honkura R., Nishimiya S., Nakamura S., Honkura R., Nishimiya S., Honkura R., Nishimiya S., Nishimiya S., et al. Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 1996;37:49–59. doi: 10.1093/oxfordjournals.pcp.a028913. [DOI] [PubMed] [Google Scholar]
- Nadeau J.A., Sack F.D., Sack F.D. Control of stomatal distribution on the Arabidopsisleaf surface. Science. 2002a;296:1697–1700. doi: 10.1126/science.1069596. [DOI] [PubMed] [Google Scholar]
- Nadeau J.A., Sack F.D., Sack F.D. Stomatal development in Arabidopsis. In: Somerville C.R., Meyerowitz E.M., Meyerowitz E.M., editors. The Arabidopsis book. American Society of Plant Biologists; Rockville, MD: 2002b. http://www.aspb.org/publications/arabidopsis. [DOI] [Google Scholar]
- Nadeau J.A., Sack F.D., Sack F.D. Stomatal development: Cross talk puts mouths in place. Trends Plant Sci. 2003;8:294–299. doi: 10.1016/S1360-1385(03)00102-X. [DOI] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht J., Brunak S., von Heijne G., Engelbrecht J., Brunak S., von Heijne G., Brunak S., von Heijne G., von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Bergmann D.C., Bergmann D.C. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell. 2006;18:2493–2505. doi: 10.1105/tpc.106.046136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L.J., Sloan D.B., Bogenschutz N.L., Torii K.U., Sloan D.B., Bogenschutz N.L., Torii K.U., Bogenschutz N.L., Torii K.U., Torii K.U. Termination of asymmetric cell division and differentiation of stomata. Nature. 2007;445:501–505. doi: 10.1038/nature05467. [DOI] [PubMed] [Google Scholar]
- Sachs T. Pattern formation in plant tissues. Cambridge University Press; Cambridge, UK: 1991. [Google Scholar]
- Scheres B., Benfey P.N., Benfey P.N. Asymmetric cell division in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:505–537. doi: 10.1146/annurev.arplant.50.1.505. [DOI] [PubMed] [Google Scholar]
- Shpak E.D., Berthiaume C.T., Hill E.J., Torii K.U., Berthiaume C.T., Hill E.J., Torii K.U., Hill E.J., Torii K.U., Torii K.U. Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development. 2004;131:1491–1501. doi: 10.1242/dev.01028. [DOI] [PubMed] [Google Scholar]
- Shpak E.D., McAbee J.M., Pillitteri L.J., Torii K.U., McAbee J.M., Pillitteri L.J., Torii K.U., Pillitteri L.J., Torii K.U., Torii K.U. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science. 2005;309:290–293. doi: 10.1126/science.1109710. [DOI] [PubMed] [Google Scholar]
- von Groll U., Berger D., Altmann T., Berger D., Altmann T., Altmann T. The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell. 2002;14:1527–1539. doi: 10.1105/tpc.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Sack F.D., Sack F.D. The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell. 1995;7:2227–2239. doi: 10.1105/tpc.7.12.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]