Abstract
LEDGF/p75 directly interacts with lentiviral integrase proteins and can modulate their enzymatic activities and chromosomal association. A novel genetic knockout model was established that allowed us for the first time to analyze HIV-1 integration in the absence of LEDGF/p75 protein. Supporting a crucial role for the cofactor in viral replication, HIV-1 vector integration and reporter gene expression were significantly reduced in LEDGF-null cells. Yet, integrase processed the viral cDNA termini normally and maintained its local target DNA sequence preference during integration. Preintegration complexes extracted from knockout cells moreover supported normal levels of DNA strand transfer activity in vitro. In contrast, HIV-1 lost its strong bias toward integrating into transcription units, displaying instead increased affinity for promoter regions and CpG islands. Our results reveal LEDGF/p75 as a critical targeting factor, commandeering lentiviruses from promoter- and/or CpG island-proximal pathways that are favored by other members of Retroviridae. Akin to yeast retrotransposons, disrupting the lentiviral targeting mechanism significantly perturbs overall integration.
Keywords: Integrase, LEDGF/p75, HIV-1, AIDS, transcription, integration
Retroviral integration, which is essential for productive infection, takes place within the context of nucleoprotein preintegration complexes (PICs) (Brown et al. 1987). Therein, the viral enzyme integrase (IN) catalyzes two activities: 3′ processing and DNA strand transfer. The 3′ ends of neo-synthesized cDNA are processed soon after reverse transcription. After nuclear entry, the recessed 3′-OH groups are utilized by IN to cut the chromosome in a staggered fashion, which at the same time joins the 3′ ends to the resultant 5′ phosphates (Engelman et al. 1991). Cellular enzymes most likely repair the single-strand gaps abutting the viral DNA 5′ ends to complete the integration process (for review, see Turlure et al. 2004).
Purified IN proteins display 3′ processing and DNA strand transfer activities in vitro (Craigie et al. 1990; Katz et al. 1990; Engelman et al. 1991), yet results of numerous studies indicate cell proteins play important roles during virus infection. More than a dozen cell factors have been shown to directly bind human immunodeficiency virus type 1 (HIV-1) IN, but genetic evidence indicating important roles for the majority of these remains scant (for review, see Vandegraaff and Engelman 2007). Recent results have highlighted a crucial role for the IN-interacting protein lens epithelium-derived growth factor (LEDGF)/p75 (Cherepanov et al. 2003; Turlure et al. 2004; Emiliani et al. 2005) in HIV-1 replication and integration (Llano et al. 2006a; Vandekerckhove et al. 2006; Zielske and Stevenson 2006). The precise function of LEDGF/p75 in viral replication, however, is unknown.
LEDGF/p75 is a hepatoma-derived growth factor-related protein (HRP) that interacts specifically with lentiviral INs (Llano et al. 2004b; Busschots et al. 2005; Cherepanov 2007) and significantly stimulates their enzymatic function in vitro (Cherepanov et al. 2003, 2004; Turlure et al. 2006; Cherepanov 2007). Binding occurs through a conserved IN-binding domain (IBD) found within the C-terminal portion of the larger p75 LEDGF splice variant (Maertens et al. 2003; Cherepanov et al. 2004; Vanegas et al. 2005). The IBD is essential for stimulation of IN activity in vitro (Cherepanov et al. 2004, 2005b; Cherepanov 2007) and for LEDGF/p75 function during HIV-1 infection (Llano et al. 2006a). LEDGF/p75 might therefore act as a critical costimulator of IN activity (for reviews, see Goff 2007; Vandegraaff and Engelman 2007). Ectopically expressed HIV-1 IN is degraded by the proteasome in human cells (Mulder and Muesing 2000; Llano et al. 2004a), and LEDGF/p75 significantly increases its stability (Maertens et al. 2003; Llano et al. 2004a). The HIV-1 PIC can be degraded by the proteasome (Schwartz et al. 1998), so the IN-LEDGF/p75 interaction might help maintain PIC integrity during infection. Consistent with these models, functional HIV-1 and feline immunodeficiency virus PICs were recovered from cytoplasmic extracts of infected cells using anti-LEDGF antibodies (Llano et al. 2004b).
Alternatively, LEDGF/p75 might function as an obligate chromatin acceptor for the PIC. Consistent with this hypothesis, LEDGF/p75 intimately associates with chromatin (Nishizawa et al. 2001; Cherepanov et al. 2003), and its N-terminal PWWP domain and AT-hook (ATh) DNA-binding motifs, which mediate chromatin binding (Llano et al. 2006b; Turlure et al. 2006), are required for HIV-1 infection (Llano et al. 2006a). Yeast retrotransposon Ty5 represents a paradigm whereby a direct interaction between IN and a chromatin-associated protein, Sir4p in this case, targets a significant fraction of overall transposition events (Xie et al. 2001). Mapping large numbers of retroviral integrations has revealed biases toward or against various host genomic DNA features, though retroviral targeting appears less prominent than highly selective elements such as Ty5 (for review, see Bushman et al. 2005). Lentiviruses in large part favor actively expressed genes, integrating into transcription units (TUs) nearly equally along their lengths (Schroder et al. 2002; Mitchell et al. 2004; Crise et al. 2005). The γ-retrovirus Moloney murine leukemia virus (MLV) modestly favors TUs, but in stark contrast to HIV-1, preferentially integrates nearby promoter regions and CpG islands (Wu et al. 2003; Mitchell et al. 2004). Simian foamy virus (SFV), a Spumaretrovirus, is slightly biased against insertion into TUs, while significantly favoring promoter regions and CpG islands, albeit to lesser extents than MLV (Nowrouzi et al. 2006; Trobridge et al. 2006). Avian sarcoma-leukosis virus (ASLV), an α-retrovirus, only modestly favors TUs, promoter regions, and CpG islands, and thus displays the least selectivity toward genomic DNA features among studied retroviruses (Mitchell et al. 2004; Narezkina et al. 2004). Collectively, these results suggest that most if not all retroviruses differentially interact with chromatin and/or transcription complexes to affect a significant fraction of their overall integration events (Bushman et al. 2005). Analyses of MLV/HIV chimerae have revealed that IN is the primary viral determinant governing integration site selection (Lewinski et al. 2006).
Intriguingly, LEDGF/p75 was shown to play a modest role in HIV-1 targeting under initial knockdown conditions (Ciuffi et al. 2005) that paradoxically failed to reveal a viral infectivity defect (Llano et al. 2004b). Subsequent RNA interference (RNAi)-based studies indicated modest roles for LEDGF/p75 in HIV-1 replication and integration (Vandekerckhove et al. 2006; Zielske and Stevenson 2006), while the most recent study utilizing lentiviral-based short hairpin RNA (shRNA) vectors highlighted an essential role for the host factor (Llano et al. 2006a). As varying efficiencies of RNAi-mediated knockdowns have yielded contrasted conclusions, we reasoned that a cell system completely devoid of LEDGF/p75 protein would be an invaluable tool for analyzing its role(s) in HIV-1 biology (Vandegraaff et al. 2006). A highly significant though nonessential role for LEDGF/p75 in HIV-1 infection and integration is described here using mouse embryo fibroblasts (MEFs) disrupted for Psip1, the gene that encodes for the highly conserved murine Ledgf/p75 ortholog of human LEDGF/p75. Using the genetic knockout, we show that HIV-1 in large part loses its ability to target TUs while maintaining its preferred local target DNA sequence at the site of insertion. Accordingly, biochemical fractionation reveals normal processing of HIV-1 cDNA 3′ ends in Ledgf-null cells and wild-type levels of knockout cell PIC integration activity in vitro. Our results pinpoint LEDGF/p75 function to a step downstream from functional PICs in the nucleus, as a component of chromatin and/or transcription complexes to affect the lentiviral-specific pattern of genomic DNA targeting.
Results
Construction of Ledgf knockout cells
Murine and human LEDGF/p75 are highly homologous, sharing 92.3% identity and 97.7% homology considering conservative amino acid substitutions. Importantly, the known functional regions (PWWP domain, AThs, and IBD) are 100% conserved between orthologs. As murine fibroblasts are readily infected with pseudotyped HIV vectors (Siva and Bushman 2002; Shun et al. 2007), MEFs were derived from Psip1 knockout mice to analyze the mechanism of LEDGF/p75 function during HIV-1 integration. Ledgf and LEDGF are used throughout to denote mouse and human orthologs, respectively.
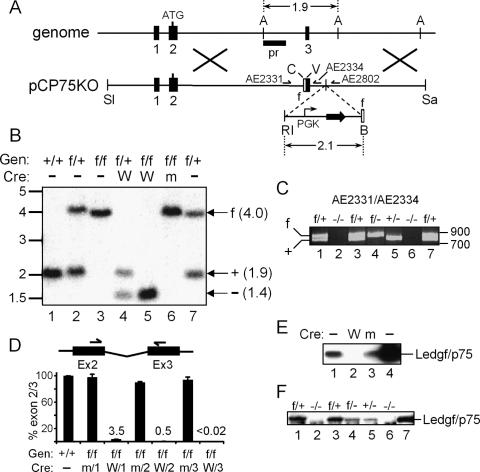
A conditional knockout strategy was utilized (Sauer 1998): DNA recombination sites for the Cre recombinase were engineered to flank exon 3, which is the second coding exon of Psip1 (Fig. 1A). Chimeric animals created following implantation of transfected embryonic stem (ES) cells were mated to C57BL/6 mice, and resultant MEFs were analyzed for flox (f) sites by Southern blotting and/or PCR (Fig. 1A–C). F1 (f/+) crosses yielded expected wild-type (+/+), heterozygous, and homozygous (f/f) progeny (Fig. 1B, lanes 1–3). For most experiments, MEFs were transformed with simian virus (SV) 40 large T antigen to effect long-term tissue culture passage. Cre was introduced into f/f cells in vitro or through selective breeding; a self-inactivating retroviral vector deleted Psip1 exon 3 (Fig. 1B, lanes 4,5) and/or its own gene (Silver and Livingston 2001), yielding ∼97% exon 3 loss at the mRNA level (Fig. 1D). Due to the residual level of intact Psip1, two additional rounds of transduction were performed, which reduced the level of exon 2/3 amplification to below the detection limit of the quantitative RT–PCR (qRT–PCR) assay (<0.02%). Exon 3 deletion shifts the open reading frame, yielding a potential N-terminal 24-residue product fused to an unrelated 11-mer tail. Accordingly, knockout (also referred to as −/−) cells failed to express detectable levels of Ledgf/p75 protein (Fig. 1E, lane 2). Importantly, f/f cells thrice transduced with a vector expressing R173K active site mutant Cre (Silver and Livingston 2001) failed to delete exon 3 (Fig. 1B, lane 6) and maintained wild-type levels of Psip1 expression (Fig. 1D,E). Most virological experiments were conducted using transformed −/− and f/f cells. Intermittent qRT–PCR analyses importantly revealed persistent, undetectable levels of exon 2/3 amplification over relatively long periods of −/− cell culturing (>35 passages). Alternatively, f/f animals were mated to Sox2Cre transgenic mice (Hayashi et al. 2002) to effect recombination in vivo. Subsequent +/− Sox2Cre, f/f crosses yielded f/+, f/-, +/−, and −/− offspring (Fig. 1C). Similar intermediate levels of Ledgf/p75 protein were detected in f/- and +/− cells (Fig. 1F, lanes 4,5), confirming that the nonrearranged f allele expressed Ledgf/p75 at normal levels. Cells derived from knockout animals predictably failed to yield detectable levels of Ledgf/p75 protein (Fig. 1F, lanes 2,6) or exon 2/3 amplification by qRT–PCR (data not shown). E1f/+ and E2−/− MEFs (Fig. 1C,F, lanes 1,2) were used in a subset of experiments; f/+ and −/− will denote cells transformed by SV40 large T antigen, whereas E1f/+ and E2−/− will refer to primary cells. We note that exon 3 loss disrupts the expression of the Ledgf/p52 splice variant (Ge et al. 1998) that does not harbor the IBD and hence does not bind IN (Maertens et al. 2003).
Figure 1.
Creation and characterization of Psip1/Ledgf knockout cells. (A) The pCP75KO targeting vector harbors ∼7.5 kb of chromosome 4 (Psip1 exons 1–3); the 2.1-kb EcoRI (RI)–BamHI (B) fragment confers resistance to neomycin (horizontal arrows). (A) AflIII; (SI) SalI; (C) ClaI; (V) EcoRV; (Sa) SacII; (pr) Southern blotting probe; (half-head arrows) PCR primers; (empty boxes) flox (f) sites. Cre deletes ∼2.6 kb, yielding an ∼1.4-kb AflIII fragment. (B) Southern analysis of AflIII-digested DNA. (Lanes 1–3) Samples from wild-type, f/+, or f/f MEFs. (Lanes 4–6) DNA prepared after three rounds of transduction with wild-type (W) or mutant (m) Cre expression vector. (Lane 7) Founder B5a ES cell DNA. Migration positions (in kilobases) of size standards are indicated at the left. (Gen) Genotype. (C) PCR analysis of indicated MEF cell DNAs. AE2331/AE2334 amplify 802-bp and 745-bp f and wild-type allele products, respectively. Recombination removes AE2334-specific DNA, yielding a novel 324-bp AE2331/AE2802 product (not shown). (Lane 7) B5a ES cell DNA; standard migrations are to the right. (D) mRNA levels in f/f and +/+ MEFs (set to 100%) following one, two, or three rounds of exposure to W or m Cre. Standard errors from duplicate qRT–PCR assays are shown. (E) Western analysis of 1.3 μg of HeLa (lane 1) or f/f cell nuclear proteins after three rounds of exposure to W (lane 2) or m (lane 3) Cre. (Lane 4) Five nanograms of purified LEDGF/p75 protein. (F) Western analysis of nuclear proteins (5 μg) from indicated primary MEF cultures. (Lane 7) One nanogram of purified protein.
Ledgf knockout cells resist infection by HIV-1 but not MLV
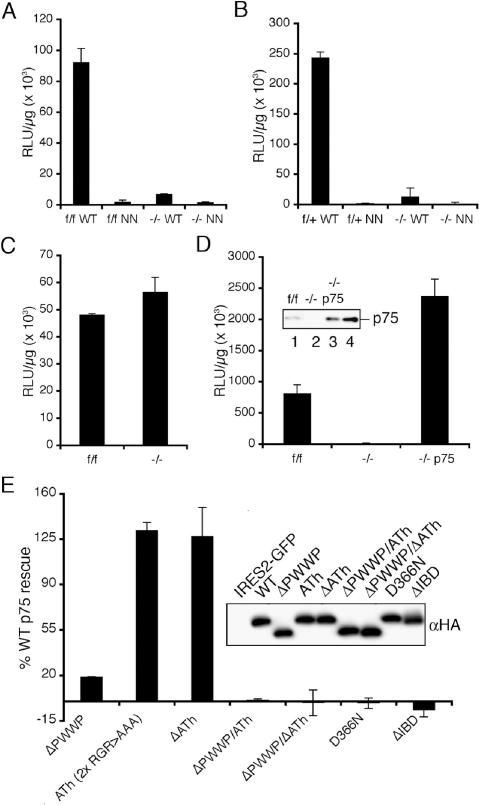
Viral vectors that express the luciferase (Luc) reporter gene (HIV-Luc, HIV-SIN-Luc, or MLV-Luc) (Shun et al. 2007) were pseudotyped with the pan-tropic vesicular stomatitis virus G (VSV-G) envelope glycoprotein, and infectivity was quantified as the level of Luc activity per microgram of total protein in cell extracts. An IN active site mutant virus (NN-Luc, containing D64N and D116N changes) yielded similar low levels of IN-independent Luc activity in control and −/− cells, indicating the knockout did not significantly affect HIV promoter function under these conditions (Fig. 2A,B). Side-by-side comparison of f/f, −/− and f/+, −/− cell pairs revealed ∼5% of integration-dependent HIV expression in Ledgf knockout as compared with control cells (Fig. 2A,B). Some variation in the level of residual HIV-Luc infectivity in −/− as compared with f/f cells was noted upon multiple experimental replicates (0.3%–9.4%; x = 1.2 ± 1.3% for n = 14). As NN-Luc was invariably less infectious (12 ± 6%, n = 14) than HIV-Luc in −/− cells, we conclude that Ledgf accounts for the vast majority, but not all, of the integration-dependent reporter gene expression under these conditions. Similar results were obtained using fluorescence-activated cell sorting (FACS) in conjunction with green fluorescent protein (GFP) reporter viruses (data not shown). Knockout and f/f cells importantly supported indistinguishable levels of MLV-Luc infectivity, revealing that the knockout does not perturb general susceptibility to retrovirus infection (Fig. 2C).
Figure 2.
LEDGF/p75 requirements for HIV-1 infection. (A) Normalized Luc values in f/f and −/− cell lysates following infection with wild-type (WT) or NN mutant virus. (B) Same as in A, except the f/+ and −/− cell pair was analyzed. (C) Normalized MLV-Luc infectivities. (D) LEDGF/p75 expression rescues HIV-1 infection in −/− cells. NN-Luc activities were subtracted from the presented values. (Inset) Western analysis of 5 μg of f/f (lane 1), −/− (lane 2), and −/− p75 (lane 3) nuclear extract. (Lane 4) One nanogram of purified protein. (E) LEDGF/p75 effector functions important for HIV-1 infection. NN-Luc-corrected values expressed as percent wild-type LEDGF/p75 activity. (Inset) Anti-HA immunoblot of 160 ng of whole-cell extract. The C-terminal HA tag reduced wild-type LEDGF/p75 function approximately twofold, but did not affect the fractional activity of any mutant protein (data not shown). (A–D) Error bars derived from duplicate Luc assays of duplicate infections. (E) Averages from a minimum of two independent experiments (duplicate Luc assays of quadruplicate infections).
LEDGF/p75 chromatin and IN-binding functions required for HIV-1 infection
Stable expression of LEDGF/p75 in −/− cells fully restored their susceptibility to HIV-1 infection under conditions wherein murine p52 expression failed to reveal an effect (Fig. 2D; data not shown). A transient expression system that utilized FACS to select for transfectants was developed to probe the requirement for LEDGF/p75 effector domains in HIV-1 infection. As expected from previous analyses (Cherepanov et al. 2004; Llano et al. 2006a), an IBD deletion mutant failed to support infection (Fig. 2E). Asp-366 within the IBD plays a critical role in mediating the viral–host interaction (Cherepanov et al. 2005a, b). The D366N missense mutant likewise failed to support infection, highlighting the importance of the specific binding interaction for HIV-1 infection (Fig. 2E). As recently reported (Llano et al. 2006a), the combined deletion of the PWWP domain and ATh DNA-binding motifs (ΔPWWPΔATh) rendered LEDGF/p75 inoperative (Fig. 2E). Separate mutations were analyzed to assess the relative contributions of these conserved elements. The ΔPWWP mutant functioned at ∼17% (n = 5) of wild-type LEDGF/p75, whereas ΔATh function was indistinguishable from wild-type (Fig. 2E). Altering the invariant Arg–Gly–Arg sequences within the AThs to Ala–Ala–Ala reduced recombinant LEDGF/p75 DNA-binding activity to ∼7% of wild-type (Turlure et al. 2006). When combined with ΔPWWP, the six Ala mutations rendered the same additive affect as the ΔATh deletion (Fig. 2E).
HIV-1 integration is defective in Ledgf knockout cells
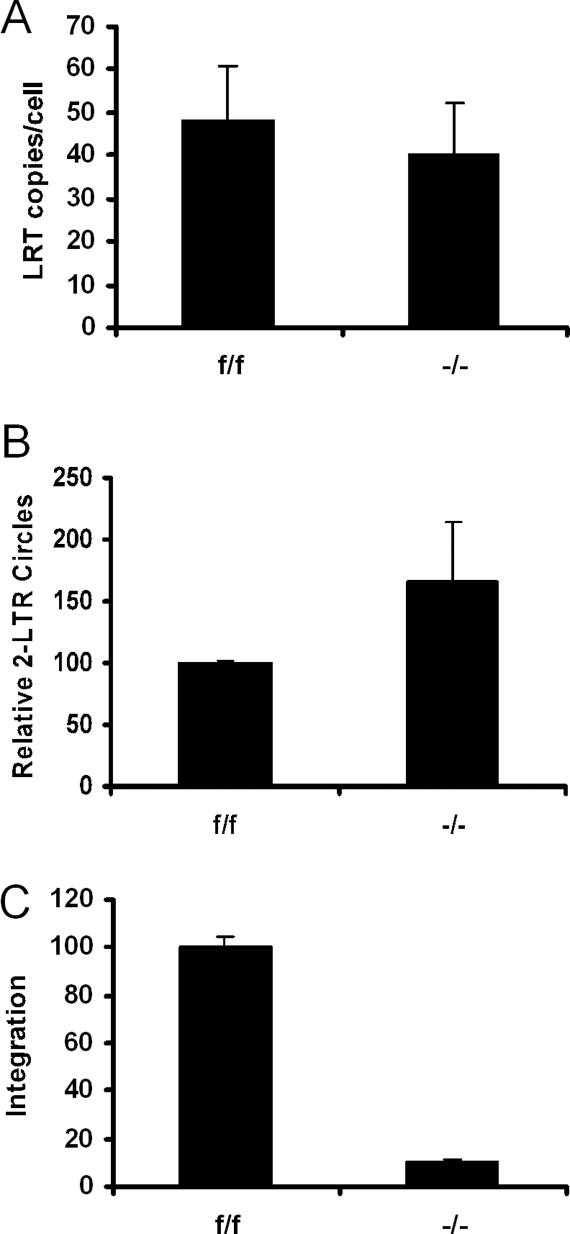
Levels of HIV-1 cDNA synthesis, nuclear migration, and integration were assessed by qPCR to characterize the infectivity block. A primer pair and Taqman probe that rely on the second template switch for amplification revealed similar levels of late reverse transcription (LRT) product formation in f/f and −/− cells (Fig. 3A). Host-mediated nonhomologous DNA end-joining acts on a small fraction of cDNA in the nucleus, yielding a circular ligation product containing two copies of the viral long terminal repeat (2-LTR circle) (Li et al. 2001). Knockout cells supported 2-LTR circle formation at levels equal to or above those observed in f/f cells (Fig. 3B), indicating that nuclear migration was unimpaired by the knockout. Levels of HIV-1 integration into human DNA can be quantified by nested real-time Alu-PCR; as Alu is a primate-specific repeat (Deininger and Batzer 1999), a novel assay utilizing mouse B1, B2, and LINE-1 repeats—referred to as BBL-PCR—was designed to measure integration. Knockout cells supported ∼11% of the level of integration detected in f/f cells (Fig. 3C).
Figure 3.
HIV-1 integration is severely reduced in Ledgf knockout cells. (A) LRT products normalized on a per cell basis. (B) 2-LTR circle levels normalized to f/f cell content (set to 100%). (C) Integration in −/− cells relative to f/f cells (set to 100%). Standard errors from duplicate qPCR assays are shown.
A critical role for Ledgf in gene-specific HIV-1 integration
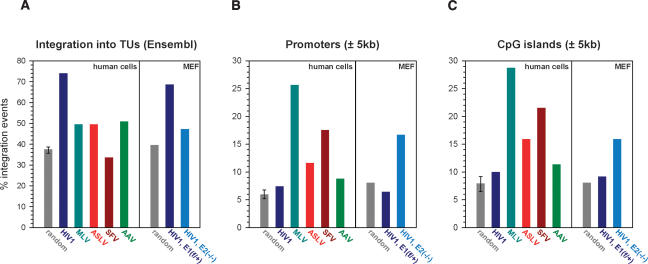
HIV-1 is strongly biased toward integration into transcriptionally active chromatin (Schroder et al. 2002; Mitchell et al. 2004). Monitoring Luc activity alongside an expanded set (n = 7) of BBL-PCR assays supported preliminary observations that knockout cells portray a larger infectivity than integration defect (∼8% integration, corresponding to ∼1.5% residual infection). This suggested that a significant proportion of knockout cell integrations might occur within unfavorable regions such as transcriptionally silent DNA. Indeed, partial depletion of LEDGF/p75 in human 293T cells yielded a modest reduction of HIV-1 integration into TUs in the absence of an accompanying infection defect (Ciuffi et al. 2005). Integration levels in Ledgf-null MEFs afforded the recovery of sufficient viral–chromosomal junctions for downstream statistical analyses. In total, 326 and 408 unique sites isolated from primary E1f/+ and E2−/− cells, respectively, were unambiguously mapped on the draft mouse genome. TUs (based on Ensembl definitions) hosted 68.7% and 47.3% of HIV-1 integration events in E1f/+ and E2−/− cells, respectively (Fig. 4A), a highly statistically significant (P = 10−8) difference. The suppression of gene targeting in Ledgf-null cells is quite remarkable, considering that the random level derived from computer simulation was 39.6%. Although E2−/− cells supported 7.7% more gene-specific integration than random (P = 0.01), this residual level was lower than those observed for ASLV (49.6%), MLV (49.6%), and adeno-associated virus (AAV) (50.9%), a parvovirus, in human cells (Fig. 4A; Supplementary Table S1).
Figure 4.
Ledgf channels HIV-1 to integrate into TUs away from promoters and CpG islands. Frequencies of integration within Ensembl TUs (A), ±5 kb of RefSeq transcription start sites (B), and ±5 kb of CpG islands (C) are shown. Random integration sites (n = 857) in the mouse genome were generated employing a computer simulation of the procedures used to clone and map the sites from infected cells. For the human genome, three random sets of sites were generated (Supplementary Table S1); the resulting average depicts the highest and lowest frequencies observed among the data sets. GenBank accession numbers for the integration sites in E1f/+ and E2−/− cells are ER870925–ER871372 and ER871373–ER872153, respectively.
Unlike MLV and SFV, HIV-1 avoids integrating in the vicinity of promoter regions and tends to disfavor CpG islands (Mitchell et al. 2004). Our scrutiny of human cell-based HIV-1, MLV, ASLV, and SFV data sets alongside HIV-1 site distributions in Ledgf-expressing E1f/+ cells confirmed these observations (Fig. 4B,C; Supplementary Table S1): 6.4% and 9.2% of HIV-1 proviruses in E1f/+ cells were found within 5 kb of a transcription start site and CpG island, respectively. These values are very close to those observed for HIV-1 (7.4% and 10.0%) and considerably lower than for MLV (25.7% and 28.8%) in human cells. In the absence of Ledgf, HIV-1 selected these elements at strikingly increased frequencies: Transcription start sites and CpG islands were targeted ∼2.6-fold (P < 0.0001) and 1.7-fold (P < 0.01) more frequently, respectively, in E2−/− as compared with E1f/+ cells (Fig. 4B,C; Supplementary Table S1). At least one in every six proviruses resided within 5 kb of a promoter in Ledgf-null cells.
The G/C content of genomic HIV-1 integration sites is significantly lower than that of other retroviruses. As noted by Berry et al. (2006), this is paradoxical, since HIV-1 primarily targets TUs, which tend to be G/C-rich. Knockdown of LEDGF/p75 in human cells increased the average G/C content of HIV-1 integration sites (Ciuffi et al. 2005). Our analyses fully confirmed this observation, as HIV-1 tended to integrate into G/C-rich regions in the absence of Ledgf (Supplementary Fig. S1).
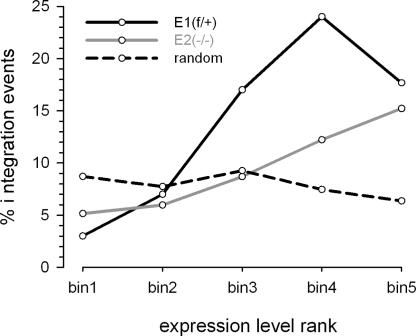
Using oligonucleotide microarrays, we determined the transcriptional profiles of three matched pairs of primary f/+ and −/− MEFs, including the E1/E2 pair used for the integration site analyses. Overall, Ledgf ablation did not lead to global changes in transcription profiles, with <200 genes significantly (>1.5-fold) and consistently (false discovery rate <5%) up- or down-regulated in knockout cells (data not shown). The similarities in gene expression profiles are in accord with high prenatal survival rates among knockout animals (Sutherland et al. 2006; J.E. Daigle and A. Engelman, unpubl.). Gene expression and HIV-1 integration data sets were cross-correlated to determine the effect of transcriptional activity on integration site selection. Ensembl TUs represented by probes on arrays were divided into five equal bins based on relative expression level, and integrations into TUs in each bin were counted (Fig. 5). In E1f/+ cells, integration strongly correlated with gene expression (P ≈ 10−15). The mouse model therefore faithfully replicates the preference of HIV-1 to integrate into active genes (Schroder et al. 2002; Mitchell et al. 2004). Interestingly, the trend appears to break for genes expressed at the highest levels (Fig. 5, f/+ bin 5). This has been noted in human cells, presumably illustrating that very high levels of gene expression can interfere with retroviral integration (Mitchell et al. 2004). Overall, integration in E2−/− cells displayed a weaker correlation with gene expression (P = 0.02 for the difference between E1f/+ and E1−/−). While not significantly influencing the frequency of integration into genes expressed at the lowest and highest levels (bins 1, 2, and 5), Ledgf ablation led to approximately twofold reductions in integration into genes expressed at medium to high levels (bins 3 and 4). Therefore, Ledgf-null cells not only hosted a larger proportion of integration events into transcriptionally silent regions, but a larger proportion of TU-specific integrations ended up in genes that were expressed to lower levels.
Figure 5.
Ledgf directs HIV-1 integration to a subset of highly expressed genes. Ensembl TUs ranked based on expression levels in E1f/+ and E2−/− MEFs were divided into five equal bins of increasing expression: bin1 < bin2 < bin3 < bin4 < bin5; TUs that hosted HIV-1 or simulated random integration sites in each bin were counted. The expression profile of E2−/− cells was used to generate the random plot (dashed line), which was almost indistinguishable from the plot generated using E1f/+ cell data (not shown).
The HIV-1 target DNA consensus site is maintained in Ledgf-null cells
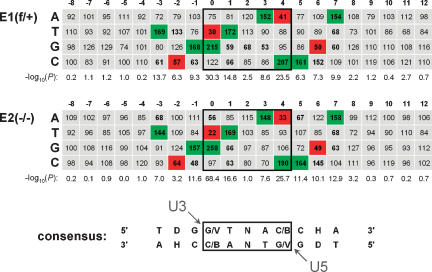
Retroviruses display short palindromic consensi in the immediate vicinity of their integration sites, which appear to be virus- and/or genus-specific (Carteau et al. 1998; Holman and Coffin 2005; Berry et al. 2006). The TDG↓GTWACCHA consensus, wherein the virus plus-strand becomes joined to the underlined nucleotide, has been elaborated for HIV-1 (Holman and Coffin 2005). Integration site sequences recovered from E1f/+ and E2−/− cells were aligned to determine if Ledgf/p75 influences the selection of local target DNA sequence. The resulting nucleotide frequencies for positions −8 to +12 are shown in Figure 6. Under both conditions, a strong bias was evident at positions −3 to +7, with positions 0 and +4 displaying the highest degree of selection (P < 10−23). The consensus derived from our alignments, TDG↓(G/V)TNA(C/B)CHA, is very similar to the reported sequence (Holman and Coffin 2005) and clearly independent of Ledgf cell content.
Figure 6.
The local HIV-1 integration site consensus sequence is preserved in Ledgf-null cells. Tabulated is the percent expected nucleotide frequency at each position. The position 0 nucleotide was joined to the processed U3 end of the LTR. Nucleotide sequences for positions 0–12 were experimentally determined by sequencing; those for positions −8 to −1 were assumed from genomic sequences upstream of mapped integration sites. Positions 0–4, which become duplicated following integration and gap repair, are boxed. Statistical significance, expressed as −log10(P), was calculated for the difference between observed and expected nucleotide frequency at each position. Frequencies <70% or >130% at positions with P < 0.001 are shown in bold; those that were used to derive the consensus are highlighted in green (>130%) or red (<70%). The arrowheads indicate the nucleotides within the target consensus sequence that become joined to the U3 and U5 ends of proviral DNA. (D) A, G, or T; (V) A, C, or G; (B) G, C, or T; (H) A, C, or T.
IN catalytic function
Recent results have highlighted that an IN multimer possessing dyad symmetry, likely a tetramer, catalyzes HIV-1 cDNA integration (Li et al. 2006). Finding the palindromic target DNA consensus sequence intact in Ledgf-null cells suggested that basic IN catalytic function might persist in the absence of the host factor. Levels of IN 3′ processing and DNA strand transfer activities were measured to directly address this supposition.
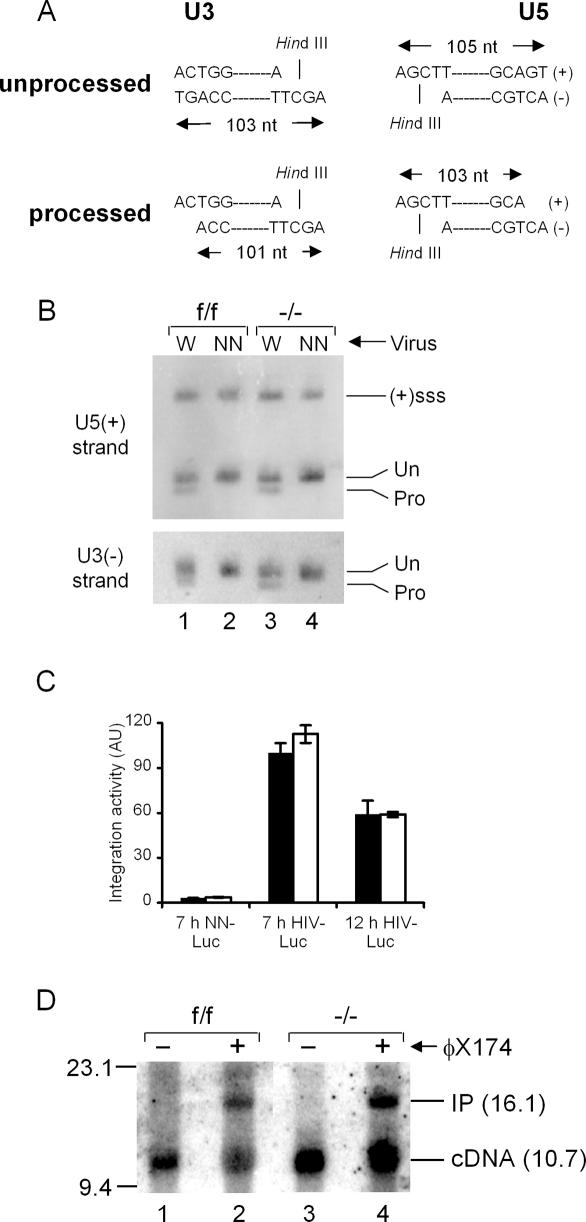
The 3′ ends of the reverse transcript must be processed by IN prior to integration. To determine 3′ processing activity, unintegrated DNA extracted from acutely infected cells was analyzed by indirect end-labeling following denaturing polyacrylamide gel electrophoresis to visualize substrate termini (Fig. 7A). The nascent U3 minus-strand, for example, is detected as 103 nucleotides (nt) after HindIII digestion; IN processing yields a 101-mer product. The NN active site mutant protein expectedly failed to detectably process either viral DNA end, regardless of Ledgf cell content (Fig. 7B, lanes 2,4). As both ends were similarly processed by wild-type IN in f/f and −/− cells (Fig. 7B, lanes 1,3), we conclude that its in vivo activity does not depend on Ledgf/p75 binding. In vitro integration assays were conducted with extracted, native PICs to assess IN DNA strand transfer activity. Most experiments utilized a sensitive qPCR design to quantify the level of U3 end integration into a circular plasmid target DNA (Engelman 2007). As expected, NN-Luc PICs supported background levels of integration activity (Fig. 7C). HIV-Luc PICs extracted from −/− cell cytoplasm at 7 h post-infection supported the same level of integration activity as f/f cell PICs. In one experiment, PICs were extracted a second time at 12 h: The f/f cell complexes were marginally less active than the corresponding 7 h samples, yet, notably, knockout cell PICs displayed the same activity as f/f cell PICs at both time points (Fig. 7C). Nuclear PICs were also analyzed, in this case by Southern blotting with a viral-specific probe to visualize the full-length DNA recombination product (Engelman 2007). The cDNA substrate is detected in its full-length 10.7-kb form, whereas integration into linearized 5.4-kb ϕX174 DNA yields a linear 16.1-kb integration product (IP) (Fig. 7D). Consistent with our interpretation that the knockout does not perturb PIC nuclear localization, cDNA levels in −/− cell nuclei equaled or exceeded those observed in control cells (Fig. 7D, lanes 1,3). Importantly, control and knockout cell nuclear PICs displayed indistinguishable levels of integration activity (Fig. 7D, lanes 2,4).
Figure 7.
IN 3′ processing and DNA strand transfer activities. (A) 3′ Processing schematic. HindIII digestion yields the depicted end-specific fragments. IN processing converts the 103-nt U3 strand to a 101-base product; the product of U5 end processing is 103 nt. (B) Strand-specific indirect end labeling of unintegrated wild-type (W) and NN-SIN-Luc DNA isolated from the indicated cells. (Un) Migration positions of unprocessed DNA strands; (Pro) 3′ processing products. Gel mobilities were confirmed via side-by-side comparison to an M13 sequencing ladder. [(+)sss] Plus-strand strong-stop DNA. (C) In vitro integration activities of wild-type and NN-Luc PICs isolated from f/f (filled bars) or −/− (empty bars) cell extracts at the indicated times post-infection. Activities plotted relative to 7-h HIV-Luc f/f PICs (set to 100%); 7-h samples, average ± standard deviation (n = 10); NN-Luc and 12-h HIV-Luc error bars, standard error of duplicate qPCR assays. (AU) Arbitrary units. (D) Integration activities of nuclear PICs extracted at 7 h post-infection. Target DNA was omitted from the reactions in lanes 1 and 3. (Lane 2) The f/f cell complexes converted 46% of the 10.7-kb cDNA substrate into the 16.1-kb integration product (IP). (Lane 4) Knockout cell PICs displayed 43% activity.
Discussion
Recent results using lentiviral-based shRNA expression vectors to deplete the bulk of cellular LEDGF/p75 revealed a critical role for the host factor in HIV-1 infection and integration (Llano et al. 2006a). Our results using genetic knockout cells support these findings. Though not of human origin, cells derived from knockout mice are powerful experimental tools and have been previously used to critically address the roles of purported host cell factors in integration and HIV-1 infection (Siva and Bushman 2002; Ariumi et al. 2005; Shun et al. 2007). As the known PWWP domain, ATh, and IBD functional determinants of human LEDGF/p75 are 100% conserved within the murine ortholog, we conclude that these cells are well suited to analyze the fundamental role(s) of LEDGF/p75 in HIV-1 integration. By including an IN active site mutant virus in our experiments, we have determined that LEDGF/p75 plays a crucial, albeit nonessential, role in integration and HIV-1 infection.
LEDGF/p75 domains and amino acid residues important for HIV-1 function
MLV efficiently infected Ledgf-null cells, and stable expression of human LEDGF/p75 fully restored their susceptibility to HIV-1 infection (Fig. 2). Moreover, the D366N missense mutant failed to complement the infectivity defect in knockout cells (Fig. 2E), revealing a critical role for the specific IN-LEDGF/p75 interaction in HIV-1 replication and integration. Of note, while the D366N mutation abrogates the IN-LEDGF/p75 interaction, it does not affect the interaction with JPO2, the cellular transcription factor that likewise associates with LEDGF/p75 through the IBD (Maertens et al. 2006). Consistent with recent results (Llano et al. 2006a), the combined ΔPWWPΔATh deletion mutant, which is defective for chromatin binding (Llano et al. 2006b), failed to rescue HIV-1 (Fig. 2E). By analyzing separate ATh and PWWP mutations, we can conclude that the PWWP domain is the dominant of the two conserved N-terminal regions in terms of HIV-1 function. The PRGR sequence comprises the heart of the ATh DNA-binding motif, and full-length LEDGF/p75 containing dual RGR > AAA substitutions supported ∼7% of residual DNA-binding activity in vitro (Turlure et al. 2006). Combining these changes with the ΔPWWP deletion rendered LEDGF/p75 inactive (Fig. 2E), consistent with the notion that these sequences are likely to function as AThs during infection. However, it is unclear if the predominant ATh function is engagement of AT-rich target DNA sequences.
Our integration site libraries revealed that HIV-1 favors integrating into G/C-rich regions in the absence of Ledgf (Supplementary Fig. S1), akin to the result reported using initial knockdown conditions (Ciuffi et al. 2005). The propensity to normally integrate within A/T-rich regions extends beyond local DNA sequence, as analyzing short (1-kb; P = 0.002), medium (5-kb; P < 0.001), and relatively long (30-kb; P = 0.008) sequence windows around the proviruses revealed shifts toward higher G/C content in all cases (Supplementary Fig. S1). LEDGF/p75 would therefore appear to direct HIV-1 to A/T-enriched regions of chromatin. As the ΔATh deletion and ATh missense mutants rescued wild-type levels of infectivity (Fig. 2E), the propensity for HIV-1 to target AT-rich sequences may very well require LEDGF/p75 interaction(s) that exceed direct ATh–DNA binding.
The mechanism of LEDGF/p75 function during HIV-1 integration
PICs isolated from knockout cells supported the same level of in vitro integration activity as matched control complexes (Fig. 7C,D). Integration site sequence alignments moreover revealed the HIV-1 target consensus site maintained in Ledgf-null cells (Fig. 6). Taken together, we conclude that IN is structurally and functionally intact in the absence of cellular LEDGF/p75 and, by extension, that the propensity for different retroviruses to integrate at weakly preferred target DNA sequences (Holman and Coffin 2005) is mechanistically separable from the targeting machineries that attract them to different chromatin structural elements.
In stark contrast, the propensity for HIV-1 to integrate into TUs was in large part countermanded by the knockout, yielding an overall frequency of gene targeting that was similar to MLV, ASLV, and AAV (Fig. 4A). Furthermore, the dramatic drop in gene-specific targeting was accompanied by a surge in integration activity nearby gene start sites and CpG islands (Fig. 4B,C; Supplementary Table S1). From these results, we conclude that LEDGF/p75 is a bona fide targeting factor for lentiviral DNA integration. In its absence, HIV-1 suffers an overall integration defect, yet is able to access promoter- and CpG island-proximal regions within chromatin to accomplish significant fractions of its residual integration events.
Compared with other retroviruses, ASLV is the least biased toward genomic DNA features (Fig. 4A–C; Mitchell et al. 2004; Narezkina et al. 2004). Rep-deficient AAV vectors, which integrate via cellular DNA double-strand breaks (Miller et al. 2004), can be considered a benchmark for random mobile DNA insertion in the human genome. The gene targeting reduction observed for HIV-1 in Ledgf-null cells, to levels normally seen for AAV and ASLV in human cells, suggests that HIV-1 PIC trafficking and resulting integration site distributions could be governed by general and/or nebulous parameters such as chromosomal DNA accessibility. We note, however, that at present we cannot rule out that additional interactions between the PIC and chromatin-associated factors dictate augmented integration nearby CpG islands and promoter regions (Fig. 4B,C) or residual levels of gene targeting (Figs. 4A, 5) in Ledgf-null cells. Our preliminary experiments revealed that when overexpressed, human HRP2 (Cherepanov et al. 2004) can rescue the block to HIV-1 infection in Ledgf knockout cells, hinting that other IN-binding proteins might possibly contribute to HIV-1 integration.
LEDGF/p75 is known to function as a molecular tether, whereby cellular JPO2 (Maertens et al. 2006) or HIV-1 IN (Maertens et al. 2003; Emiliani et al. 2005; Vanegas et al. 2005) bound to the IBD is tethered to chromatin via the N-terminal PWWP domain and AThs (Llano et al. 2006b; Turlure et al. 2006). The IN–IBD interaction, also critical for HIV-1 infection (Fig. 2E; Llano et al. 2006a), is well understood at the molecular level (Cherepanov et al. 2005a), but the mechanism of chromatin engagement is far from clear. Although a considerable body of evidence implicates LEDGF/p75 in transcriptional processes (Ge et al. 1998; Shinohara et al. 2002), a clear model for its cellular function has not been formulated. Our results strongly indicate that LEDGF/p75 would associate with transcriptionally active genes and, moreover, that a significant fraction of the protein could be expected to footprint approximately equally along their lengths (Mitchell et al. 2004). LEDGF/p75 has been proposed to interact directly with RNA polymerase II subunits (Ge et al. 1998). Alternatively, specific histone modifications are associated with transcriptional elongation (for review, see Saunders et al. 2006) and the PWWP domain, which plays an important role in HIV-1 infection (Fig. 2E), is structurally analogous to other modular protein domains like Tudor and Chromo that interact with specific histone tail modifications (Kim et al. 2006). Available evidence strongly indicates that reverse transcription, PIC assembly, and nuclear import proceed normally in the absence of LEDGF/p75 (Figs. 3, 7; Llano et al. 2006a; Vandekerckhove et al. 2006; Zielske and Stevenson 2006). We propose that upon engaging TU-associated LEDGF/p75, HIV-1 is encouraged to integrate into a nearby region. Although IN DNA strand transfer activity does not absolutely require the cofactor, the interaction sufficiently biases resulting integration site distributions toward LEDGF/p75-associated regions. Accordingly, recombinant HIV-1 IN protein is active in the absence of cellular factors (Engelman et al. 1991), and its activity can be drastically increased by LEDGF/p75 (Cherepanov et al. 2003, 2004; Turlure et al. 2006). LEDGF/p75 engagement, however, is unlikely to be an all-or-none decision. The conservation of the integration site consensus in Ledgf-null cells suggests that IN has sufficient opportunity to select its appropriate target DNA sequence independent of LEDGF/p75 binding. This model explains why a certain percentage of HIV-1 proviruses are normally found in gene-poor regions, and also why partial LEDGF/p75 knockdown could reveal a difference in integration site distribution without a concomitant decrease in infectivity (Ciuffi et al. 2005). Ledgf ablation would appear to leave the PIC little choice but to rely on basal IN strand transfer activity, leading to ∼10-fold reductions in overall integration (Fig. 3). While hijacking a cellular factor to direct most of their integrations into active TUs, lentiviruses have avoided a complete dependence on it for IN catalytic function. Such behavior has important implications for both disease progression and viral latency.
Our results establish LEDGF/p75 as a critical lentiviral-specific integration targeting factor. Retrotransposons have evolved largely site-specific integration mechanisms to help maintain the integrity of the host cell genome (for review, see Lesage and Todeschini 2005). The molecular mechanism of targeted integration is best understood for Ty5, whereby heterochromatin-associated Sir4p interacts with the C terminus of Ty5 IN to direct integration. Akin to the results reported here, disruption of the IN-tethering factor interaction significantly reduced overall levels of Ty5 transposition (Xie et al. 2001). Evolution has apparently ensured that yeast and human retroelement INs maintain close relationships with their respective tethering factors. An exciting avenue of research will be to identify cellular factors that commandeer the integration machineries of other retroviruses, such as MLV or SFV, whose telltale profiles hint of specific targeting mechanisms at play (Fig. 4A–C; Wu et al. 2003; Mitchell et al. 2004; Nowrouzi et al. 2006; Trobridge et al. 2006). Additional research into the mechanisms of retroelement targeting will help to assess and improve the safety of viral-based gene therapy vectors, and may lead to novel antiviral therapies.
Materials and methods
Plasmids
Plasmids were constructed using standard techniques, details of which are given in the Supplemental Material. Regions of all plasmids that underwent PCR amplification were verified by DNA sequencing.
Cells, viruses, and infections
Linearized pCP75KO was electroporated into TC1 ES cells (derived from the 129SvEv strain), and G418-resistant clones carrying successfully targeted Psip1/f alleles were identified by Southern blotting after prescreening by PCR with AE2331/AE2334 (primer sequences listed in Supplementary Table S2). One clone, B5a, microinjected into C57BL/6 blastocysts generated high-percentage chimera mice, which were subsequently bred to C57BL/6 animals. MEFs were isolated from 13.5-d-old embryos.
HIV-Luc is a near-full-length derivative of HIV-1NL4–3 that expresses Luc from the viral nef position, whereas HIV-SIN-Luc is a minimal transfer vector that achieves expression from a heterologous cytomegalovirus immediate early promoter (Shun et al. 2007). MLV-Luc, derived from pFB-Luc (Stratagene), expresses Luc from the MLV promoter. Viral supernatants were produced from transfected 293T cells as described (Shun et al. 2007). HIV-Luc and HIV-SIN-Luc titered using a 32P-based RT assay were treated for 1 h at 37°C with 40 U/mL Turbo DNase (Ambion). MEFs (4 × 104 per well) plated in 12-well trays 16 h before infection were infected with 1–4 × 106 RT-cpm for 8 h. MLV vectors (10 mL) were concentrated to 300 μL by ultracentrifugation, and cells were infected with 800 μL of a 1:20 dilution of concentrated virus. At 44 h post-infection, infected cells were processed for determination of Luc activity as described (Shun et al. 2007). Cells transduced with VSV-G-pseudotyped pLPCX-LEDGF/p75 or pLPCX-Ledgf/p52-HA were selected in puromycin. Cells transfected with pIRES2-eGFP expression vectors and sorted by FACS were lysed for Western blotting or plated for infection, which was conducted 10 h after seeding.
DNA and RNA analyses
The Southern blotting probe in Figure 1 was constructed by PCR using AE2772/AE2773. Relative DNA content in extrachromosomal and chromosomal DNA fractions (Vandegraaff et al. 2001) was determined by qPCR using mouse mitochondrial (AE2507/AE2508) and superoxide dismutase-specific (AE2697/AE2698) primers, respectively.
LRT products in extrachromosomal DNA fractions at 7 h post-infection were analyzed by qPCR using MH531/MH532 primers, LRT-P probe, and pTY-CMVLuc (Shun et al. 2007) to generate the standard curve. 2-LTR circle formation at 24 h post-infection was quantified using primers AE2621/AE2622 and AE2623 probe. Chromosomal DNA integration at 24 h post-infection was quantified using BBL-PCR: First-round products amplified essentially as described (Vandegraaff et al. 2001) using AE2257 and a mixture of target primers annealing to murine B1 (AE2604; AE2605), B2 (AE2606; AE2607), and LINE-1 (AE2608; AE2609) repeats were diluted 1:2000 for qPCR using AE989/AE990 primers and AE995 probe. Serial dilutions of f/f cell Hirt supernatant and pellet DNAs were utilized to generate 2-LTR circle and BBL-PCR standard curves, respectively. HindIII-digested Hirt supernatant DNA was separated through sequencing gels and transferred to Duralon-UV membrane (Stratagene) in 0.3× TBE for 1 h at 50 mA using a TransBlot SD Cell (Bio-Rad) to visualize HIV-1 3′ ends. EcoRI-linearized pCR2.1-U3 and pCR2.1-U5 were used to generate strand-specific riboprobes. PICs were isolated from cells essentially as previously described (Brown et al. 1987); see the Supplemental Material for details.
RNA extracted using the RNeasy Mini Kit (Qiagen) and quantified by spectrophotometry was analyzed by qRT–PCR using Psip1 exon 2/3 primers AE2624/AE2625. Standard curves were constructed by analyzing serial fivefold dilutions of RNA extracted from Ledgf/p75-expressing cells as described (Vandegraaff et al. 2006).
Western blotting
Proteins were extracted from whole cells (Vandegraaff et al. 2006) or isolated nuclei (Maertens et al. 2006) as described, and concentrations were determined using Dc Protein or Bradford Assays (Bio-Rad), respectively. Anti-Ledgf/p75 antibodies were from BD Biosciences, Abnova, or Bethyl; HA antibodies were from Roche.
Gene expression and integration site cloning analyses
Total RNA extracted from primary MEFs with Trizol (Invitrogen) was purified on RNeasy spin columns (Qiagen). Four independent RNA samples were isolated in parallel from each of two MEF lines, and Cy5-labeled probes were prepared using Agilent cRNA linear amplification, labeling, and fragmentation reagents. Cy3-labeled internal control cRNA was made using pooled Standard Mouse RNA (Stratagene). Cy5- and Cy3-labeled probes were cohybridized to eight 44k Whole-Mouse Genome oligonucleotide arrays (4 × 44k format, Agilent). The arrays were processed according to the manufacturer’s instructions and scanned using a G2505B Microarray Scanner (Agilent). Feature Extraction software version 9.5 was used to process the expression data. Spike-in controls included in amplification reactions indicated linear dose signal response in both channels. For analysis of differential gene expression, microarray data for E1f/+ and E2−/− cells were normalized and compared using GeneSpring GX software (Agilent). Signal intensity in the red channel was used to estimate relative abundance of transcripts within the samples. The Cy5 signal for each probe was normalized on the mean Cy5 signal over the whole array and averaged among four arrays. Probe IDs were matched to the corresponding Ensembl IDs using BioMart (http://www.ensembl.org). All Ensembl transcripts detected on the 44k arrays were ranked based on their expression level (averaged signal was taken for transcripts represented by two or more probes) and divided into five equal bins, each containing ∼4965 transcripts. TUs that hosted HIV-1 integration or simulated random integration were counted in each bin to generate Figure 5.
HIV-1 integration sites were cloned via ligation-mediated (LM)-PCR essentially as previously described (Schroder et al. 2002). Genomic DNA digested with a mixture of AvrII, NheI, and SpeI was ligated overnight with AE2844/AE2845 linker. Sequences amplified by PCR with AE2814/SB-76 were nested using AE2815/ASB-1. Resulting products subcloned into pCR4.1-TOPO (Invitrogen) were sequenced (SeqWright) using AE2865. Sequences were parsed to remove U3 and linker-derived portions; low-quality sequences and sequences that did not contain the processed 5′-TTAGCCCTTCCA-3′ U3 terminus, or with <16 base pair (bp) of genomic DNA between the processed U3 end and beginning of the linker sequence, were discarded. Details of integration site sequence analyses are given in the Supplemental Material.
Acknowledgments
Plasmids pCEP4-Tat and pHP-dI-N/A were obtained through the AIDS Research and Reference Reagent Program, from Dr. Lung-Ji Chang. We are grateful to L.-J. Chang for sharing the pHP-dI-N/A sequence, J. Walsh and R. Mulligan for pCG-gagpol and pCG-VSV-G, D. Borger and J. DeCaprio for pLB(N)CX-large T, D. Silver for HR-MMPCreGFP and HR-MMPCreGFPR173K, D. Gabuzda for pNL4-3env-GFP, and J. Skaar for help in preparing MEF cells. This work was supported by NIH grant AI39394 (to A.E.) and UK Medical Research Council (MRC) grant G0600009 (to P.C).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1565107
References
- Ariumi Y., Turelli P., Masutani M., Trono D., Turelli P., Masutani M., Trono D., Masutani M., Trono D., Trono D. DNA damage sensors ATM, ATR, DNA-PKcs, and PARP-1 are dispensable for human immunodeficiency virus type 1 integration. J. Virol. 2005;79:2973–2978. doi: 10.1128/JVI.79.5.2973-2978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C., Hannenhalli S., Leipzig J., Bushman F.D., Hannenhalli S., Leipzig J., Bushman F.D., Leipzig J., Bushman F.D., Bushman F.D. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput. Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P.O., Bowerman B., Varmus H.E., Bishop J.M., Bowerman B., Varmus H.E., Bishop J.M., Varmus H.E., Bishop J.M., Bishop J.M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- Bushman F., Lewinski M., Ciuffi A., Barr S., Leipzig J., Hannenhalli S., Hoffmann C., Lewinski M., Ciuffi A., Barr S., Leipzig J., Hannenhalli S., Hoffmann C., Ciuffi A., Barr S., Leipzig J., Hannenhalli S., Hoffmann C., Barr S., Leipzig J., Hannenhalli S., Hoffmann C., Leipzig J., Hannenhalli S., Hoffmann C., Hannenhalli S., Hoffmann C., Hoffmann C. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- Busschots K., Vercammen J., Emiliani S., Benarous R., Engelborghs Y., Christ F., Debyser Z., Vercammen J., Emiliani S., Benarous R., Engelborghs Y., Christ F., Debyser Z., Emiliani S., Benarous R., Engelborghs Y., Christ F., Debyser Z., Benarous R., Engelborghs Y., Christ F., Debyser Z., Engelborghs Y., Christ F., Debyser Z., Christ F., Debyser Z., Debyser Z. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 2005;280:17841–17847. doi: 10.1074/jbc.M411681200. [DOI] [PubMed] [Google Scholar]
- Carteau S., Hoffmann C., Bushman F., Hoffmann C., Bushman F., Bushman F. Chromosome structure and human immunodeficiency virus type 1 cDNA integration: Centromeric alphoid repeats are a disfavored target. J. Virol. 1998;72:4005–4014. doi: 10.1128/jvi.72.5.4005-4014.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 2007;35:113–124. doi: 10.1093/nar/gkl885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z., Engelborghs Y., De Clercq E., Debyser Z., De Clercq E., Debyser Z., Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- Cherepanov P., Devroe E., Silver P.A., Engelman A., Devroe E., Silver P.A., Engelman A., Silver P.A., Engelman A., Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- Cherepanov P., Ambrosio A.L., Rahman S., Ellenberger T., Engelman A., Ambrosio A.L., Rahman S., Ellenberger T., Engelman A., Rahman S., Ellenberger T., Engelman A., Ellenberger T., Engelman A., Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl. Acad. Sci. 2005a;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P., Sun Z.-Y., Rahman S., Maertens G., Wagner G., Engelman A., Sun Z.-Y., Rahman S., Maertens G., Wagner G., Engelman A., Rahman S., Maertens G., Wagner G., Engelman A., Maertens G., Wagner G., Engelman A., Wagner G., Engelman A., Engelman A. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat. Struct. Mol. Biol. 2005b;12:526–532. doi: 10.1038/nsmb937. [DOI] [PubMed] [Google Scholar]
- Ciuffi A., Llano M., Poeschla E., Hoffmann C., Leipzig J., Shinn P., Ecker J.R., Bushman F., Llano M., Poeschla E., Hoffmann C., Leipzig J., Shinn P., Ecker J.R., Bushman F., Poeschla E., Hoffmann C., Leipzig J., Shinn P., Ecker J.R., Bushman F., Hoffmann C., Leipzig J., Shinn P., Ecker J.R., Bushman F., Leipzig J., Shinn P., Ecker J.R., Bushman F., Shinn P., Ecker J.R., Bushman F., Ecker J.R., Bushman F., Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- Craigie R., Fujiwara T., Bushman F., Fujiwara T., Bushman F., Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- Crise B., Li Y., Yuan C., Morcock D.R., Whitby D., Munroe D.J., Arthur L.O., Wu X., Li Y., Yuan C., Morcock D.R., Whitby D., Munroe D.J., Arthur L.O., Wu X., Yuan C., Morcock D.R., Whitby D., Munroe D.J., Arthur L.O., Wu X., Morcock D.R., Whitby D., Munroe D.J., Arthur L.O., Wu X., Whitby D., Munroe D.J., Arthur L.O., Wu X., Munroe D.J., Arthur L.O., Wu X., Arthur L.O., Wu X., Wu X. Simian immunodeficiency virus integration preference is similar to that of human immunodeficiency virus type 1. J. Virol. 2005;79:12199–12204. doi: 10.1128/JVI.79.19.12199-12204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P.L., Batzer M.A., Batzer M.A. Alu repeats and human disease. Mol. Genet. Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- Emiliani S., Mousnier A., Busschots K., Maroun M., Van Maele B., Tempe D., Vandekerckhove L., Moisant F., Ben-Slama L., Witvrouw M., Mousnier A., Busschots K., Maroun M., Van Maele B., Tempe D., Vandekerckhove L., Moisant F., Ben-Slama L., Witvrouw M., Busschots K., Maroun M., Van Maele B., Tempe D., Vandekerckhove L., Moisant F., Ben-Slama L., Witvrouw M., Maroun M., Van Maele B., Tempe D., Vandekerckhove L., Moisant F., Ben-Slama L., Witvrouw M., Van Maele B., Tempe D., Vandekerckhove L., Moisant F., Ben-Slama L., Witvrouw M., Tempe D., Vandekerckhove L., Moisant F., Ben-Slama L., Witvrouw M., Vandekerckhove L., Moisant F., Ben-Slama L., Witvrouw M., Moisant F., Ben-Slama L., Witvrouw M., Ben-Slama L., Witvrouw M., Witvrouw M., et al. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 2005;280:25517–25523. doi: 10.1074/jbc.M501378200. [DOI] [PubMed] [Google Scholar]
- Engelman A. Isolation and analysis of HIV-1 preintegration complexes. In: Prasad V.R., Kalpana G.V., Kalpana G.V., editors. HIV protocols, 2nd edition. Humana Press; Totowa, NJ: 2007. (in press) [Google Scholar]
- Engelman A., Mizuuchi K., Craigie R., Mizuuchi K., Craigie R., Craigie R. HIV-1 DNA integration: Mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- Ge H., Si Y., Roeder R.G., Si Y., Roeder R.G., Roeder R.G. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 1998;17:6723–6729. doi: 10.1093/emboj/17.22.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.P. Host factors exploited by retroviruses. Nat. Rev. Microbiol. 2007;5:253–263. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- Hayashi S., Lewis P., Pevny L., McMahon A.P., Lewis P., Pevny L., McMahon A.P., Pevny L., McMahon A.P., McMahon A.P. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 2002;119 (Suppl. 1):S97–S101. doi: 10.1016/S0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Holman A.G., Coffin J.M., Coffin J.M. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc. Natl. Acad. Sci. 2005;102:6103–6107. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R.A., Merkel G., Kulkosky J., Leis J., Skalka A.M., Merkel G., Kulkosky J., Leis J., Skalka A.M., Kulkosky J., Leis J., Skalka A.M., Leis J., Skalka A.M., Skalka A.M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- Kim J., Daniel J., Espejo A., Lake A., Krishna M., Xia L., Zhang Y., Bedford M.T., Daniel J., Espejo A., Lake A., Krishna M., Xia L., Zhang Y., Bedford M.T., Espejo A., Lake A., Krishna M., Xia L., Zhang Y., Bedford M.T., Lake A., Krishna M., Xia L., Zhang Y., Bedford M.T., Krishna M., Xia L., Zhang Y., Bedford M.T., Xia L., Zhang Y., Bedford M.T., Zhang Y., Bedford M.T., Bedford M.T. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage P., Todeschini A.L., Todeschini A.L. Happy together: The life and times of Ty retrotransposons and their hosts. Cytogenet. Genome Res. 2005;110:70–90. doi: 10.1159/000084940. [DOI] [PubMed] [Google Scholar]
- Lewinski M.K., Yamashita M., Emerman M., Ciuffi A., Marshall H., Crawford G., Collins F., Shinn P., Leipzig J., Hannenhalli S., Yamashita M., Emerman M., Ciuffi A., Marshall H., Crawford G., Collins F., Shinn P., Leipzig J., Hannenhalli S., Emerman M., Ciuffi A., Marshall H., Crawford G., Collins F., Shinn P., Leipzig J., Hannenhalli S., Ciuffi A., Marshall H., Crawford G., Collins F., Shinn P., Leipzig J., Hannenhalli S., Marshall H., Crawford G., Collins F., Shinn P., Leipzig J., Hannenhalli S., Crawford G., Collins F., Shinn P., Leipzig J., Hannenhalli S., Collins F., Shinn P., Leipzig J., Hannenhalli S., Shinn P., Leipzig J., Hannenhalli S., Leipzig J., Hannenhalli S., Hannenhalli S., et al. Retroviral DNA integration: Viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Olvera J.M., Yoder K.E., Mitchell R.S., Butler S.L., Lieber M., Martin S.L., Bushman F.D., Olvera J.M., Yoder K.E., Mitchell R.S., Butler S.L., Lieber M., Martin S.L., Bushman F.D., Yoder K.E., Mitchell R.S., Butler S.L., Lieber M., Martin S.L., Bushman F.D., Mitchell R.S., Butler S.L., Lieber M., Martin S.L., Bushman F.D., Butler S.L., Lieber M., Martin S.L., Bushman F.D., Lieber M., Martin S.L., Bushman F.D., Martin S.L., Bushman F.D., Bushman F.D. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 2001;20:3272–3281. doi: 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Mizuuchi M., Burke T.R., Craigie R., Mizuuchi M., Burke T.R., Craigie R., Burke T.R., Craigie R., Craigie R. Retroviral DNA integration: Reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M., Delgado S., Vanegas M., Poeschla E.M., Delgado S., Vanegas M., Poeschla E.M., Vanegas M., Poeschla E.M., Poeschla E.M. Lens epithelium-derived growth factor/p75 prevents proteasomal degradation of HIV-1 integrase. J. Biol. Chem. 2004a;279:55570–55577. doi: 10.1074/jbc.M408508200. [DOI] [PubMed] [Google Scholar]
- Llano M., Vanegas M., Fregoso O., Saenz D., Chung S., Peretz M., Poeschla E.M., Vanegas M., Fregoso O., Saenz D., Chung S., Peretz M., Poeschla E.M., Fregoso O., Saenz D., Chung S., Peretz M., Poeschla E.M., Saenz D., Chung S., Peretz M., Poeschla E.M., Chung S., Peretz M., Poeschla E.M., Peretz M., Poeschla E.M., Poeschla E.M. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 2004b;78:9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M., Saenz D.T., Meehan A., Wongthida P., Peretz M., Walker W.H., Teo W., Poeschla E.M., Saenz D.T., Meehan A., Wongthida P., Peretz M., Walker W.H., Teo W., Poeschla E.M., Meehan A., Wongthida P., Peretz M., Walker W.H., Teo W., Poeschla E.M., Wongthida P., Peretz M., Walker W.H., Teo W., Poeschla E.M., Peretz M., Walker W.H., Teo W., Poeschla E.M., Walker W.H., Teo W., Poeschla E.M., Teo W., Poeschla E.M., Poeschla E.M. An essential role for LEDGF/p75 in HIV integration. Science. 2006a;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- Llano M., Vanegas M., Hutchins N., Thompson D., Delgado S., Poeschla E.M., Vanegas M., Hutchins N., Thompson D., Delgado S., Poeschla E.M., Hutchins N., Thompson D., Delgado S., Poeschla E.M., Thompson D., Delgado S., Poeschla E.M., Delgado S., Poeschla E.M., Poeschla E.M. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 2006b;360:760–773. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- Maertens G., Cherepanov P., Pluymers W., Busschots K., De Clercq E., Debyser Z., Engelborghs Y., Cherepanov P., Pluymers W., Busschots K., De Clercq E., Debyser Z., Engelborghs Y., Pluymers W., Busschots K., De Clercq E., Debyser Z., Engelborghs Y., Busschots K., De Clercq E., Debyser Z., Engelborghs Y., De Clercq E., Debyser Z., Engelborghs Y., Debyser Z., Engelborghs Y., Engelborghs Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- Maertens G.N., Cherepanov P., Engelman A., Cherepanov P., Engelman A., Engelman A. Transcriptional co-activator p75 binds and tethers the c-Myc-interacting protein JPO2 to chromatin. J. Cell Sci. 2006;119:2563–2571. doi: 10.1242/jcs.02995. [DOI] [PubMed] [Google Scholar]
- Miller D.G., Petek L.M., Russell D.W., Petek L.M., Russell D.W., Russell D.W. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat. Genet. 2004;36:767–773. doi: 10.1038/ng1380. [DOI] [PubMed] [Google Scholar]
- Mitchell R.S., Beitzel B.F., Schroder A.R., Shinn P., Chen H., Berry C.C., Ecker J.R., Bushman F.D., Beitzel B.F., Schroder A.R., Shinn P., Chen H., Berry C.C., Ecker J.R., Bushman F.D., Schroder A.R., Shinn P., Chen H., Berry C.C., Ecker J.R., Bushman F.D., Shinn P., Chen H., Berry C.C., Ecker J.R., Bushman F.D., Chen H., Berry C.C., Ecker J.R., Bushman F.D., Berry C.C., Ecker J.R., Bushman F.D., Ecker J.R., Bushman F.D., Bushman F.D. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:e234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder L.C., Muesing M.A., Muesing M.A. Degradation of HIV-1 integrase by the N-end rule pathway. J. Biol. Chem. 2000;275:29749–29753. doi: 10.1074/jbc.M004670200. [DOI] [PubMed] [Google Scholar]
- Narezkina A., Taganov K.D., Litwin S., Stoyanova R., Hayashi J., Seeger C., Skalka A.M., Katz R.A., Taganov K.D., Litwin S., Stoyanova R., Hayashi J., Seeger C., Skalka A.M., Katz R.A., Litwin S., Stoyanova R., Hayashi J., Seeger C., Skalka A.M., Katz R.A., Stoyanova R., Hayashi J., Seeger C., Skalka A.M., Katz R.A., Hayashi J., Seeger C., Skalka A.M., Katz R.A., Seeger C., Skalka A.M., Katz R.A., Skalka A.M., Katz R.A., Katz R.A. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa Y., Usukura J., Singh D.P., Chylack L.T., Shinohara T., Usukura J., Singh D.P., Chylack L.T., Shinohara T., Singh D.P., Chylack L.T., Shinohara T., Chylack L.T., Shinohara T., Shinohara T. Spatial and temporal dynamics of two alternatively spliced regulatory factors, lens epithelium-derived growth factor (ledgf/p75) and p52, in the nucleus. Cell Tissue Res. 2001;305:107–114. doi: 10.1007/s004410100398. [DOI] [PubMed] [Google Scholar]
- Nowrouzi A., Dittrich M., Klanke C., Heinkelein M., Rammling M., Dandekar T., von Kalle C., Rethwilm A., Dittrich M., Klanke C., Heinkelein M., Rammling M., Dandekar T., von Kalle C., Rethwilm A., Klanke C., Heinkelein M., Rammling M., Dandekar T., von Kalle C., Rethwilm A., Heinkelein M., Rammling M., Dandekar T., von Kalle C., Rethwilm A., Rammling M., Dandekar T., von Kalle C., Rethwilm A., Dandekar T., von Kalle C., Rethwilm A., von Kalle C., Rethwilm A., Rethwilm A. Genome-wide mapping of foamy virus vector integrations into a human cell line. J. Gen. Virol. 2006;87:1339–1347. doi: 10.1099/vir.0.81554-0. [DOI] [PubMed] [Google Scholar]
- Sauer B. Inducible gene targeting in mice using the Cre/lox system. Methods. 1998;14:381–392. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- Saunders A., Core L.J., Lis J.T., Core L.J., Lis J.T., Lis J.T. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Schroder A.R., Shinn P., Chen H., Berry C., Ecker J.R., Bushman F., Shinn P., Chen H., Berry C., Ecker J.R., Bushman F., Chen H., Berry C., Ecker J.R., Bushman F., Berry C., Ecker J.R., Bushman F., Ecker J.R., Bushman F., Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Schwartz O., Marechal V., Friguet B., Arenzana-Seisdedos F., Heard J.-M., Marechal V., Friguet B., Arenzana-Seisdedos F., Heard J.-M., Friguet B., Arenzana-Seisdedos F., Heard J.-M., Arenzana-Seisdedos F., Heard J.-M., Heard J.-M. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J. Virol. 1998;72:3845–3850. doi: 10.1128/jvi.72.5.3845-3850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara T., Singh D.P., Fatma N., Singh D.P., Fatma N., Fatma N. LEDGF, a survival factor, activates stress-related genes. Prog. Retin. Eye Res. 2002;21:341–358. doi: 10.1016/s1350-9462(02)00007-1. [DOI] [PubMed] [Google Scholar]
- Shun M.-C., Daigle J.E., Vandegraaff N., Engelman A., Daigle J.E., Vandegraaff N., Engelman A., Vandegraaff N., Engelman A., Engelman A. Wild-type levels of human immunodeficiency virus type 1 infectivity in the absence of cellular emerin protein. J. Virol. 2007;81:166–172. doi: 10.1128/JVI.01953-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver D.P., Livingston D.M., Livingston D.M. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol. Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- Siva A.C., Bushman F., Bushman F. Poly(ADP-ribose) polymerase 1 is not strictly required for infection of murine cells by retroviruses. J. Virol. 2002;76:11904–11910. doi: 10.1128/JVI.76.23.11904-11910.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland H.G., Newton K., Brownstein D.G., Holmes M.C., Kress C., Semple C.A., Bickmore W.A., Newton K., Brownstein D.G., Holmes M.C., Kress C., Semple C.A., Bickmore W.A., Brownstein D.G., Holmes M.C., Kress C., Semple C.A., Bickmore W.A., Holmes M.C., Kress C., Semple C.A., Bickmore W.A., Kress C., Semple C.A., Bickmore W.A., Semple C.A., Bickmore W.A., Bickmore W.A. Disruption of Ledgf/Psip1 results in perinatal mortality and homeotic skeletal transformations. Mol. Cell. Biol. 2006;26:7201–7210. doi: 10.1128/MCB.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobridge G.D., Miller D.G., Jacobs M.A., Allen J.M., Kiem H.-P., Kaul R., Russell D.W., Miller D.G., Jacobs M.A., Allen J.M., Kiem H.-P., Kaul R., Russell D.W., Jacobs M.A., Allen J.M., Kiem H.-P., Kaul R., Russell D.W., Allen J.M., Kiem H.-P., Kaul R., Russell D.W., Kiem H.-P., Kaul R., Russell D.W., Kaul R., Russell D.W., Russell D.W. Foamy virus vector integration sites in normal human cells. Proc. Natl. Acad. Sci. 2006;103:1498–1503. doi: 10.1073/pnas.0510046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlure F., Devroe E., Silver P.A., Engelman A., Devroe E., Silver P.A., Engelman A., Silver P.A., Engelman A., Engelman A. Human cell proteins and human immunodeficiency virus DNA integration. Front. Biosci. 2004;9:3187–3208. doi: 10.2741/1472. [DOI] [PubMed] [Google Scholar]
- Turlure F., Maertens G., Rahman S., Cherepanov P., Engelman A., Maertens G., Rahman S., Cherepanov P., Engelman A., Rahman S., Cherepanov P., Engelman A., Cherepanov P., Engelman A., Engelman A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin invivo. Nucleic Acids Res. 2006;34:1663–1675. doi: 10.1093/nar/gkl052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandegraaff N., Engelman A., Engelman A. Molecular mechanism of HIV integration and therapeutic intervention. Expert Rev. Mol. Med. 2007;9:1–19. doi: 10.1017/S1462399407000257. [DOI] [PubMed] [Google Scholar]
- Vandegraaff N., Kumar R., Hocking H., Burke T.R., Mills J., Rhodes D., Burrell C.J., Li P., Kumar R., Hocking H., Burke T.R., Mills J., Rhodes D., Burrell C.J., Li P., Hocking H., Burke T.R., Mills J., Rhodes D., Burrell C.J., Li P., Burke T.R., Mills J., Rhodes D., Burrell C.J., Li P., Mills J., Rhodes D., Burrell C.J., Li P., Rhodes D., Burrell C.J., Li P., Burrell C.J., Li P., Li P. Specific inhibition of human immunodeficiency virus type 1 (HIV-1) integration in cell culture: putative inhibitors of HIV-1 integrase. Antimicrob. Agents Chemother. 2001;45:2510–2516. doi: 10.1128/AAC.45.9.2510-2516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandegraaff N., Devroe E., Turlure F., Silver P.A., Engelman A., Devroe E., Turlure F., Silver P.A., Engelman A., Turlure F., Silver P.A., Engelman A., Silver P.A., Engelman A., Engelman A. Biochemical and genetic analyses of integrase-interacting proteins lens epithelium-derived growth factor (LEDGF)/p75 and hepatoma-derived growth factor related protein 2 (HRP2) in preintegration complex function and HIV-1 replication. Virology. 2006;346:415–426. doi: 10.1016/j.virol.2005.11.022. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove L., Christ F., Van Maele B., De Rijck J., Gijsbers R., Van den Haute C., Witvrouw M., Debyser Z., Christ F., Van Maele B., De Rijck J., Gijsbers R., Van den Haute C., Witvrouw M., Debyser Z., Van Maele B., De Rijck J., Gijsbers R., Van den Haute C., Witvrouw M., Debyser Z., De Rijck J., Gijsbers R., Van den Haute C., Witvrouw M., Debyser Z., Gijsbers R., Van den Haute C., Witvrouw M., Debyser Z., Van den Haute C., Witvrouw M., Debyser Z., Witvrouw M., Debyser Z., Debyser Z. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 2006;80:1886–1896. doi: 10.1128/JVI.80.4.1886-1896.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanegas M., Llano M., Delgado S., Thompson D., Peretz M., Poeschla E., Llano M., Delgado S., Thompson D., Peretz M., Poeschla E., Delgado S., Thompson D., Peretz M., Poeschla E., Thompson D., Peretz M., Poeschla E., Peretz M., Poeschla E., Poeschla E. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 2005;118:1733–1743. doi: 10.1242/jcs.02299. [DOI] [PubMed] [Google Scholar]
- Wu X., Li Y., Crise B., Burgess S.M., Li Y., Crise B., Burgess S.M., Crise B., Burgess S.M., Burgess S.M. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Xie W., Gai X., Zhu Y., Zappulla D.C., Sternglanz R., Voytas D.F., Gai X., Zhu Y., Zappulla D.C., Sternglanz R., Voytas D.F., Zhu Y., Zappulla D.C., Sternglanz R., Voytas D.F., Zappulla D.C., Sternglanz R., Voytas D.F., Sternglanz R., Voytas D.F., Voytas D.F. Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol. Cell. Biol. 2001;21:6606–6614. doi: 10.1128/MCB.21.19.6606-6614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielske S.P., Stevenson M., Stevenson M. Modest but reproducible inhibition of human immunodeficiency virus type 1 infection in macrophages following LEDGFp75 silencing. J. Virol. 2006;80:7275–7280. doi: 10.1128/JVI.02470-05. [DOI] [PMC free article] [PubMed] [Google Scholar]