Abstract
Meiotic recombination between homologous chromosomes ensures their proper segregation at the first division of meiosis and is the main force shaping genetic variation of genomes. The HOP2 and MND1 genes are essential for this recombination: Their disruption results in severe defects in homologous chromosome synapsis and an early-stage failure in meiotic recombination. The mouse Hop2 and Mnd1 proteins form a stable heterodimer (Hop2/Mnd1) that greatly enhances Dmc1-mediated strand invasion. In order to elucidate the mechanism by which Hop2/Mnd1 stimulates Dmc1, we identify several intermediate steps in the homologous pairing reaction promoted by Dmc1. We show that Hop2/Mnd1 greatly stimulates Dmc1 to promote synaptic complex formation on long duplex DNAs, a step previously revealed only for bacterial homologous recombinases. This synaptic alignment is a consequence of the ability of Hop2/Mnd1 to (1) stabilize Dmc1–single-stranded DNA (ssDNA) nucleoprotein complexes, and (2) facilitate the conjoining of DNA molecules through the capture of double-stranded DNA by the Dmc1–ssDNA nucleoprotein filament. To our knowledge, Hop2/Mnd1 is the first homologous recombinase accessory protein that acts on these two separate and critical steps in mammalian meiotic recombination.
Keywords: DNA repair, Dmc1 recombinase, homologous recombination, strand invasion, synaptic complex
Homologous recombination serves a critical function in the repair of DNA double-strand breaks (DSBs) and in the proper segregation of homologous chromosomes in meiosis (Kleckner 1996; Roeder 1997). Failure to establish a physical connection through chiasmata causes missegregation of chromosomes at prophase I and results in meiotic cell apoptosis or aneuploid gametes. The Dmc1 recombinase, a eukaryotic homolog of the bacterial RecA protein, is expressed exclusively in meiotic cells and is a major player in meiotic homologous recombination. It promotes the search for homology and catalyzes the invasion of a single-stranded end generated by the 5′ resection of DSBs introduced by Spo11 into a homologous unbroken double-stranded DNA (dsDNA) to form joint molecules through strand invasion (D-loop formation) (Li et al. 1997; Masson et al. 1999; Hong et al. 2001; Masson and West 2001; Neale and Keeney 2006). The interaction between Hop2, Mnd1, and Dmc1 and/or Rad51, the ubiquitously expressed eukaryotic homolog of RecA, is crucial for the progression of meiotic homologous recombination. Biochemical studies have shown that the Hop2/Mnd1 complex physically interacts with and stimulates Dmc1 and Rad51 strand invasion activity (Chen et al. 2004; Petukhova et al. 2005; Pezza et al. 2006). The cooperation between Dmc1/Rad51 and Hop2/Mnd1 is likely to be crucial in vivo, since without Hop2 and/or Mnd1, in yeast (Leu et al. 1998; Gerton and DeRisi 2002; Tsubouchi and Roeder 2002, 2003; Zierhut et al. 2004; Henry et al. 2006), Arabidopsis thaliana (Domenichini et al. 2006; Kerzendorfer et al. 2006; Panoli et al. 2006), and mouse (Petukhova et al. 2003) recombination proceeds properly up to the point when Dmc1 and Rad51 are loaded onto the ends of DSBs, but further progression is impaired. Finally, the genetic interaction of DMC1 with HOP2/MND1 appears to be particularly important, as in all the genomes examined to date HOP2 and MND1 are found only in those organisms that have DMC1. For example, the genomes of Drosophila melanogaster and Caenorhabditis elegans do not contain HOP2, MND1, and DMC1, but RAD51 is present. Revealing the mechanism by which Hop2/Mnd1 stimulates Dmc1 is fundamental to understand the exquisitely regulated pathway of meiotic recombination.
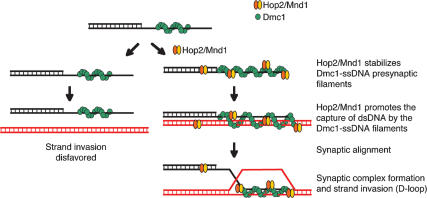
In this study we unraveled the mechanism by which the Hop2/Mnd1 complex stimulates Dmc1. We show that Hop2/Mnd1 greatly increases the homologous pairing promoted by Dmc1, as evidenced by the formation of the protein-decorated product of synaptic alignment (synaptic complex). This large stimulation can be accounted for by the effect of Hop2/Mnd1 on two consecutive intermediate steps in homologous pairing promoted by Dmc1: (1) The Hop2/Mnd1 heterodimer promotes the conjoining of DNA molecules through the capture of dsDNA to Dmc1–single-stranded DNA (Dmc1–ssDNA) nucleoprotein filaments, and (2) in an earlier step, Hop2/Mnd1 stabilizes Dmc1–ssDNA presynaptic filaments. We propose that Hop2/Mnd1 increases the stability of the Dmc1–ssDNA filament found on the resected DSBs and promotes the capture of potential partner chromosomes to facilitate the search for homology. Thus, our results indicate both a presynaptic and synaptic role for Hop2/Mnd1 in meiotic recombination. To our knowledge, this is the first time that the mechanism for an accessory protein for Dmc1 has been delineated. Hop2/Mnd1 is the only accessory homologous recombination protein that acts on these two critical and separate steps in mammalian meiotic recombination.
Results
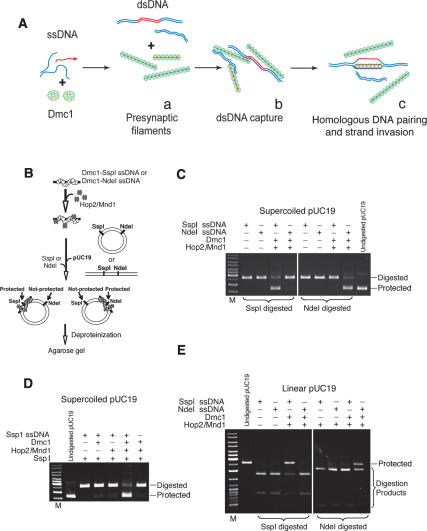
We have previously shown that the Hop2/Mnd1 heterodimer interacts physically with Dmc1 and stimulates its strand invasion activity by >30-fold (Petukhova et al. 2005; Pezza et al. 2006). In addition, we have shown that neither of the individual proteins, Hop2 or Mnd1, can stimulate Dmc1 and that the stimulation by Hop2/Mnd1 depends on the creation of a novel interface found in the heterodimer as only the heterodimer, but neither Hop2 nor Mnd1 alone can interact with Dmc1 (Pezza et al. 2006). In trying to dissect the mechanism by which Hop2/Mnd1 stimulates Dmc1-promoted strand invasion it is useful to distinguish at least three consecutive steps that have been revealed through the study of RecA-mediated strand invasion (Fig. 1A). The first two steps, presynaptic filament formation and the conjoining of DNAs, do not require homology but are prerequisites for the homologous pairing in the third step. In previous studies on the bacterial RecA recombinase and eukaryotic recombinase by us and others, this first homology-dependent step is revealed by the detection of protein-free D-loops stabilized on supercoiled duplex DNA. The superhelicity of the DNA stabilizes the ssDNA assimilated into the duplex (the D-loop) so that the ssDNA does not branch migrate out of the duplex. Protein-free D-loops on supercoiled DNA are related to, but quite distinct from, the biologically relevant first product of homologous pairing that includes the ssDNA, the attendant proteins, and the target duplex DNA (not necessarily topologically underwound or supercoiled). In this study, we describe these synaptic complexes on long dsDNA for Dmc1.
Figure 1.
Hop2/Mnd1 and Dmc1 proteins are required for the formation of a synaptic complex at sites of DNA homology. (A) Formation of joint molecules promoted by Dmc1. (Panel a) The presynaptic polymerization of Dmc1 protein on ssDNA. (Panel b) Conjunction of ssDNA and dsDNA without homologous alignment. (Panel c) Homologous DNA pairing and strand invasion. Deproteinization of the complex in panel c in the context of a supercoiled target duplex DNA results in a stable D-loop. (B) Experimental design. ssDNA oligonucleotide was used as a binding substrate for Dmc1 and Hop2/Mnd1 proteins. The nucleoprotein complexes were incubated with pUC19 dsDNA and treated with SspI or NdeI restriction endonucleases, followed by deproteinization and electrophoresis in 1% agarose gel. (C) Hop2/Mnd1 and the Dmc1–ssDNA nucleoprotein complex are targeted to sites in which ssDNA and supercoiled dsDNA are homologous, thus protecting dsDNA from endonuclease degradation. (D) Both Dmc1 and Hop2/Mnd1 proteins are required for the protection of dsDNA. (E) Linear pUC19 dsDNA is a substrate for synaptic complex formation. After Dmc1 and Hop2/Mnd1 were assembled on the ssDNA oligonucleotide, linear pUC19 dsDNA was added to the reaction and the nucleoprotein complexes were treated with SspI or NdeI restriction endonucleases.
The Hop2/Mnd1 complex is required for the synaptic complex formation promoted by Dmc1
The first product of homologous pairing on long dsDNA shown in Figure 1A (homologous DNA pairing) has so far never been delineated for a eukaryotic recombinase without the use of deproteinization and supercoiled duplex DNA. Before strand invasion (D-loop formation), RecA promotes homologous alignment of ssDNA and dsDNA molecules (Hsieh et al. 1992). In this reaction, a presynaptic filament of ssDNA and recombination proteins is allowed to find its homologous target embedded in an otherwise heterologous DNA, and the product is the synaptic complex (Fig. 1B). The formation of this recombinational intermediate is accomplished by incubating duplex DNA, a homologous oligonucleotide, and Dmc1 either in the presence or absence of Hop2/Mnd1. In this version of the experiment, the homologous oligonucleotide spans either one of two restriction endonuclease recognition sites in the duplex DNA (SspI and NdeI). Formation of a synaptic complex involving the proteins, duplex DNA, and one of the homologous oligonucleotides renders the duplex resistant to cleavage by one of the restriction endonucleases (Fig. 1B; Hsieh et al. 1992). The combination of the ssDNA oligonucleotide spanning the restriction endonuclease site being probed, dsDNA, Dmc1, and Hop2/Mnd1 protects dsDNA from the restriction endonuclease (Fig. 1C,D), and under our experimental conditions, Hop2/Mnd1 is required to form these synaptic complexes (Fig. 1D). In the absence of ATP, we observed a complete digestion of dsDNA, suggesting that the assembly of synaptic complexes depends on the formation of active Dmc1–ssDNA presynaptic filaments (data not shown). We also tested the formation of synaptic complexes using linear dsDNA, obtaining similar results to those described above (Fig. 1E).
These results indicate that neither supercoiling of the target duplex nor deproteinization is required to detect the first product of homologous pairing. In aggregate, we find that the Hop2/Mnd1 heterodimer greatly enhances the alignment of homologous sequences promoted by Dmc1.
The Hop2/Mnd1 heterodimer promotes the capture of duplex DNA by the Dmc1–ssDNA filament
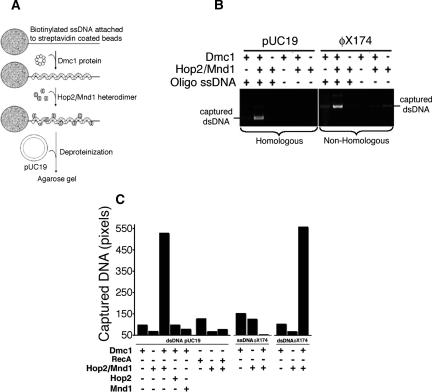
Because the Hop2/Mnd1 heterodimer binds dsDNA (Chen et al. 2004; Petukhova et al. 2005; Enomoto et al. 2006; Pezza et al. 2006) and physically interacts with Dmc1 nucleated on ssDNA (Supplementary Fig. S1A), the question arises as to whether Hop2/Mnd1 can bring dsDNA to preformed Dmc1–ssDNA nucleoprotein filaments, an essential prerequisite for alignment of homologous sequences. First, we confirmed using electron microscopy that Dmc1 forms filaments on ssDNA in the presence of ATP (data not shown). To obtain physical evidence for this capture mechanism, we evaluated the extent of the binding of dsDNA to immobilized Dmc1–ssDNA filaments (Fig. 2A–C). Notably, we observed that the addition of Hop2/Mnd1 to Dmc1–ssDNA nucleoprotein filaments leads to a large increase in the dsDNA retained by the beads. The addition of either Hop2 or Mnd1 alone to Dmc1–ssDNA filaments shows no effect on the capture of dsDNA. The observations that the addition of Hop2/Mnd1 to either RecA or to a defective Dmc1–ssDNA nucleoprotein filament (generated using a mutant Dmc1 deficient in DNA binding, A272P) (Bannister et al. 2007) results in a marginal increase of dsDNA in the beads (Fig. 2C; Supplementary Fig. S1B, respectively) suggest that the effect observed is specific for both the Hop2/Mnd1 complex and the Dmc1–ssDNA nucleoprotein filament. We further confirm the requirement for an active Dmc1–ssDNA filament in the capture of duplex DNA. After incubation of Dmc1 with ssDNA-beads in the presence of ATP (leading to the formation of active helical filaments on DNA) or in the absence of nucleotide factor (Dmc1 binds DNA but not as active filaments), we analyzed the fraction corresponding to Dmc1 bound to the ssDNA-beads and scored the extent of duplex capture. Whereas we observed similar quantities of Dmc1 and Hop2/Mnd1 bound to the ssDNA in both conditions tested, only Dmc1 in the presence of both Hop2/Mnd1 and ATP captures duplex DNA (Supplementary Fig. S1C,D). In a different experiment, we show that soluble Dmc1 does not functionally interact with Hop2/Mnd1. Here, we preincubate a molar excess of soluble Dmc1 with limited amounts of Hop2/Mnd1 and found that this mixture still stimulates the capture of duplex DNA by a preformed Dmc1–ssDNA filament (Supplementary Fig. S1E). In sum, these results show that the capture of duplex DNA stimulated by Hop2/Mnd1 is mediated by active Dmc1–ssDNA nucleoprotein filaments.
Figure 2.
The Hop2/Mnd1 heterodimer stimulates the capture of duplex DNA by the Dmc1–ssDNA nucleoprotein complex. (A) The experimental outline. Biotinylated DNA, immobilized on streptavidin-agarose beads, was used as a binding substrate for Dmc1 and Hop2/Mnd1 proteins. After incubation with pUC19 dsDNA, nucleoprotein complexes were separated from unbound proteins and dsDNA by centrifugation, and products were deproteinized before analysis by gel electrophoresis. (B) The products of dsDNA capture revealed by agarose gel electrophoresis. There is an increase in the homologous (pUC19) and nonhomologous (ϕX174, RFI) dsDNA captured by Dmc1–ssDNA nucleoprotein complexes in the presence of Hop2/Mnd1. In control reactions, the oligonucleotide ssDNA substrate was replaced by standard binding buffer. (C) Quantitation of the ethidium-bromide signals from gels similar to those shown in B and plotted as the number of pixels per band.
We also found that neither homology (Fig. 2B,C) nor supercoiling (data not shown) is required for the capture of a duplex DNA by the Dmc1–ssDNA filament. Finally, we asked whether Dmc1 filaments on dsDNA might also capture dsDNA. Supplementary Figure S1, F and G, show that the Hop2/Mnd1 heterodimer does not stimulate the capture of dsDNA by Dmc1–dsDNA complexes. Together with the results that Hop2/Mnd1 does not stimulate capture of ssDNA by the Dmc1–ssDNA nucleoprotein complex (Fig. 2C), our results indicate that only filaments of Dmc1 on ssDNA form ternary complexes with duplex DNA.
The Hop2/Mnd1 heterodimer stabilizes the Dmc1–ssDNA nucleoprotein filament against dissociation
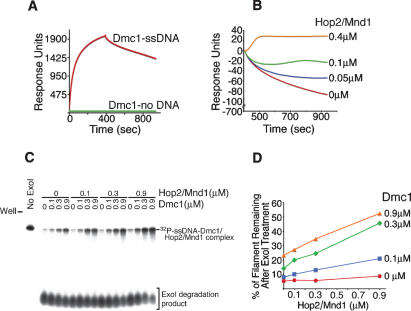
We asked whether Hop2/Mnd1 has an additional role prior to duplex DNA capture. We used surface plasmon resonance experiments (Fig. 3A) to show the stabilization of Dmc1–ssDNA filaments by Hop2/Mnd1 (Fig. 3B). In a different experiment, we verified that the surface plasmon resonance signals corresponding to direct binding of Hop2/Mnd1 to ssDNA, or to its association with the Dmc1–ssDNA nucleoprotein filament, does not account for the flattening of the slope seen in the dissociation phase after Hop2/Mnd1 is added (Supplementary Fig. S2). The addition of Hop2/Mnd1 has no significant effect on the association phase of the Dmc1–ssDNA filaments (Supplementary Fig. S2).
Figure 3.
The Hop2/Mnd1 protein stabilizes presynaptic Dmc1 filaments formed on ssDNA. (A) Real-time interactions of Dmc1 with ssDNA were analyzed using surface plasmon resonance. (B) Real-time analyses of the dissociation phase of the Dmc1–ssDNA filament with increasing amounts of Hop2/Mnd1. (C) The binding of Hop2/Mnd1 protein to the Dmc1–ssDNA nucleoprotein filament protects ssDNA against exonuclease I degradation. Complexes of a 32P-labeled 100-mer oligonucleotide ssDNA plus Dmc1 and varying amounts of Hop2/Mnd1 were formed as described in Materials and Methods. The complex was treated with ExoI, followed by electrophoresis on a polyacrylamide gel. (D) Quantitation of the radioactive signal corresponding to protected ssDNA in Dmc1–ssDNA/Hop2/Mnd1 complexes.
We then used an independent approach to confirm the stabilization effect of Hop2/Mnd1 on Dmc1–ssDNA filaments. The Hop2/Mnd1 heterodimer was added to the Dmc1–ssDNA nucleoprotein filaments preformed on a 5′-radiolabeled 100-mer oligonucleotide, and then the ternary complexes were treated with exonuclease I (Lusetti et al. 2004). The binding of either Dmc1 or Hop2/Mnd1 alone provided only a slight protection of ssDNA against degradation by exonuclease I. However, the addition of Hop2/Mnd1 to the Dmc1–ssDNA filament resulted in a Hop2/Mnd1 concentration-dependent protection of the ssDNA against exonuclease I degradation (up to 53% of protection) (Fig. 3C,D). These results are consistent with Hop2/Mnd1 stabilizing the Dmc1–ssDNA nucleoprotein complex. Using these two independent approaches, we demonstrated that the stability of preformed Dmc1–ssDNA nucleoprotein filaments is increased through interaction with the Hop2/Mnd1 heterodimer.
Hop2/Mnd1 stimulates two separate critical steps in Dmc1-promoted homologous pairing
In principle, all of the stimulation we observed by Hop2/Mnd1 on the intermediate steps downstream from presynaptic filament formation, up to and including strand invasion, could be a consequence of the robust stabilization of the Dmc1–ssDNA filament by the heterodimer. In order to address whether this is in fact the case, we sought conditions to stabilize the presynaptic filament in the absence of Hop2/Mnd1 and asked whether this stabilization was sufficient to stimulate Dmc1-promoted strand invasion or synaptic complex formation.
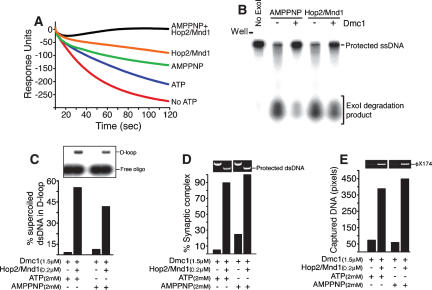
It has been shown that Hop2/Mnd1 can stimulate the activity of Dmc1 for forming D-loops in the presence of the nonhyrolyzable ATP analog, AMPPNP (Petukhova et al. 2005). To allow comparison of the results obtained for filament stabilization (surface plasmon resonance experiments) with those for D-loop formation, synaptic complex formation, and duplex capture, we used the same biotinylated oligonucleotide, oligonucleotide #3 (Supplementary Table S1). While the presynaptic filament is highly stable in the presence of AMPPNP and in the absence of the Hop2/Mnd1 heterodimer (Fig. 4A,B), the yield of D-loops (Sehorn et al. 2004) and the level of synaptic complex formation is much lower than with Hop2/Mnd1 (Fig. 4C,D). Similar results were obtained when Ca++ was used to stabilize the Dmc1–ssDNA filaments (Bugreev et al. 2005; data not shown).
Figure 4.
The stabilization of the Dmc1–ssDNA presynaptic filament is not sufficient to stimulate Dmc1-promoted strand invasion. (A) The real-time dissociation phase of Dmc1 from ssDNA in the presence of AMPPNP plus Hop2/Mnd1, Hop2/Mnd1, AMPPNP, and buffer with or without ATP was analyzed using surface plasmon resonance. (B) Hop2/Mnd1 and Dmc1–ssDNA complexes protect DNA against exonuclease I degradation in the presence of AMPPNP. (C) D-loop formation carried out with Dmc1 (1.5 μM) allowed to bind a 57-mer oligonucleotide (#3, ssDNA, 4.5 μM nucleotide), in a buffer containing 2.5 mM MgCl2, before the addition of Hop2/Mnd1 (0.2 μM) and supercoiled pUC19 (10 μM bp). (D) The extent of synaptic complex formation by the Dmc1 filament with and without Hop2/Mnd1 in the presence of ATP or AMPPNP. (E) Bars represent the quantitation of the ethidium-bromide signal corresponding to the dsDNA captured by the Dmc1–ssDNA filament alone or in the presence of Hop2/Mnd1.
These results show that the stabilization of the Dmc1–ssDNA filament by Hop2/Mnd1 is not sufficient to account for most of the stimulation of homologous pairing. Thus, Hop2/Mnd1 must also have an effect on the second intermediate step, duplex capture (Fig. 4E).
Discussion
The Dmc1 recombinase pairs single-stranded or partially single-stranded DNA with homologous duplex DNA to form joint molecules by strand invasion. Here we show direct evidence that, as previously described for RecA and other RecA-related proteins (Camerini-Otero and Hsieh 1995; Bianco et al. 1998; Cox 2003), the Dmc1 homologous pairing reaction proceeds in at least three sequential steps (Figs. 1A, 5).
Figure 5.
The Hop2/Mnd1 complex functions by stabilizing recombinational intermediates promoted by Dmc1. The proposed model summarizes observations of this and previous studies. Hop2/Mnd1 assists Dmc1 in the formation of recombinational intermediates. The stabilization of homologous pairing intermediates allows Dmc1 to find regions of DNA homology, leading to strand invasion. Hop2/Mnd1 protein has two actions: One stabilizes Dmc1 nucleated on processed ends of DSBs (ssDNA), and the second enhances the conjunction of DNA molecules through the specific capture of dsDNA to Dmc1–ssDNA nucleoprotein filaments.
Synaptic complexes as intermediates in the homologous pairing promoted by Dmc1
Traditionally, the homologous DNA pairing activity of a recombinase is scored by the formation of D-loops that result from the trapping of an ssDNA (Shibata et al. 1979) or oligonucleotide (Hsieh et al. 1992) by a homologous region of supercoiled plasmid DNA after deproteinization. In order to analyze the formation of Dmc1 and Hop2/Mnd1-mediated homology-dependent and protein-containing synaptic complexes, we adopted a restriction endonuclease protection approach originally developed by us for use with Escherichia coli RecA (Ferrin and Camerini-Otero 1991; Hsieh et al. 1992). Using this method, we were able to detect a synaptic complex as an intermediate of the homologous pairing reaction, both on superhelical and linear target DNAs, and to examine the requirements for Dmc1-mediated homologous alignment (Fig. 1). Our results indicate that the detection of the in vitro pairing that we observe promoted by Dmc1 and stimulated by Hop2/Mnd1 requires neither supercoiling of the target duplex to stabilize the complex nor the deproteinization of the pairing intermediate to induce some structural rearrangements. Using an oligonucleotide-based assay, it has been previously shown that Dmc1 is able to form such synaptic complex intermediates (Gupta et al. 2001). Here, we present an alternative assay, in that long duplex DNA is used. Our results indicate that Dmc1 is able to find homology in the presence of a great excess of nonhomologous DNA, leading to the formation of the first product of homologous pairing.
The ability of Dmc1 and Hop2/Mnd1 to target an oligonucleotide to a specific site on a large duplex DNA suggests that, in vivo, the interplay between these proteins is required to achieve homologous-specific pairing in a complex genome such as that of eukaryotes. Indeed, the characterization of the product of homologous alignment shown in this work (synaptic complexes) will allow us to study both the kinetics and the sequence requirements for the search for homology that is arguably the central problem in the process of chromosome pairing during meiosis (Ferrin and Camerini-Otero 1991; Yancey-Wrona and Camerini-Otero 1995).
Hop2/Mnd1 promotes the capture of duplex DNA by the Dmc1–ssDNA filament in a homology-independent manner
Recent biochemical studies have attracted attention to the role of accessory proteins on the mechanism of action of Dmc1 (Chen et al. 2004; Sehorn et al. 2004; Petukhova et al. 2005). For the efficient homologous alignment promoted by Dmc1, the essential prerequisites are the formation of stable presynaptic Dmc1–ssDNA filaments and the conjoining of ssDNA and dsDNA in a homology-independent manner. Hop2/Mnd1 greatly enhances the ability of Dmc1 to bring ssDNA and dsDNA together to form a ternary complex composed of Dmc1–ssDNA nucleoprotein filaments and duplex DNA. While Hop2/Mnd1 stimulates Dmc1–ssDNA filaments to capture duplex DNA in a homology-independent manner (Fig. 2B,C), this step is of paramount importance for the subsequent homologous pairing. Such a capture is proposed to be an important step in the reaction of homologous DNA pairing by the prototypic recombinase, E. coli RecA (Gonda and Radding 1983; Flory et al. 1984), and it might be an obligatory process in the homologous pairing pathways governed by all RecA-like proteins. Our data on DNA capture show that Dmc1 promotes the capture of both homologous and heterologous DNA (increased in the presence of Hop2/Mnd1), suggesting that Dmc1 promotes homologous alignment as observed for RecA (Gonda and Radding 1983; Flory et al. 1984; Julin et al. 1986; Hsieh et al. 1992), in which DNA molecules would first form an unaligned ternary complex and then align within the ternary complex.
Hop2/Mnd1 stabilizes the Dmc1–ssDNA filament
In an earlier step, Hop2/Mnd1 contributes to homologous DNA pairing by stabilizing the presynaptic filament formed by Dmc1 on ssDNA (Fig. 3B,D). In this regard it is interesting to note that the 5′ strand of DSBs are hyperresected by exonucleases when Dmc1 and/or Rad51 is missing (Bishop et al. 1992; Shinohara et al. 1992; Schwacha and Kleckner 1997). This finding indicates that nuclease and Dmc1 are somehow competing for DNA substrate. Consistent with our observation that Hop2/Mnd1 stabilizes presynaptic Dmc1–ssDNA filaments, previous data in yeast Mnd1- and Hop2-null mutants show that the ends of DSBs are more extensively degraded (Leu et al. 1998; Gerton and DeRisi 2002; Zierhut et al. 2004; Henry et al. 2006).
Again, it is important to note that, although both the stability of the Dmc1–ssDNA filament and the capture of the duplex DNA are promoted by Hop2/Mnd1, a stable filament is not per se a better captor of duplexes (Fig. 4), and Hop2/Mnd1 must actively participate in the capture process.
A presynaptic and synaptic role for Hop2/Mnd1 in meiotic recombination
In several steps of the recombination pathway, accessory proteins are required for recombinase function. For RecA (Kowalczykowski et al. 1994; Papavinasasundaram et al. 1997; Voloshin et al. 2001) and Rad51 (Sung et al. 2003; Krogh and Symington 2004) a number of well-characterized factors have been described. For Dmc1, however, the detailed biochemical mechanisms of its accessory proteins are unknown (Chen et al. 2004; Hayase et al. 2004; Sehorn et al. 2004; Petukhova et al. 2005; Haruta et al. 2006; Pezza et al. 2006; Sarai et al. 2006). Both genetics and biochemical studies have demonstrated the functional interaction between Dmc1 and Hop2/Mnd1 proteins. In yeast (Leu et al. 1998; Gerton and DeRisi 2002; Tsubouchi and Roeder 2002, 2003; Zierhut et al. 2004; Henry et al. 2006), A. thaliana (Domenichini et al. 2006; Kerzendorfer et al. 2006; Panoli et al. 2006), and mouse (Petukhova et al. 2003) Hop2- or Mnd1-null mutants, recombination proceeds properly up to the point when Dmc1 and Rad51 are loaded onto the ends of processed DSBs, but further progression is impaired. These results are closely coupled with findings from biochemical studies. Purified mammalian and yeast Hop2/Mnd1 complexes all stimulate D-loop formation (Chen et al. 2004; Petukhova et al. 2005; Pezza et al. 2006) and early steps in the strand exchange reaction (Enomoto et al. 2006) promoted by the Dmc1 protein. Until this study, the mechanism of action of Hop2/Mnd1 was unclear (Neale and Keeney 2006). The data shown here indicate that Hop2/Mnd1 greatly enhances the formation and/or stabilizes intermediates in Dmc1-mediated homologous pairing consistent with both a presynaptic and synaptic role for Hop2/Mnd1 (Fig. 5). In an accompanying paper, Chi et al. (2007) show that Hop2/Mnd1 has similar effects on the activities of Rad51.
In line with our biochemical findings, the role of Hop2/Mnd1 as a synaptic effector of Dmc1 has strong support from cytological and genetic data. In yeast and mice, Hop2- and Mnd1-null mutants show a near-complete block to recombination and in yeast, DNA intermediates of recombination, either Holliday junctions or stable strand invasion products, are completely absent (Gerton and DeRisi 2002; Tsubouchi and Roeder 2002; Henry et al. 2006). Based on our observations, we can imagine that in the absence of Hop2/Mnd1, Dmc1 will form a relatively unstable (perhaps shorter) filament on ssDNA that will not be able to initiate the synaptic phase through the formation of a ternary complex of the Dmc1–ssDNA filament with duplex DNA (failure to capture dsDNA). Consequently, a delay in the progression of meiosis will cause chromosomes to synapse nonhomologously as observed in yeast, or mostly not at all, as observed in mice.
The enhancement of these sequential steps in homologous pairing by Hop2/Mnd1 might help Dmc1 process a variety of structural barriers in vivo or increase the efficiency of processing of the large number of DSBs (several hundred in mice) introduced by Spo11 during the initiation of meiotic homologous recombination.
In summary, we propose that in meiotic recombination Hop2/Mnd1 promotes chromosome synapsis by stabilizing Dmc1 on the resected DNA at DSBs and by participating directly, presumably both by protein–protein and protein–DNA interactions, in the capture of potential partner chromosomes.
Materials and methods
Proteins, DNAs, and oligonucleotides
The mouse Hop2 (Petukhova et al. 2005) and Mnd1 (Pezza et al. 2006) proteins and the Hop2/Mnd1 (Petukhova et al. 2005) complex were purified as described previously. The human Dmc1 was purified according to a published protocol (Masson et al. 1999). pUC19 (except for that used to form D-loops) and ϕX174 were purchased from New England BioLabs and full-length linear duplex DNA was generated by the digestion of ϕX174 dsDNA (RFI) with XhoI. Oligonucleotides, one of which (as specified) was modified at the 3′ end by a biotin residue, were from MWG Biotech. The sequence of the oligonucleotides is given in Supplementary Table Sl. The concentration of ssDNA and dsDNA were determined by absorbance at 260 nm, using 36 μg/mL and 50 μg/mL A260, respectively, as conversion factors, and are expressed as molar concentrations of nucleotides or base pairs for ssDNA or dsDNA, respectively.
Synaptic complex formation
In a reaction volume of 10 μL, Dmc1 protein (4 μM) was incubated with ssDNA (3 μM nucleotide) (oligonucletides #1, SspI; or #2 NdeI; or #3, ScaI) (Supplementary Table S1) in standard binding buffer (20 mM Tris-HCl at pH 7.4, 70 mM NaCl, 2.5 mM MgCl2, 1 mM DTT, 2 mM ATP, 7.5 mM creatine phosphate, 30 U/mL creatine kinase). For experiments shown in Figure 4, the concentration of Dmc1 was 1.5 μM. Samples containing the Dmc1–ssDNA complexes were mixed with Hop2/Mnd1 (0.5 μM) and pUC19 dsDNA (15 μM bp), and DNA cleavage was initiated by the addition of SspI or NdeI restriction endonucleases (1.25U and 5 U, respectively). The reactions were carried out for 5 min at 37°C, and products were deproteinized, resolved on 1% agarose, and visualized with ethidium bromide.
Immobilization of biotinylated DNA on agarose steptavidin beads
Aliquots of a 50% (w/v) bead stock (Invitrogen) were mixed with biotinylated ssDNA or dsDNA (#3 or #3/4, respectively) (Supplementary Table S1) in 10 mM Tris-HCl (pH 7.4) and 20 mM NaCl, and immobilization was carried out for 30 min at 28°C. We used radioactive oligonucleotides to confirm that all of the DNA incubated with the beads was immobilized (data not shown). After immobilization, the beads were equilibrated in the original volume in standard binding buffer.
DNA capture assay
Figure 2A illustrates our experimental procedure for measuring the Hop2/Mnd1-promoted capture of duplex DNA by Dmc1–ssDNA nucleoprotein filaments. Unless indicated otherwise, Dmc1 (22 μM) was preincubated with DNA beads (60 μM nucleotides) in the standard binding buffer for 5 min at 37°C followed by the addition of Hop2/Mnd1 (0.4 μM), and was incubated for an additional 5 min after which 75 μM DNA (pUC19 dsDNA, ssDNA ϕX174, or dsDNA ϕX174) was added and the reactions were incubated for an additional 10 min. After completion of the reaction, beads were washed twice with 1 vol of standard binding buffer and resuspended in 25 μL of the same buffer, followed by deproteinization (0.5% [w/v] SDS, 1 mg/mL proteinase K) for 20 min at 37°C. The products were resolved on 1% agarose. The bands were visualized by ethidium-bromide staining, quantitated using a BAS 2500 Bio-Imaging Analysis System (Fuji Medical System), and expressed as the number of total pixels. For samples for protein analyses, beads were resuspended in 20 μL of LDS–polyacrylamide gel loading buffer and analyzed by electrophoresis in 4%–12% SDS–polyacrylamide gels. Protein bands were visualized by staining with Coomassie Brilliant Blue.
Surface plasmon resonance
Experiments were performed on a BIAcore 3000 instrument (BIAcore) at 25°C. The flow buffer was 20 mM Tris-HCl (pH 7.5), 70 mM NaCl, 6 mM MgCl2, 1 mM DTT, 1 mM ATP, and 0.005% surfactant P-20 (purchased from BIAcore). Biotinylated ssDNA (#3 for ssDNA) (Supplementary Table S1) were immobilized on an SA chip to give a signal of ∼100 response units. In order to form Dmc1–ssDNA filaments, 3 μM Dmc1 was injected at a flow rate of 10 μL/min for a contact time of 5 min. The binding phase of the experiment was followed by a dissociation step, where Dmc1–ssDNA complexes were allowed to dissociate in running buffer or in the presence of the Hop2/Mnd1 complex, Dmc1, or Dmc1 plus Hop2/Mnd1. The surfaces were regenerated with consecutive injections, 10 μL each, of 3 M NaCl, 2 M MgCl2, and 3M NaCl. The sensograms were normalized by subtracting the signal from the flow cell containing no immobilized DNA.
Exonuclease I protection assay
Dmc1–ssDNA filaments were formed by incubating Dmc1 with 5′ 32P-labeled phosphorylated synthetic 100-mer single-stranded fragment (#5) (Supplementary Table S1). The buffer conditions were identical to ones used in BIAcore experiments, except for surfactant P20 that was omitted. The volume of the reaction was 20 μL, and the incubation temperature was 37°C. After an initial 10 min of incubation, the Hop2/Mnd1 complex was added to the concentrations indicated and the reaction was allowed to proceed for an additional 10 min. The 3′ → 5′ digestion of DNA was initiated by addition of 1 μL of ExoI nuclease (10 U; United States Biochemicals). The digestion reaction was stopped after 20 min by the addition of 10 μL of a solution containing 60 mM EDTA, 6% SDS, and 30% glycerol. Ten microliters of the samples were electrophoresed on a 20% SequaGel (National Diagnostics) denaturing gel, and the radioactive signal was quantitated using a BAS 2500 Bio-Imaging Analysis System.
D-loop formation
D-loop formations were performed according to a published protocol with modifications (Petukhova et al. 2005).
Acknowledgments
We express our great appreciation to Peter Chi and Patrick Sung for sharing unpublished results and materials, as well as for their valuable ideas, helpful discussions, and a critical reading of this manuscript. Thanks also to P. Hsieh and E. Egelman for a critical reading of our manuscript. The electron microscopy of Dmc1–ssDNA filaments was carried out in the laboratory of Ed Egelman. The Intramural Research Program of the NIH, NIDDK, supported this research. We are grateful to Linda Robinson and Lorraine Moore for their assistance.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1562907
References
- Bannister L.A., Pezza R.J., Donaldson J.R., de Rooij D.G., Schimenti K.J., Camerini-Otero R.D., Schimenti J.C., Pezza R.J., Donaldson J.R., de Rooij D.G., Schimenti K.J., Camerini-Otero R.D., Schimenti J.C., Donaldson J.R., de Rooij D.G., Schimenti K.J., Camerini-Otero R.D., Schimenti J.C., de Rooij D.G., Schimenti K.J., Camerini-Otero R.D., Schimenti J.C., Schimenti K.J., Camerini-Otero R.D., Schimenti J.C., Camerini-Otero R.D., Schimenti J.C., Schimenti J.C. A dominant, recombination-defective allele of Dmc1 causing male-specific sterility. PLoS Biol. 2007;10:e105. doi: 10.1371/journal.pbio.0050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P.R., Tracy R.B., Kowalczykowski S.C., Tracy R.B., Kowalczykowski S.C., Kowalczykowski S.C. DNA strand exchange proteins: A biochemical and physical comparison. Front. Biosci. 1998;3:570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- Bishop D.K., Park D., Xu L., Kleckner N., Park D., Xu L., Kleckner N., Xu L., Kleckner N., Kleckner N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Bugreev D.V., Golub E.I., Stasiak A.Z., Stasiak A., Mazin A.V., Golub E.I., Stasiak A.Z., Stasiak A., Mazin A.V., Stasiak A.Z., Stasiak A., Mazin A.V., Stasiak A., Mazin A.V., Mazin A.V. Activation of human meiosis-specific recombinase Dmc1 by Ca2+ J. Biol. Chem. 2005;280:26886–26895. doi: 10.1074/jbc.M502248200. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R.D., Hsieh P., Hsieh P. Homologous recombination proteins in prokaryotes and eukaryotes. Annu. Rev. Genet. 1995;29:509–552. doi: 10.1146/annurev.ge.29.120195.002453. [DOI] [PubMed] [Google Scholar]
- Chen Y.K., Leng C.H., Olivares H., Lee M.H., Chang Y.C., Kung W.M., Ti S.C., Lo Y.H., Wang A.H., Chang C.S., Leng C.H., Olivares H., Lee M.H., Chang Y.C., Kung W.M., Ti S.C., Lo Y.H., Wang A.H., Chang C.S., Olivares H., Lee M.H., Chang Y.C., Kung W.M., Ti S.C., Lo Y.H., Wang A.H., Chang C.S., Lee M.H., Chang Y.C., Kung W.M., Ti S.C., Lo Y.H., Wang A.H., Chang C.S., Chang Y.C., Kung W.M., Ti S.C., Lo Y.H., Wang A.H., Chang C.S., Kung W.M., Ti S.C., Lo Y.H., Wang A.H., Chang C.S., Ti S.C., Lo Y.H., Wang A.H., Chang C.S., Lo Y.H., Wang A.H., Chang C.S., Wang A.H., Chang C.S., Chang C.S., et al. Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc. Natl. Acad. Sci. 2004;101:10572–10577. doi: 10.1073/pnas.0404195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P., San Filippo J., Sehorn M.G., Petukhova G.V., Sung P., San Filippo J., Sehorn M.G., Petukhova G.V., Sung P., Sehorn M.G., Petukhova G.V., Sung P., Petukhova G.V., Sung P., Sung P. Bipartite stimulatory action of the Hop2–Mnd1 complex on the Rad51 recombinase. Genes & Dev. 2007 doi: 10.1101/gad.1563007. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M.M. The bacterial RecA protein as a motor protein. Annu. Rev. Microbiol. 2003;57:551–577. doi: 10.1146/annurev.micro.57.030502.090953. [DOI] [PubMed] [Google Scholar]
- Domenichini S., Raynaud C., Ni D.A., Henry Y., Bergounioux C., Raynaud C., Ni D.A., Henry Y., Bergounioux C., Ni D.A., Henry Y., Bergounioux C., Henry Y., Bergounioux C., Bergounioux C. Atmnd1-δ1 is sensitive to γ-irradiation and defective in meiotic DNA repair. DNA Repair (Amst.) 2006;5:455–464. doi: 10.1016/j.dnarep.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Enomoto R., Kinebuchi T., Sato M., Yagi H., Kurumizaka H., Yokoyama S., Kinebuchi T., Sato M., Yagi H., Kurumizaka H., Yokoyama S., Sato M., Yagi H., Kurumizaka H., Yokoyama S., Yagi H., Kurumizaka H., Yokoyama S., Kurumizaka H., Yokoyama S., Yokoyama S. Stimulation of DNA strand exchange by the human TBPIP/Hop2/Mnd1 complex. J. Biol. Chem. 2006;281:5575–5581. doi: 10.1074/jbc.M506506200. [DOI] [PubMed] [Google Scholar]
- Ferrin L.J., Camerini-Otero R.D., Camerini-Otero R.D. Selective cleavage of human DNA: RecA-assisted restriction endonuclease (RARE) cleavage. Science. 1991;254:1494–1497. doi: 10.1126/science.1962209. [DOI] [PubMed] [Google Scholar]
- Flory S.S., Tsang J., Muniyappa K., Bianchi M., Gonda D., Kahn R., Azhderian E., Egner C., Shaner S., Radding C.M., Tsang J., Muniyappa K., Bianchi M., Gonda D., Kahn R., Azhderian E., Egner C., Shaner S., Radding C.M., Muniyappa K., Bianchi M., Gonda D., Kahn R., Azhderian E., Egner C., Shaner S., Radding C.M., Bianchi M., Gonda D., Kahn R., Azhderian E., Egner C., Shaner S., Radding C.M., Gonda D., Kahn R., Azhderian E., Egner C., Shaner S., Radding C.M., Kahn R., Azhderian E., Egner C., Shaner S., Radding C.M., Azhderian E., Egner C., Shaner S., Radding C.M., Egner C., Shaner S., Radding C.M., Shaner S., Radding C.M., Radding C.M. Intermediates in homologous pairing promoted by RecA protein and correlations of recombination in vitro and in vivo. Cold Spring Harb. Symp. Quant. Biol. 1984;49:513–523. doi: 10.1101/sqb.1984.049.01.058. [DOI] [PubMed] [Google Scholar]
- Gerton J.L., DeRisi J.L., DeRisi J.L. Mnd1p: An evolutionarily conserved protein required for meiotic recombination. Proc. Natl. Acad. Sci. 2002;99:6895–6900. doi: 10.1073/pnas.102167899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda D.K., Radding C.M., Radding C.M. By searching processively RecA protein pairs DNA molecules that share a limited stretch of homology. Cell. 1983;34:647–654. doi: 10.1016/0092-8674(83)90397-5. [DOI] [PubMed] [Google Scholar]
- Gupta R.C., Golub E., Bi B., Radding C.M., Golub E., Bi B., Radding C.M., Bi B., Radding C.M., Radding C.M. The synaptic activity of HsDmc1, a human recombination protein specific to meiosis. Proc. Natl. Acad. Sci. 2001;98:8433–8439. doi: 10.1073/pnas.121005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta N., Kurokawa Y., Murayama Y., Akamatsu Y., Unzai S., Tsutsui Y., Iwasaki H., Kurokawa Y., Murayama Y., Akamatsu Y., Unzai S., Tsutsui Y., Iwasaki H., Murayama Y., Akamatsu Y., Unzai S., Tsutsui Y., Iwasaki H., Akamatsu Y., Unzai S., Tsutsui Y., Iwasaki H., Unzai S., Tsutsui Y., Iwasaki H., Tsutsui Y., Iwasaki H., Iwasaki H. The Swi5–Sfr1 complex stimulates Rhp51/Rad51 and Dmc1-mediated DNA strand exchange in vitro. Nat. Struct. Mol. Biol. 2006;3:823–830. doi: 10.1038/nsmb1136. [DOI] [PubMed] [Google Scholar]
- Hayase A., Takagi M., Miyazaki T., Oshiumi H., Shinohara M., Shinohara A., Takagi M., Miyazaki T., Oshiumi H., Shinohara M., Shinohara A., Miyazaki T., Oshiumi H., Shinohara M., Shinohara A., Oshiumi H., Shinohara M., Shinohara A., Shinohara M., Shinohara A., Shinohara A. A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell. 2004;119:927–940. doi: 10.1016/j.cell.2004.10.031. [DOI] [PubMed] [Google Scholar]
- Henry J.M., Camahort R., Rice D.A., Florens L., Swanson S.K., Washburn M.P., Gerton J.L., Camahort R., Rice D.A., Florens L., Swanson S.K., Washburn M.P., Gerton J.L., Rice D.A., Florens L., Swanson S.K., Washburn M.P., Gerton J.L., Florens L., Swanson S.K., Washburn M.P., Gerton J.L., Swanson S.K., Washburn M.P., Gerton J.L., Washburn M.P., Gerton J.L., Gerton J.L. Mnd1/Hop2 facilitates Dmc1-dependent interhomolog crossover formation in meiosis of budding yeast. Mol. Cell. Biol. 2006;26:2913–2923. doi: 10.1128/MCB.26.8.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E.L., Shinohara A., Bishop D.K., Shinohara A., Bishop D.K., Bishop D.K. Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J. Biol. Chem. 2001;276:41906–41912. doi: 10.1074/jbc.M105563200. [DOI] [PubMed] [Google Scholar]
- Hsieh P., Camerini-Otero C.S., Camerini-Otero R.D., Camerini-Otero C.S., Camerini-Otero R.D., Camerini-Otero R.D. The synapsis event in the homologous pairing of DNAs: RecA recognizes and pairs less than one helical repeat of DNA. Proc. Natl. Acad. Sci. 1992;89:6492–6496. doi: 10.1073/pnas.89.14.6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julin D.A., Riddles P.W., Lehman I.R., Riddles P.W., Lehman I.R., Lehman I.R. On the mechanism of pairing of single- and double-stranded DNA molecules by the recA and single-stranded DNA-binding proteins of Escherichia coli. J. Biol. Chem. 1986;261:1025–1030. [PubMed] [Google Scholar]
- Kerzendorfer C., Vignard J., Pedrosa-Harand A., Siwiec T., Akimcheva S., Jolivet S., Sablowski R., Armstrong S., Mercier R., Vignard J., Pedrosa-Harand A., Siwiec T., Akimcheva S., Jolivet S., Sablowski R., Armstrong S., Mercier R., Pedrosa-Harand A., Siwiec T., Akimcheva S., Jolivet S., Sablowski R., Armstrong S., Mercier R., Siwiec T., Akimcheva S., Jolivet S., Sablowski R., Armstrong S., Mercier R., Akimcheva S., Jolivet S., Sablowski R., Armstrong S., Mercier R., Jolivet S., Sablowski R., Armstrong S., Mercier R., Sablowski R., Armstrong S., Mercier R., Armstrong S., Mercier R., Mercier R., et al. The Arabidopsis thaliana MND1 homologue plays a key role in meiotic homologous pairing, synapsis and recombination. J. Cell Sci. 2006;119:2486–2496. doi: 10.1242/jcs.02967. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Meiosis: How could it work? Proc. Natl. Acad. Sci. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski S.C., Dixon D.A., Eggleston A.K., Lauder S.D., Rehrauer W.M., Dixon D.A., Eggleston A.K., Lauder S.D., Rehrauer W.M., Eggleston A.K., Lauder S.D., Rehrauer W.M., Lauder S.D., Rehrauer W.M., Rehrauer W.M. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh B.O., Symington L.S., Symington L.S. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Leu J.Y., Chua P.R., Roeder G.S., Chua P.R., Roeder G.S., Roeder G.S. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell. 1998;94:375–386. doi: 10.1016/s0092-8674(00)81480-4. [DOI] [PubMed] [Google Scholar]
- Li Z., Golub E.I., Gupta R., Radding C.M., Golub E.I., Gupta R., Radding C.M., Gupta R., Radding C.M., Radding C.M. Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein. Proc. Natl. Acad. Sci. 1997;94:11221–11226. doi: 10.1073/pnas.94.21.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusetti S.L., Voloshin O.N., Inman R.B., Camerini-Otero R.D., Cox M.M., Voloshin O.N., Inman R.B., Camerini-Otero R.D., Cox M.M., Inman R.B., Camerini-Otero R.D., Cox M.M., Camerini-Otero R.D., Cox M.M., Cox M.M. The DinI protein stabilizes RecA protein filaments. J. Biol. Chem. 2004;279:30037–30046. doi: 10.1074/jbc.M403064200. [DOI] [PubMed] [Google Scholar]
- Masson J.Y., West S.C., West S.C. The Rad51 and Dmc1 recombinases: A non-identical twin relationship. Trends Biochem. Sci. 2001;26:131–136. doi: 10.1016/s0968-0004(00)01742-4. [DOI] [PubMed] [Google Scholar]
- Masson J.Y., Davies A.A., Hajibagheri N., Van Dyck E., Benson F.E., Stasiak A.Z., Stasiak A., West S.C., Davies A.A., Hajibagheri N., Van Dyck E., Benson F.E., Stasiak A.Z., Stasiak A., West S.C., Hajibagheri N., Van Dyck E., Benson F.E., Stasiak A.Z., Stasiak A., West S.C., Van Dyck E., Benson F.E., Stasiak A.Z., Stasiak A., West S.C., Benson F.E., Stasiak A.Z., Stasiak A., West S.C., Stasiak A.Z., Stasiak A., West S.C., Stasiak A., West S.C., West S.C. The meiosis-specific recombinase hDmc1 forms ring structures and interacts with hRad51. EMBO J. 1999;18:6552–6560. doi: 10.1093/emboj/18.22.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M.J., Keeney S., Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panoli A.P., Ravi M., Sebastian J., Nishal B., Reddy T.V., Marimuthu M.P., Subbiah V., Vijaybhaskar V., Siddiqi I., Ravi M., Sebastian J., Nishal B., Reddy T.V., Marimuthu M.P., Subbiah V., Vijaybhaskar V., Siddiqi I., Sebastian J., Nishal B., Reddy T.V., Marimuthu M.P., Subbiah V., Vijaybhaskar V., Siddiqi I., Nishal B., Reddy T.V., Marimuthu M.P., Subbiah V., Vijaybhaskar V., Siddiqi I., Reddy T.V., Marimuthu M.P., Subbiah V., Vijaybhaskar V., Siddiqi I., Marimuthu M.P., Subbiah V., Vijaybhaskar V., Siddiqi I., Subbiah V., Vijaybhaskar V., Siddiqi I., Vijaybhaskar V., Siddiqi I., Siddiqi I. AtMND1 is required for homologous pairing during meiosis in Arabidopsis. BMC Mol. Biol. 2006;27:7–24. doi: 10.1186/1471-2199-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavinasasundaram K.G., Movahedzadeh F., Keer J.T., Stoker N.G., Colston M.J., Davis E.O., Movahedzadeh F., Keer J.T., Stoker N.G., Colston M.J., Davis E.O., Keer J.T., Stoker N.G., Colston M.J., Davis E.O., Stoker N.G., Colston M.J., Davis E.O., Colston M.J., Davis E.O., Davis E.O. Mycobacterial recA is cotranscribed with a potential regulatory gene called recX. Mol. Microbiol. 1997;24:141–153. doi: 10.1046/j.1365-2958.1997.3441697.x. [DOI] [PubMed] [Google Scholar]
- Petukhova G.V., Romanienko P.J., Camerini-Otero R.D., Romanienko P.J., Camerini-Otero R.D., Camerini-Otero R.D. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev. Cell. 2003;5:927–936. doi: 10.1016/s1534-5807(03)00369-1. [DOI] [PubMed] [Google Scholar]
- Petukhova G.V., Pezza R.J., Vanevski F., Ploquin M., Masson J.Y., Camerini-Otero R.D., Pezza R.J., Vanevski F., Ploquin M., Masson J.Y., Camerini-Otero R.D., Vanevski F., Ploquin M., Masson J.Y., Camerini-Otero R.D., Ploquin M., Masson J.Y., Camerini-Otero R.D., Masson J.Y., Camerini-Otero R.D., Camerini-Otero R.D. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat. Struct. Mol. Biol. 2005;12:449–453. doi: 10.1038/nsmb923. [DOI] [PubMed] [Google Scholar]
- Pezza R.J., Petukhova G.V., Ghirlando R., Camerini-Otero R.D., Petukhova G.V., Ghirlando R., Camerini-Otero R.D., Ghirlando R., Camerini-Otero R.D., Camerini-Otero R.D. Molecular activities of meiosis-specific proteins Hop2, Mnd1, and the Hop2/Mnd1 complex. J. Biol. Chem. 2006;281:18426–18434. doi: 10.1074/jbc.M601073200. [DOI] [PubMed] [Google Scholar]
- Roeder G.S. Meiotic chromosomes: It takes two to tango. Genes & Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- Sarai N., Kagawa W., Kinebuchi T., Kagawa A., Tanaka K., Miyagawa K., Ikawa S., Shibata T., Kurumizaka H., Yokoyama S., Kagawa W., Kinebuchi T., Kagawa A., Tanaka K., Miyagawa K., Ikawa S., Shibata T., Kurumizaka H., Yokoyama S., Kinebuchi T., Kagawa A., Tanaka K., Miyagawa K., Ikawa S., Shibata T., Kurumizaka H., Yokoyama S., Kagawa A., Tanaka K., Miyagawa K., Ikawa S., Shibata T., Kurumizaka H., Yokoyama S., Tanaka K., Miyagawa K., Ikawa S., Shibata T., Kurumizaka H., Yokoyama S., Miyagawa K., Ikawa S., Shibata T., Kurumizaka H., Yokoyama S., Ikawa S., Shibata T., Kurumizaka H., Yokoyama S., Shibata T., Kurumizaka H., Yokoyama S., Kurumizaka H., Yokoyama S., Yokoyama S. Stimulation of Dmc1-mediated DNA strand exchange by the human Rad54B protein. Nucleic Acids Res. 2006;34:4429–4437. doi: 10.1093/nar/gkl562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N., Kleckner N. Interhomolog bias during meiotic recombination: Meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;19:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Sehorn M.G., Sigurdsson S., Bussen W., Unger V.M., Sung P., Sigurdsson S., Bussen W., Unger V.M., Sung P., Bussen W., Unger V.M., Sung P., Unger V.M., Sung P., Sung P. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature. 2004;429:433–437. doi: 10.1038/nature02563. [DOI] [PubMed] [Google Scholar]
- Shibata T., DasGupta C., Cunningham R.P., Radding C.M., DasGupta C., Cunningham R.P., Radding C.M., Cunningham R.P., Radding C.M., Radding C.M. Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc. Natl. Acad. Sci. 1979;76:1638–1642. doi: 10.1073/pnas.76.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A., Ogawa H., Ogawa T., Ogawa H., Ogawa T., Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Sung P., Krejci L., Van Komen S., Sehorn M.G., Krejci L., Van Komen S., Sehorn M.G., Van Komen S., Sehorn M.G., Sehorn M.G. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H., Roeder G.S., Roeder G.S. The Mnd1 protein forms a complex with Hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol. Cell. Biol. 2002;22:3078–3088. doi: 10.1128/MCB.22.9.3078-3088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H., Roeder G.S., Roeder G.S. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev. Cell. 2003;5:915–925. doi: 10.1016/s1534-5807(03)00357-5. [DOI] [PubMed] [Google Scholar]
- Voloshin O.N., Ramirez B.E., Bax A., Camerini-Otero R.D., Ramirez B.E., Bax A., Camerini-Otero R.D., Bax A., Camerini-Otero R.D., Camerini-Otero R.D. A model for the abrogation of the SOS response by an SOS protein: A negatively charged helix in DinI mimics DNA in its interaction with RecA. Genes & Dev. 2001;15:415–427. doi: 10.1101/gad.862901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey-Wrona J.E., Camerini-Otero R.D., Camerini-Otero R.D. The search for DNA homology does not limit stable homologous pairing promoted by RecA protein. Curr. Biol. 1995;5:1149–1158. doi: 10.1016/s0960-9822(95)00231-4. [DOI] [PubMed] [Google Scholar]
- Zierhut C., Berlinger M., Rupp C., Shinohara A., Klein F., Berlinger M., Rupp C., Shinohara A., Klein F., Rupp C., Shinohara A., Klein F., Shinohara A., Klein F., Klein F. Mnd1 is required for meiotic interhomolog repair. Curr. Biol. 2004;14:752–762. doi: 10.1016/j.cub.2004.04.030. [DOI] [PubMed] [Google Scholar]