Abstract
Increased osteoclastic resorption leads to many bone diseases, including osteoporosis and rheumatoid arthritis. While rapid progress has been made in characterizing osteoclast differentiation signaling pathways, how receptor activator of nuclear factor κB (NF-κB) ligand (RANKL) evokes essential [Ca2+]i oscillation signaling remains unknown. Here, we characterized RANKL-induced signaling proteins and found regulator of G-protein signaling 10 (RGS10) is predominantly expressed in osteoclasts. We generated RGS10-deficient (RGS10−/−) mice that exhibited severe osteopetrosis and impaired osteoclast differentiation. Our data demonstrated that ectopic expression of RGS10 dramatically increased the sensitivity of osteoclast differentiation to RANKL signaling; the deficiency of RGS10 resulted in the absence of [Ca2+]i oscillations and loss of NFATc1; ectopic NFATc1 expression rescues impaired osteoclast differentiation from deletion of RGS10; phosphatidylinositol 3,4,5-trisphosphate (PIP3) is essential to PLCγ activation; and RGS10 competitively interacts with Ca2+/calmodulin and PIP3 in a [Ca2+]i-dependent manner to mediate PLCγ activation and [Ca2+]i oscillations. Our results revealed a mechanism through which RGS10 specifically regulates the RANKL-evoked RGS10/calmodulin–[Ca2+]i oscillation–calcineurin–NFATc1 signaling pathway in osteoclast differentiation using an in vivo model. RGS10 provides a potential therapeutic target for the treatment of bone diseases.
Keywords: RGS10, osteopetrosis, [Ca2+]i oscillation, osteoclast differentiation, RANKL signaling pathway, therapeutic target
Osteoclasts are the sole bone-resorbing cells. These cells are essential for skeletal development and bone remodeling throughout life. Deficiency of osteoclasts leads to osteopetrosis, a disease manifested by increased nonremodeled bone mass. On the other hand, increased number and activity of osteoclasts under certain pathologic conditions causes accelerated bone resorption and may lead to osteoporosis and osteolytic diseases. To better understand the mechanisms underlying osteoclast-based diseases and to design relevant therapies, it is necessary to unveil the molecular basis of osteoclast differentiation and function as well as the regulatory mechanisms of osteoclast signaling.
The past several years have witnessed important insights into osteoclast formation and function (Boyle et al. 2003; Zhao et al. 2007). The binding of receptor activator of nuclear factor κB (NF-κB) ligand (RANKL) to its receptor, RANK, results in the recruitment of tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6), which activates the NF-κB, c-Fos, Jun N-terminal kinase (JNK), and p38 pathways (Boyle et al. 2003). Recently, Takayanagi et al. (2002) reported that RANKL-evoked [Ca2+]i oscillations play a switch-on role in osteoclast differentiation through the nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (NFATc1) activation pathway that triggers osteoclast-specific gene expression. Their observation suggests that sustained [Ca2+]i oscillations, rather than transient activation of a Ca2+ spike, is necessary for the sustained NFATc1 activation during osteoclastogenesis (Takayanagi et al. 2002). However, it remained unclear how RANKL activates calcium signals and [Ca2+]i oscillations leading to the induction and nuclear localization of NFATc1. Most recently, Koga et al. (2004) and Mao et al. (2006) reported that RANKL-mediated costimulatory signals initiated by immunoreceptor tyrosine-based activation motifs (ITAMs) of DNAX-activating protein 12 (DAP12) and Fc receptor γ polypeptide (FcRγ) regulate osteoclast differentiation through the phospholipase Cγ (PLCγ) phosphorylation-[Ca2+]i oscillation–NFATc1 pathway. Their working model may explain how RANKL evokes transient activation of a Ca2+ spike, but still could not explain how RANKL signaling regulates sustained [Ca2+]i oscillations to ensure the NFATc1-mediated transcriptional program in RANKL-stimulated bone marrow-derived monocytes (BMMs).
The regulator of G-protein signaling (RGS) proteins are a family of 21 proteins, RGS1–14 and RGS16–22, all containing the RGS domain. RGS proteins have been reported to be involved in cell proliferation and differentiation (Schwable et al. 2005; Appleton et al. 2006). The regulation of RGS proteins in [Ca2+]i oscillations has been studied previously in the immune (Kehrl 1998), neural (Sinnarajah et al. 2001), and cardiovascular systems (Ishii et al. 2002). Ca2+/calmodulin directly binds to RGS4 in a Ca2+-dependent manner and Ca2+/calmodulin competes with phosphatidylinositol 3,4,5-trisphosphate (PIP3) for binding to RGS4 (Popov et al. 2000). Calmodulin and PIP3 both bind to the C-terminal portion of helix 4 of the RGS domain of RGS4 (Ishii et al. 2005). This binding site is well conserved in different RGS proteins, suggesting that reciprocal regulation by PIP3 and Ca2+/calmodulin may be important for the physiological control of multiple RGS subtypes (Abramow-Newerly et al. 2006). In addition, a number of RGS proteins—including RGS2, RGS3, and RGS4—have been shown to block PLCβ activation (Hepler et al. 1997; Heximer et al. 1997; Saugstad et al. 1998; Cunningham et al. 2001). RGS10 is currently known to be densely expressed in rat brain, where it plays a role in determining signaling pathways and synaptic activity (Gold et al. 1997).
In this study, we demonstrated that RGS10 is prominently expressed in osteoclasts; RGS10−/− mice exhibit severe osteopetrosis; and disruption of RGS10 impairs osteoclast differentiation due to the absence of [Ca2+]i oscillations and loss of NFATc1 expression. Ectopic expression of RGS10 markedly enhanced RANKL-induced osteoclast differentiation. RGS10 competitively binds Ca2+/calmodulin and PIP3 in a [Ca2+]i-dependent manner and PIP3 is essential to PLCγ activation. Our results suggest that RGS10 is a key regulator of [Ca2+]i oscillations during osteoclast differentiation.
Results
RGS10 is predominantly expressed in osteoclasts
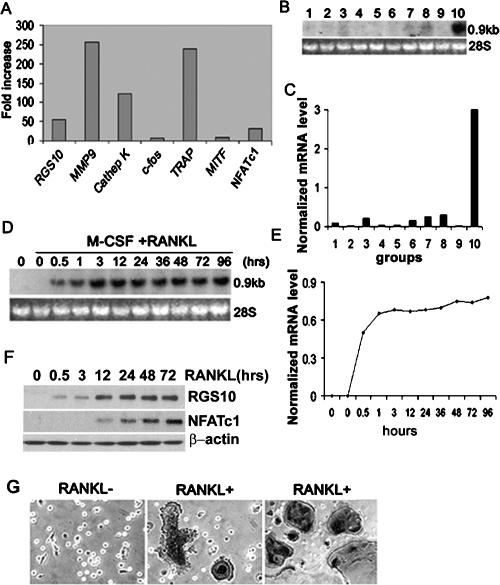
We hypothesized that signal protein(s) induced by RANKL and expressed in osteoclasts regulate [Ca2+]i oscillations in the RANKL-[Ca2+]i oscillation–NFATc1 pathway. To identify the signal protein(s) specifically expressed in osteoclasts, we performed a genome-wide screening of mRNAs in human osteoclastoma, compared with that of human stromal cells, using an RNA expression profile array (Affymetrix GeneChip). We found that RGS10 was highly expressed in human osteoclastoma (Fig. 1A). The results were confirmed by Northern blot in different human tissues and cell lines (Fig. 1B,C). RGS10 was most highly expressed in human osteoclastoma. There was also expression observed in the brain, liver, kidneys, and Hep-2, but to a far lesser extent. The expression of RGS10 was undetectable in the other tissues and cell lines (Fig. 1B,C), indicating that RGS10 was predominantly and selectively expressed in osteoclasts. To characterize RANKL-induced RGS10 expression in mouse osteoclasts, we analyzed a time course of RGS10 mRNA expression in RANKL-induced mouse BMMs by Northern blot. We found that RGS10 was also highly expressed in preosteoclasts and osteoclasts derived from BMMs (Fig. 1D,E) and the dominant expression of RGS10 started at 0.5 h after RANKL induction and continued to increase until osteoclast formation. The time course of RGS10 expression was confirmed by Western blot (Fig. 1F). NFATc1 was expressed in RANKL-induced BMMs beginning at 12 h after RANKL induction and continued to increase through 72 h (Fig. 1F). At 96 h after RANKL induction, essentially all of the tartrate-resistant acid phosphatase-positive (TRAP+) multinucleated cells (MNCs) became strongly positive for RGS10 protein (Fig. 1G). Thus, there is a close correlation between RGS10 expression and osteoclast differentiation. In addition, we compared expression of RGS10 in osteoblast and osteoclast during development using Northern blot (Supplementary Fig. 1A,B) and in situ immunostaining (Supplementary Fig. 1C). RGS10 mRNA expression was detected both in osteoclasts and in mononuclear, TRAP+ preosteoclasts. However, RGS10 mRNA was not detected in osteoblasts or preosteoblasts (Supplementary Fig. 1A). RGS10 mRNA was also not detected in BMMs treated with macrophage colony-stimulating factor (M-CSF) alone in the absence of RANKL (Supplementary Fig. 1B). RGS10 protein was expressed in the vertebrae of embryonic day 14 (E14) and E19 mice and in the brain of E19 mice (Supplementary Fig. 1C), as shown in previous studies (Gold et al. 1997).
Figure 1.
RGS10 is prominently expressed in osteoclasts and osteoclast precursors induced by RANKL/M-CSF. (A) Genome-wide screening of osteoclast-specific genes by GeneChip. mRNAs of osteoclast marker genes matrix metalloprotease (MMP9), cathepsin K, TRAP, and NFATc1 were strongly expressed in osteoclasts, confirming the validity of our screening protocol. RGS10 was prominently expressed in human osteoclasts. (B) Northern blot analysis of RGS10 mRNA. Total RNA was extracted from human tissues and cell lines. (Lane 1) U-937. (Lane 2) HOS-TE85. (Lane 3) Hep-2. (Lane 4) HSB-2. (Lane 5) Skeletal muscle. (Lane 6) Liver. (Lane 7) Kidney. (Lane 8) Brain. (Lane 9) Human stromal cells. (Lane 10) Human osteoclastoma. (C) Normalized mRNA level of B. (D) Time-course Northern blot analysis of mouse RGS10 mRNA expression in preosteoclasts and osteoclasts derived from BMMs induced with RANKL/M-CSF at 0, 0.5, 1, 3, 12, 24, 36, 48, 72, and 96 h. RGS10 mRNA was undetectable in BMMs and BMMs treated with only M-CSF. After RANKL/M-CSF induction, the dominant expression of RGS10 starts at 0.5 h and continues to increase until 3 h, and then the expression of RGS10 remains at the same level. (E) Normalized mRNA level of D. (F) Time-course Western blot analysis of RGS10 expression in preosteoclasts and osteoclasts derived from BMMs induced with RANKL/M-CSF at 0, 0.5, 3, 12, 24, 48, and 72 h confirmed that expression of RGS10 starts at 0.5 h and remains level after 3 h. Expression of NFATc1 starts at 12 h. (G) RGS10 is strongly expressed in RANKL-induced MNCs detected by anti-RGS10 immunostaining (middle panel) as compared with BMMs without RANKL induction (left panel). (Right panel) These MNCs were confirmed to be TRAP+ osteoclasts by TRAP staining.
RGS10−/− mice have normal embryonic development but show severe growth retardation
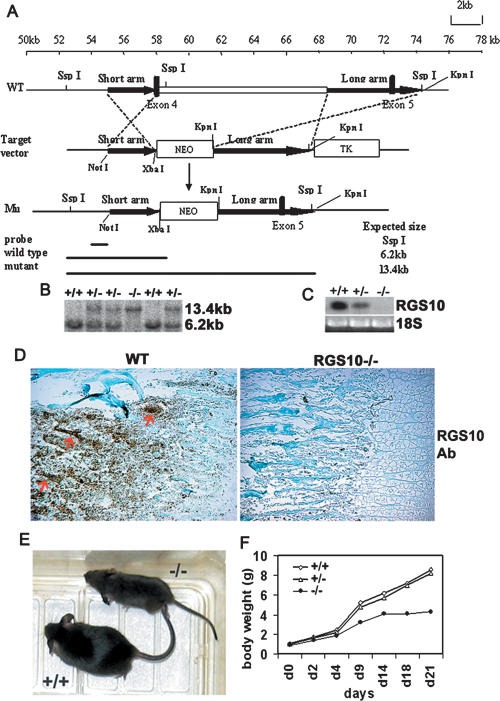
RGS proteins were reported to be involved in cell proliferation and differentiation (Schwable et al. 2005; Appleton et al. 2006). To gain insights into the biological function of RGS10 in osteoclast proliferation, differentiation, and function in vivo, mice with null mutations of RGS10 were generated by homologous recombination. A targeting vector for deletion of RGS10 was constructed as shown in Figure 2A and described in Materials and Methods. We used two heterozygous mutant embryonic stem (ES) cells to generate chimeric mice and backcrossed chimeric to C57BL/6 mice. Heterozygous RGS10+/− mice, which are healthy and fertile, were then intercrossed to generate homozygous RGS10−/− mice. Mice were born with the expected Mendelian ratios from intercrosses of heterozygous mutants. The null mutation of RGS10 was genotyped by Southern blot (Fig. 2B) and confirmed to be the absence of RGS10 expression as determined by Northern blot (Fig. 2C) and immunostaining analysis (Fig. 2D). Heterozygous RGS10+/− animals were indistinguishable from wild-type mice, but RGS10−/− animals had an identical, severe phenotype. About 25% of E18 embryos from heterozygous matings were RGS10−/− embryos. Apparent abnormalities were not observed by gross macroscopic or histological examination of the embryos, indicating that mouse embryonic development is not affected by deficiency of RGS10 (data not shown). RGS10−/− mice were smaller and had short limbs (Fig. 2E). Growth retardation became apparent during the first or second postnatal week (Fig. 2F). RGS10−/− mice survived up to 3 wk, which is consistent with the malignant osteopetrosis phenotype, as described previously (Li et al. 1999).
Figure 2.
Generation of RGS10-null mice. (A) Targeting vector for RGS10 was constructed by inserting a 1.5-kb PCR fragment as the short arm and a 5-kb KpnI fragment as the long arm, which are flanked by the neomycin-resistance cassette. Targeted allele cells were produced by replacing 10 kb of RGS10 (including exon 4, which encodes amino acids 85–133, covering most of the RGS domain) with the PGK-neo cassette to delete the RGS10 domain for null mutation. (B) Genotype analysis of mice by Southern blot. The presence of a single 13.4-kb fragment indicates a homozygous RGS10−/− genotype. (C) Northern blot analysis of RNA isolated from long bone of 3-d-old wild-type and homozygous mutant littermates. The mRNA was detectable in long bone of wild-type and heterozygous mice, but undetectable in the homozygous mutant mice, using RGS10 cDNA as a probe. (D) Anti-RGS10 immunostaining of 10-d-old wild-type and RGS10−/− tibiae. RGS10 was expressed in wild-type osteoclasts (arrows), but not in RGS10−/− cells. (E) Appearance of RGS10−/− mice at day 20. (F) Growth curve of RGS10−/− mice compared with littermates.
Severe osteopetrosis in RGS10−/− mice
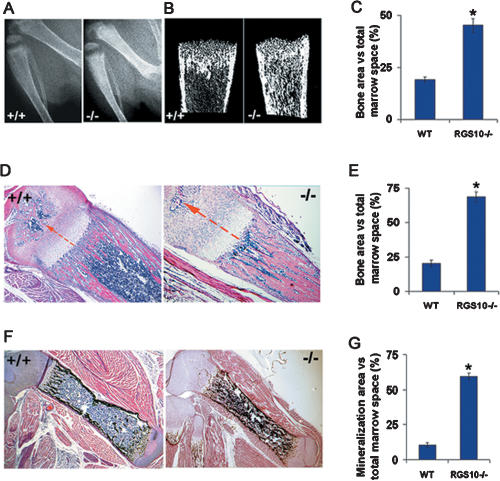
To determine whether disruption of RGS10 leads to actual changes in bone physiology, we studied tibia radiographs of 10-d-old RGS10+/+ and RGS10−/− littermates. X-ray analysis showed a significant increase in the density of trabecular and cortical bone area in RGS10−/− mice compared with RGS10+/+ littermates (Fig. 3A). The result was confirmed by Micro-CT analysis (Fig. 3B). Bone volume in the tibiae of RGS10−/− mice was 2.4-fold more than that in the RGS10+/+ mice (Fig. 3C), suggesting an increase in bone mass and the occurrence of severe osteopetrosis in RGS10 mutant mice. Histological H&E staining analysis confirmed that the long bones were ostepetrotic in appearance, with an abundance of bone and cartilage trabeculae, obliterating >80% of the marrow space in RGS10−/− mice compared with that in wild-type mice (Fig. 3D). We also observed that the growth plates of RGS10−/− mice at 10 d had an extended zone of calcified cartilage and the zone of the hypertrophic chondrocytes was increased (Fig. 3D). Von Kossa staining of 10-d-old RGS10−/− mouse tibiae showed an increase in mineralization and mature bone formation compared with wild-type mouse tibiae (Fig. 3F). Consistent with an increase in bone and cartilage trabeculae in RGS10−/− mice, bone and mineralization area over the total marrow space increased in tibiae from these animals (Fig. 3E,G). The quantitative histomorphometric analysis is described in the Supplemental Material (Supplementary Table 1). Histomorphometric analysis showed normal ratios of number of osteoblasts per bone perimeter, indicating that the loss of RGS10 had no apparent influence on osteoblast development and function (Supplementary Table 1).
Figure 3.
Increased bone mass in RGS10−/− mice. (A) Radiographic analysis of 10-d-old RGS10+/+ and RGS10−/− mice. Tibiae from RGS10−/− mice show increased density of both cortical and trabecular bones. (B) Micro-CT images of tibiae from RGS10+/+ and RGS10−/− mice. Note that increased trabecular and cortical bone mass was observed. (C) Quantitative analysis of the ratio of bone area to total marrow space at different positions of tibiae isolated from wild-type and RGS10−/− mice. Increased cortical and trabecular bone mass was indicated. (*) P < 0.01, significant difference from wild type (student’s t-test). (D,F) Histomorphologic analysis of sections of tibiae from 10-d-old RGS10+/+ and RGS10−/− mice. Histologic sections of tibiae were stained with H&E (D) or Von Kossa staining (F) to visualize bone mass. The growth plates of RGS10−/− mice at 10 d had an extended zone of calcified cartilage compared with RGS10+/+ controls, and the zone of the hypertrophic chondrocytes was increased (arrows in D). (E,G) Quantitative analysis of bone of wild-type and RGS10−/− mice expressed as the percentage of bone area (E) or mineralization area (G) versus total marrow space. N = 3; (*) P < 0.01, significant difference from wild type (student’s t-test).
Null mutation of RGS10 impairs osteoclast differentiation, but does not affect macrophage differentiation
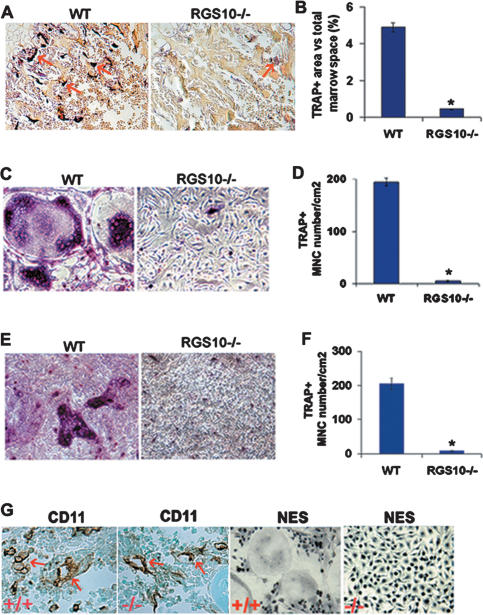
Severe osteopetrosis could be due to either the failure of osteoclast formation during development, such as in mice lacking c-Fos, PU.1, NF-κB, p50, and p52 (Wang et al. 1992; Iotsova et al. 1997; Tondravi et al. 1997), or a deficiency in osteoclast function and activation, such as in mice lacking c-Src (Soriano et al. 1991) and Atp6i (Li et al. 1999). We examined whether differentiation morphology properties of RGS10−/− osteoclasts were changed in RGS10−/− mice. Histochemical stains of 10-d-old RGS10−/− mouse tibiae for the osteoclast enzyme, TRAP, showed few MNCs with very weak TRAP activity (Fig. 4A), indicating that the osteopetrosis in RGS10−/− mice is caused by impaired osteoclast differentiation and the essential role of RGS10 in osteoclast differentiation. Consistent with the decrease in osteoclasts in RGS10−/− mice, the percentage of TRAP+ area (the area includes the MNCs with very weak TRAP activity in RGS10−/− mice) versus the total bone marrow space in RGS10−/− tibiae decreased 10-fold below that in RGS10+/+ tibiae (Fig. 4B; Supplementary Fig. 2). Despite the severe osteopetrosis in RGS10−/− mice, these mice have no obvious defect in tooth eruption (data not shown). The tooth phenotype in RGS10−/− mice is similar to that in mice lacking ITAM-harboring adaptors FcRγ and DAP12, which exhibit severe osteopetrosis owing to impaired osteoclast differentiation, but have normal tooth eruption (Koga et al. 2004).
Figure 4.
Defective osteoclastogenesis in RGS10−/− BMMs and preosteoclasts. (A) Histologic sections of 10-d-old tibiae were stained for TRAP activity. The results showed few osteoclasts and weak TRAP activity in RGS10−/− mice (arrows). (B) Quantitative analysis of TRAP+-stained area in wild-type and very weak TRAP+-stained area in RGS10−/− mice tibia sections expressed as the percentage of TRAP+-stained area versus total marrow space. N = 3; (*) P < 0.01, significant difference from wild type (student’s t-test). (C–F) BMMs from wild-type and RGS10−/− mice were incubated with RANKL/M-CSF (C,D) or with coculture system (E,F) as described in Materials and Methods. TRAP+ MNCs could be detected at 96 h in the wild-type cell culture by TRAP staining analysis, while scarcely any were detected in RGS10−/− cells. (G) Immunostaining of CD11 (Mac-1) and nonspecific esterase (NES), two monocyte/macrophage precursor cell marker genes, in RGS10+/+ and RGS10−/− tibiae. RGS10−/− mice have the same normal monocyte/macrophage as RGS10+/+ mice (arrows).
In an osteoblast-free culture system, no TRAP+ MNCs were formed in RANKL-induced RGS10−/− BMMs (Fig. 4C,D). However, 80% of BMMs from wild-type mice were differentiated to TRAP+ MNCs (Fig. 4C,D), indicating that osteoclast differentiation is abrogated by the deficiency of RGS10. To identify whether coculture with wild-type osteoblasts is able to rescue the defective osteoclastogenesis, we performed wild-type osteoblast coculture with RGS10−/− BMMs, as compared with wild-type BMMs, and found that there were a few TRAP+ mononuclear cells in RANKL-induced RGS10−/− BMMs. However, hardly any RGS10−/− BMMs differentiated into mature osteoclasts (Fig. 4E,F). These results indicate that the coculture was unable to rescue the defective osteoclastogenesis, and suggest that RGS10 is indispensable for RANKL-induced osteoclastogenesis both in vivo and in vitro.
To determine the effect of RGS10 on osteoclast gene expression, we examined the expression of osteoclast marker genes, cathepsin K and Atp6i, in RANKL-induced BMMs from RGS10+/+ or RGS10−/− mice (Supplementary Fig. 3). We first confirmed that the expression of RGS10 was detected in RANKL-induced RGS10+/+ BMMs and absent in RANKL-induced RGS10−/− BMMs. Cathepsin K and Atp6i were strongly expressed in RGS10+/+ mice, but were absent in RGS10−/− mice.
Osteoclast cells are myeloid-derived cells, closely related to macrophage cells. To determine whether deletion of RGS10 prevents macrophage differentiation as well, we assayed expression of CD11 (Mac-1) and nonspecific esterase (NES), two monocyte/macrophage precursor cell marker genes (Takahashi et al. 1994). The results showed that RGS10−/− mice have the same normal monocyte/macrophage cells as the RGS10+/+ mice (Fig. 4G), indicating that deletion of RGS10 does not affect macrophage differentiation and that RGS10 plays its role in osteoclast differentiation starting from the preosteoclast stage.
Essential role of RGS10 in the RANKL-[Ca2+]i oscillation–NFATc1 signaling pathway
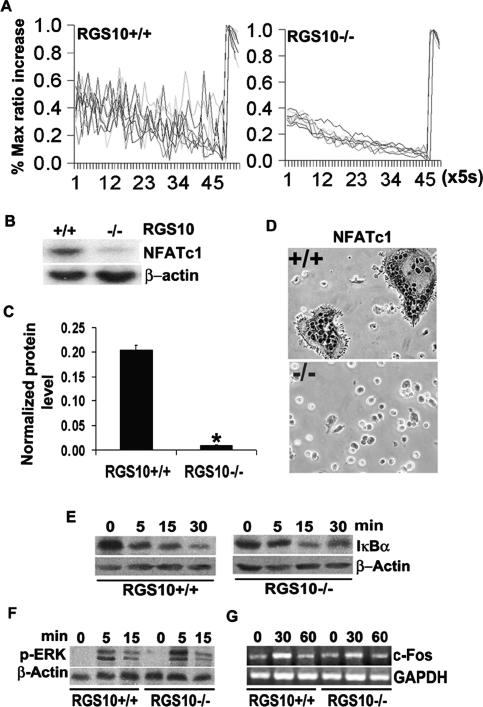
RANKL-induced [Ca2+]i oscillations are essential for osteoclastogenesis (Takayanagi et al. 2002). We therefore examined calcium signaling in RANKL-induced BMMs derived from RGS10−/− mice. Interestingly, as shown in Figure 5A, [Ca2+]i oscillations induced by RANKL were not observed in RGS10−/− cells, which suggests that RGS10 may be an essential regulator of [Ca2+]i oscillations during osteoclast differentiation.
Figure 5.
Impaired [Ca2+]i oscillations and NFATc1 expression in RGS10−/− BMMs induced by RANKL. (A) [Ca2+]i changes were traced in RGS10−/− or RGS10+/+ BMMs treated with RANKL/M-CSF for 72 h [Ca2+]i. Changes were estimated as the ratio of fluorescence intensity of fluo-4 to fura red, plotted at 5-sec intervals. Each color indicates a different cell in the same field. [Ca2+]i oscillations are impaired in RGS10−/− cells. (B–D) BMMs from RGS10−/− or RGS10+/+ mice were stimulated with RANKL/M-CSF for 96 h. (B) Western blot analysis showed weak signals of NFATc1 protein detected in RGS10−/− cells as compared with RGS10+/+ cells. (C) The bands in B were quantified. NFATc1 signals in RGS10−/− cells are 18-fold lower than those in RGS10+/+ cells. Data are presented as mean ± SD. N = 3 (student’s t-test). (*) P < 0.001, RGS10−/− versus RGS10+/+. (D) Immunostaining revealed that the expression of NFATc1 was impaired in RGS10−/− cells. (E–G) RANKL- and M-CSF-induced NF-κB, Erk, and c-Fos signaling are unaltered in RGS10−/− osteoclastic cells. (E) NF-κB activation in response to RANKL was assessed by Western blot analysis of IκB-α degradation in BMMs derived from RGS10+/+ or RGS10−/− mice. β-actin levels were used as loading control (n = 3). (F) M-CSF signaling in BMMs derived from RGS10+/+ or RGS10−/− mice was determined by Western blot analysis of the phosphorylated p42/p44 form of Erk at the indicated times. β-actin levels were used as loading control (n = 3). (G) Expression of c-Fos was determined by RT–PCR in day 2 osteoclasts stimulated with M-CSF for the indicated time. GAPDH levels were used as loading control (n = 3).
RANKL-induced [Ca2+]i oscillations are essential for autoamplification of NFATc1 during osteoclastogenesis (Takayanagi et al. 2002). To determine whether impaired [Ca2+]i oscillations caused by RGS10 deficiency affect NFATc1 expression, we examined NFATc1 expression in RANKL-induced RGS10−/− and RGS10+/+ BMMs. As shown in Figure 5, B and C, the expression of NFATc1 was very weak in RANKL-induced RGS10−/− BMMs compared with that in RANKL-induced RGS10+/+ BMMs. The normalized protein level of NFATc1 in RGS10−/− cells was 18-fold lower than that in RGS10+/+ cells (Fig. 5C). This result was confirmed by immunostaining (Fig. 5D), in which RGS10 is undetectable, indicating that RGS10 plays an essential role in autoamplification of NFATc1 during osteoclastogenesis.
Null mutation of RGS10 does not affect other RANKL-induced pathways and apoptosis
To examine whether impaired osteoclast differentiation in RGS10−/− mice results from deficiencies in other signaling pathways evoked by RANKL and M-CSF, we characterized the activation of these pathways. We found that RANKL activation of NF-κB, assessed by IκBα degradation (Fig. 5E), M-CSF-driven Erk-1/2 phosphorylation (Fig. 5F), and transcriptional induction of c-Fos (Fig. 5G), all of which are required for efficient osteoclastogenesis (Boyle et al. 2003; Faccio et al. 2003, 2005), were normal in all RGS10-deficient BMMs tested. This suggests that RGS10 disruption did not affect these signaling pathways, and RGS10 is specifically involved in the [Ca2+]i oscillation-NFATc1 signaling pathway.
These results provided convincing evidence for the role of RGS10 in RANKL-mediated osteoclast differentiation. However, the question remained whether the impaired osteoclast differentiation is due to preosteoclast apoptosis resulting from RGS10 knockout. Accordingly, using Hoechst 33258 staining, we detected characteristic apoptotic changes in the nuclei. The results showed that deficiency of RGS10 does not affect cell survival of preosteoclasts (Supplementary Fig. 4), which excludes the possibility that RGS10 is involved in apoptosis to impair osteoclast differentiation.
Reintroduction of RGS10 rescued osteoclast differentiation and ectopic expression of RGS10 increased sensitivity to RANKL signaling
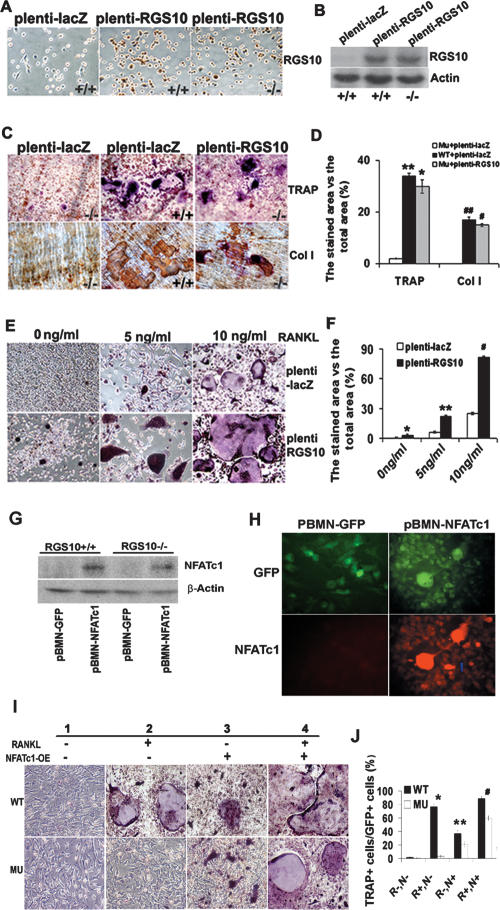
To ensure that the observed RGS10−/− mouse phenotype is solely a result of RGS10 deficiency, we determined whether impaired osteoclastogenesis in RGS10−/− mice could be rescued by reintroduction of RGS10. BMMs from RGS10−/− and RGS10+/+ mice were infected with an RGS10-expressing lentivirus (denoted as plenti-RGS10) or control virus expressing LacZ (plenti-LacZ). Expression of RGS10 protein was confirmed by immunostaining (Fig. 6A) and Western blot (Fig. 6B) in plenti-RGS10-transfected BMMs. As expected, re-expression of RGS10 fully restored the ability of RGS10−/− BMMs to differentiate into mature osteoclasts in the presence of RANKL (Fig. 6C,D), indicating that the RGS10−/− phenotype only resulted from the null mutation of RGS10.
Figure 6.
Rescue of RGS10−/− osteoclast differentiation by reintroduction of RGS10 and NFATc1 and increase in sensitivity of osteoclast differentiation to RANKL signaling by RGS10 overexpression. (A) The BMMs from RGS10+/+ and RGS10−/− mice were infected with plenti-LacZ and plenti-RGS10. Immunostaining results showed that 90% of the cells expressed RGS10 in plenti-RGS10-infected RGS10+/+ and RGS10−/− cells. (B) Western blot results confirmed the expression of RGS10 in plenti-RGS10-infected RGS10+/+ and RGS10−/− BMMs. (C) The BMMs from RGS10+/+ and RGS10−/− mice were infected with plenti-LacZ as a positive control and negative control, respectively. plenti-RGS10-infected BMMs and two controls were induced with RANKL/M-CSF as described in Materials and Methods. The cells were stained for TRAP activity. Note that some TRAP+ cells are multinucleated in plenti-RGS10-transfected RGS10−/− cells and have bone resorption activity on dentine slices (immunostaining of anti-collagen I protein; bone resorption areas become brown or dark brown), indicating that overexpression of RGS10 could rescue osteoclastogenesis. (D) Quantitative analysis of TRAP or collagen I-positive area in C expressed as the percentage of the positive stained area versus total area. Data are presented as mean ± SD. N = 3. For TRAP staining, P < 0.001 (*), Mu-plenti-LacZ versus Mu-plenti-RGS10; P > 0.05 (**), WT-plenti-LacZ versus Mu-plenti-RGS10. For immunostaining of collagen I protein, P < 0.001 (#), Mu-plenti-LacZ versus Mu-plenti-RGS10; P > 0.05 (##), WT-plenti-LacZ versus Mu-plenti-RGS10 (ANOVA). (E) The BMMs from wild-type mice were infected with plenti-LacZ or plenti-RGS10 and then treated with 0, 5, and 10 ng/mL RANKL in the presence of 10 ng/mL M-CSF for 96 h. (Bottom left panel) Without RANKL induction, 8% of precursor cells differentiated into mononuclear TRAP+ cells in plenti-RGS10-infected cells. In the presence of 5 or 10 ng/mL RANKL and 10 ng/mL M-CSF (middle and right panels), there are 3.6-fold and 3.2-fold more mononuclear and mature multinuclear TRAP+ cells, respectively, in the plenti-RGS10 group (bottom panels) compared with the plenti-LacZ group (top panels). (F) Quantitative analysis of TRAP+ cells in E. N = 3 (student’s t-test). plenti-LacZ versus plenti-RGS10 at 0 ng/mL ([*] P < 0.05), 5 ng/mL ([**] P < 0.001), and 10 ng/mL ([#] P < 0.001). (G) Western blot of NFATc1 protein in RGS10+/+ and RGS10−/− BMMs expressing pBMN-NFATc1 or control pBMN-GFP. Overexpression of NFATc1 rescues its expression in RGS10−/− BMMs. (H) NFATc1 and GFP expression in BMMs transfected with pBMN-NFATc1 or pBMN-GFP. Ninety-eight percent of transfected cells become GFP+ cells. pBMN-NFATc1 transfection induces expression of NFATc1 without RANKL induction. (I) TRAP stain of wild-type and RGS10 mutant (MU) BMMs with (panels 2,4) or without (panels 1,3) RANKL induction and with (panels 3,4) or without (panels 1,2) transfection with pBMN-NFATc1. (Bottom, panels 3,4) Overexpression of NFATc1 rescues osteoclast formation with or without RANKL induction. (J) Quantitative analysis of TRAP+ cells in I. Data are presented as mean ± SD. N = 3. (*) P < 0.001, WT−R+,N− versus Mu− R+,N−; (**) P < 0.05, WT−R−,N+ versus Mu− R−,N+; (#) P < 0.05, WT−R+,N+ versus Mu− R+,N+. (R) RANKL; (N) overexpression of NFATc1; (+) presence; (−) absence.
To identify whether RGS10 is sufficient to initiate osteoclast differentiation without RANKL stimulation, we analyzed plenti-RGS10-infected BMMs from wild-type mice and found that as many as 8% of the cells spontaneously differentiated into TRAP+ mononuclear cells and low multinuclear cells with a maximum of four nuclei in the absence of RANKL (Fig. 6E,F). However, functional assay showed that, in the absence of RANKL, these osteoclasts did not exhibit the typical functions of bone resorption and acidification (Supplementary Fig. 5). When the RGS10+/+ cells were treated with 5 or 10 ng/mL of RANKL in the presence of 10 ng/mL of M-CSF for 96 h and stained for TRAP+ cells, the percentage of TRAP+-stained area to total area in the plenti-RGS10-infected groups were increased 3.6-fold and 3.2-fold, respectively, over that in the plenti-LacZ-infected groups (Fig. 6E,F). The percentage of area expressing cathepsin K, Atp6i, and NFATc1 to the total area was 4.3-fold, 3.7-fold, and 3.8-fold higher, respectively, than that in plenti-LacZ-infected cells (Supplementary Fig. 6A,B). These results demonstrated that although ectopic expression of RGS10 could not fully initiate osteoclast differentiation without RANKL stimulation, it could increase the sensitivity of osteoclast differentiation to RANKL signaling in osteoclast precursor cells.
Ectopic NFATc1 expression could partially rescue impaired osteoclast differentiation from deletion of RGS10
In order to further clarify the role of NFATc1 in the RGS10-related signaling pathway, we examined the effect of ectopic NFATc1 expression in RGS10−/− BMMs. We constructed a retrovirus vector, pBMN-NFATc1, which is engineered to express both NFATc1 and green fluorescence protein (GFP). Then, BMMs were infected with the NFATc1-expressing virus or with a control virus (pBMN-GFP) as described (Takayanagi et al. 2002). In both RGS10+/+ and RGS10−/− BMMs expressing pBMN-NFATc1, expression of NFATc1 protein from the endogenous NFATc1 gene was detected (Fig. 6G,H). Notably, the expression of NFATc1 in BMMs could partially rescue impaired osteoclast differentiation from deletion of RGS10 even without RANKL stimulation, as shown in Figure 6, I and J. As many as 21% of the RGS10 mutant cells positive for NFATc1 overexpression in GFP+ cells underwent differentiation into TRAP+ cells. Seventy-five percent of the RGS10+/+ cells are positive without RANKL induction. In the presence of RANKL, ∼60% of the RGS10 mutant cells from GFP+ cells positive for NFATc1 overexpression underwent differentiation into TRAP+ cells. However, in wild-type cells, ∼89% are positive for TRAP (Fig. 6I,J). These results indicate that NFATc1 overexpression activated a parallel pathway to induce osteoclast differentiation without RGS10. However, osteoclast differentiation is not completely rescued by NFATc1 overexpression in RGS10 mutant cells, indicating that RGS10 has a function that cannot be fully replaced by NFATc1.
RGS10 acts downstream from ITAM and upstream of calcineurin in the RANKL-[Ca2+]i oscillation–calcineurin–NFATc1 pathway
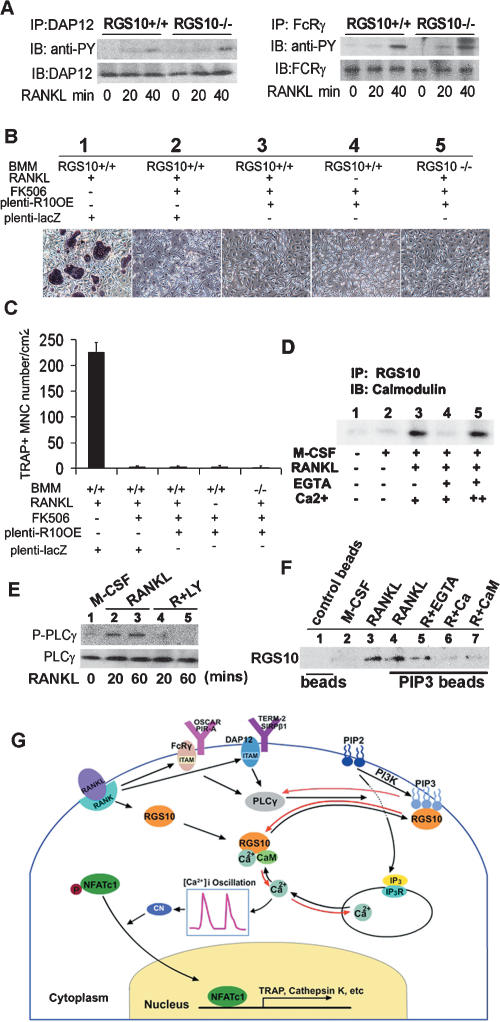
Involvement of NFATc1 directly implicates Ca2+ signaling in osteoclastogenesis, since NFAT activation and subsequent nuclear translocation is directed by the Ca2+/calmodulin-dependent serine/threonine phosphatase calcineurin (Hogan et al. 2003). In RANKL-induced BMMs from RGS10−/− mice, we were unable to observe [Ca2+]i oscillations and NFATc1 expression, indicating that RGS10 may be a key regulator of [Ca2+]i oscillations and NFATc1 expression during osteoclast differentiation. To further clarify the position of RGS10 in this pathway, we analyzed whether knockout of RGS10 effected activation of ITAM molecules DAP12 and FcRγ by RANKL, as demonstrated previously (Koga et al. 2004). Our analysis revealed that phosphorylation of DAP12 and FcRγ was the same in RGS10+/+ and RGS10−/− BMMs (Fig. 7A), indicating that RGS10 functions downstream from ITAM.
Figure 7.
Calmodulin and PIP3 competitively bind with RGS10 in a Ca2+-dependent manner and RGS10 acts downstream from ITAM and upstream of calcineurin in the RANKL-PLCγ-[Ca2+]i oscillation–calcineurin–NFATc1 pathway. (A) Western blot analysis of activation of ITAM molecules DAP12 and FcRγ by RANKL. Phosphorylation of DAP12 and FcRγ was the same in RGS10+/+ and RGS10−/− BMMs. (B) The effect of FK506 on osteoclastogenesis induced by ectopic RGS10 expression in RGS10+/+ and RGS10−/− BMMs. FK506 (1 μg/mL) inhibited osteoclast differentiation from RANKL-induced RGS10+/+ BMMs infected by plenti-LacZ (panel 2) as compared with the culture without FK506 (panel 1). (Panel 3) As a control, FK506 inhibited the formation of TRAP+ mononuclear cells in RGS10+/+ BMMs driven by RGS10 ectopic expression with RANKL induction. (Panel 4) FK506 inhibited the formation of TRAP+ mononuclear cells as shown in the bottom left panel of Figure 6E in RGS10+/+ BMMs driven by RGS10 ectopic expression without RANKL induction. (Panel 5) FK506 inhibited the rescue effect of RGS10 reintroduction as shown in the right panel of Figure 6C in RANKL-induced RGS10−/− BMMs infected with plenti-RGS10. (C) Quantitative analysis of TRAP+ cells in A. (D) Coimmunoprecipitation of RGS10 and calmodulin. (Lanes 1,2) No interaction was observed without RANKL induction. (Lane 3) Calmodulin bound to RGS10 in the presence of 1 mM CaCl2. (Lane 4) This interaction was blocked by 0.5 mM EGTA. (Lane 5) With an additional 2 mM CaCl2, the interaction between calmodulin and RGS10 was rescued. (E) Western blot analysis of activation of PLCγ with RANKL induction or with RANKL and PI3 kinase inhibitor LY294002. (Lanes 4,5) Phosphorylation of PLCγ does not occur in BMMs treated with LY294002. (F) PIP3 bead-binding assay. RGS10 was not detected with control beads without PIP3 (negative control, lane 1) or with M-CSF induction alone (lane 2). (Lane 3) RGS10 was detected with RANKL induction (positive control). (Lane 4) RGS10 was detected after PIP3 pull-down. (Lane 5) The addition of 2 mM EGTA, which removes free Ca2+, had no effect on PIP3/RGS10 binding. (Lane 6) When 1mM CaCl2 was added to the assay, allowing it to form a complex with calmodulin, PIP3/RGS10 binding was blocked. (Lane 7) The addition of 10 μM calmodulin also blocked PIP3/RGS10 binding. (G) The working model for RGS10-mediated modification of intracellular [Ca2+]i oscillations in osteoclast differentiation.
FK506 is a specific inhibitor of calcineurin (Liu et al. 1991; Sun et al. 2006) that also inhibits RANKL-induced osteoclast differentiation from BMMs in a dose-dependent manner (Takayanagi et al. 2002). To test whether RGS10 acts upstream of calcineurin in the RANKL-[Ca2+]i oscillation–calcineurin–NFATc1 pathway, we analyzed whether FK506 could block the rescue of defective osteoclastogenesis by the reintroduction of RGS10 expression. We found that FK506 inhibited the rescue effect of RGS10 reintroduction in RANKL-induced RGS10−/− BMMs (Fig. 7B,C). Notably, FK506 also inhibited the formation of TRAP+ cells from RGS10+/+ BMMs, as described in Figure 6, E and F, driven by RGS10 ectopic expression without RANKL induction (Fig. 7B,C), indicating that FK506 completely blocked RGS10 signaling. Our data showed that RGS10 acts upstream of calcineurin in the RANKL-[Ca2+]i oscillation–calcineurin–NFATc1 pathway. Our results also confirmed that RANKL evokes [Ca2+]i oscillations that lead to calcineurin-mediated activation of NFATc1, and therefore triggers a sustained NFATc1-dependent transcriptional program during osteoclast differentiation (Takayanagi et al. 2002).
Calmodulin binds with RGS10 in a Ca2+-dependent manner and competes with RGS10 binding to PIP3 to regulate [Ca2+]i oscillations and PLCγ activation
To gain insight into the molecular mechanisms linking RGS10 and [Ca2+]i oscillations, we examined the interaction between RGS10 and calmodulin using coimmunoprecipitation of lysate from BMMs (Fig. 7D). When BMMs were induced with M-CSF alone, no preosteoclasts or osteoclasts were formed, and thus RGS10 and calmodulin were not detected. As we expected, calmodulin bound to RGS10 with RANKL/M-CSF induction. To test whether calmodulin binds RGS10 in a Ca2+-dependent manner, we added 0.5 mM EGTA (a chemical compound with high affinity for Ca2+) to remove free Ca2+. Interestingly, in the absence of the Ca2+ environment, calmodulin lost the ability to bind to RGS10. To further confirm that Ca2+ is essential for the interaction between RGS10 and calmodulin, we examined their binding in the presence of additional 2 mM Ca2+. Notably, there was a strong interaction between RGS10 and calmodulin, indicating Ca2+ is essential for the interaction between RGS10 and calmodulin. Since calmodulin is a Ca2+-binding protein, in the presence of Ca2+, RGS10 should bind calmodulin as a complex of Ca2+/calmodulin/RGS10, and the Ca2+/calmodulin complex competes with PIP3 for binding to RGS10 and regulates [Ca2+]i oscillations. Our results indicate that RGS10 may regulate [Ca2+]i oscillations through interacting with calmodulin.
PI3 kinase is the enzyme that phosphorylates the position 3 of the inositol ring of PIP2 generating PIP3, and PIP3 has been shown to be a crucial player in PLCγ activation (Maffucci and Falasca 2007). To confirm that PIP3 is involved in PLCγ activation in the PLCγ phosphorylation-[Ca2+]i oscillation–NFATc1 pathway, we analyzed whether LY294002, an inhibitor of PI3 kinase (Singleton et al. 2005), could block PLCγ phosphorylation. We found that LY294002 inhibited phosphorylation of PLCγ (Fig. 7E), indicating that without PI3 kinase to generate PIP3, PLCγ cannot be activated. Our result confirms that PIP3 activates PLCγ in osteoclasts.
To determine whether RGS10 binds PIP3 and the Ca2+/calmodulin complex competes with PIP3 for binding to RGS10, as shown previously for RGS4 (Ishii et al. 2005), we performed a PIP3 bead-binding assay (Fig. 7F). RGS10 was not detected with control beads without PIP3 or with M-CSF induction alone. RGS10 was detected after PIP3 pull-down, showing that PIP3 binds to RGS10. The addition of EGTA, which removes free Ca2+, had no affect on PIP3/RGS10 binding. When calcium was added to the assay, allowing it to form a complex with calmodulin, PIP3/RGS10 binding was blocked. The addition of calmodulin also blocked PIP3/RGS10 binding, indicating that the Ca2+/calmodulin complex competes with PIP3 for binding to RGS10. Our results showed that RGS10 provides biochemical control over RANKL-evoked [Ca2+]i oscillations through dual interaction with PIP3 and calmodulin in a Ca2+-dependent manner, as described in the RGS10 working model in the Discussion (Fig. 7G).
Discussion
Our results, for the first time, establish the close dependency of osteoclast differentiation on the expression of RGS10, demonstrate that RGS10 is an essential component that acts upstream of the [Ca2+]i oscillation–calcineurin–NFATc1 signaling pathway, and reveal the mechanism underlying how RGS10 may mediate the modification of intracellular [Ca2+]i oscillations evoked by RANKL for terminal differentiation of osteoclasts. We demonstrated that RGS10 is prominently expressed in human osteoclasts and mouse preosteoclasts and osteoclasts, and that disruption of RGS10 impairs osteoclast differentiation. RGS10 knockout mice exhibit a severe osteopetrotic phenotype due to impairment of osteoclast formation. Consistent with this finding, bone marrow-derived osteoclast precursor cells do not differentiate into osteoclasts in vitro. RGS10 deficiency results in impaired activation of [Ca2+]i oscillations and subsequent NFATc1 induction, which are essential for osteoclast differentiation. In addition, ectopic expression of RGS10 partially induced formation of TRAP-positive cells, even in the absence of RANKL, and markedly enhanced RANKL-induced osteoclast differentiation. Thus, we propose that RGS10 is a critical regulator of RANKL-induced osteoclast differentiation through linking RANKL to the [Ca2+]i oscillation–NFATc1 pathway.
Ectopic expression of RGS10 in BMMs only resulted in as many as 8% of the cells spontaneously differentiating into TRAP+ cells in the absence of RANKL, but dramatically increased the sensitivity of osteoclast differentiation to RANKL signaling. Our results also showed that the expression of RGS10 starts to increase at 0.5 h after RANKL induction. However, [Ca2+]i oscillations appeared between 24 and 72 h after RANKL induction, indicating that there may be other component(s) that are required for full function of RGS10 in osteoclast differentiation. Our results provide the evidence that RGS10 does not affect phosphorylation of DAP12 and FCRγ (Fig. 7A,B), indicating that RGS10 is downstream from ITAM. Therefore, a possible situation is that the transient activation of a Ca2+ spike initiated by RANKL-ITAM-mediated PLCγ (Koga et al. 2004) is necessary before the [Ca2+]i oscillations regulated by RGS10.
Our results indicate that NFATc1 expression in RANKL-induced RGS10−/− BMMs was very weak when compared with that in RANKL-induced RGS10+/+ BMMs. The normalized protein level of NFATc1 in RGS10−/− cells was 18-fold lower than that in RGS10+/+ cells. It was suggested that autoamplification of NFATc1 is regulated by calcineurin that was activated by RANKL-induced [Ca2+]i oscillations during osteoclastogenesis (Takayanagi et al. 2002). Our results show that the calcineurin-specific inhibitor FK506 inhibited the rescue effect of RGS10 reintroduction in RANKL-induced RGS10−/− BMMs, and the formation of TRAP+ cells from RGS10+/+ BMMs in the absence of RANKL indicates that calcineurin acts downstream from RGS10 signaling. The very low expression of NFATc1 in RANKL-induced RGS10−/− BMMs that we observed fits well with our conclusion that RGS10 is an essential regulator in the RANKL-evoked [Ca2+]i oscillation–calcineurin–NFATc1 pathway. Interestingly, NFATc1 overexpression rescued the RGS10 defect, indicating that NFATc1 is downstream from RGS10. However, the rescue experiment showed that osteoclast formation is ∼30% lower in RGS10−/− BMMs with or without RANKL compared with that in wild type, indicating that the role of RGS10 in [Ca2+]i oscillations following calcineurin activation cannot be fully rescued by NFATc1 overexpression, suggesting a parallel pathway regulating osteoclast differentiation through NFATc1 expression. An important extension of our study would be to genetically confirm that RGS10 acts upstream of NFATc1. This could be done by studying mice lacking one allele of RGS10 and one allele of calcineurin or NFATc1 to test whether they develop an osteopetrosis phenotype.
Our data demonstrated that in the presence of Ca2+, RGS10 should bind calmodulin as a complex of Ca2+/calmodulin/RGS10 (Fig. 7D), and the Ca2+/calmodulin complex competes with PIP3 for binding to RGS10 and regulates [Ca2+]i oscillations in a [Ca2+]i-dependent manner (Fig. 7F). PI3 kinase is the enzyme that phosphorylates the position 3 of the inositol ring of PIP2 generating PIP3, and PIP3 has been shown to be a crucial player in PLCγ activation (Maffucci and Falasca 2007). We found that LY294002, an inhibitor of PI3 kinase, blocked phosphorylation of PLCγ in osteoclasts, indicating that without PI3 kinase to generate PIP3, PLCγ cannot be activated, showing that PIP3 is essential to PLCγ activation (Fig. 7E).
Based on our data, we proposed an RGS10 working model (Fig. 7G): RANKL mediates DAP12 and FcRγ, the membrane adaptor molecules that contain an ITAM motif and that activate PLCγ. PLCγ hydrolyzes PIP2 to generate inositol 3-phosphate (IP3). IP3 then triggers a transient initial release of Ca2+ from intracellular stores. Intracellular Ca2+ release allows an increase in intracellular Ca2+ to reach peak concentration and leads to formation of the Ca2+/calmodulin complex. The Ca2+/calmodulin complex competes for the PIP3-binding site on RGS10 and frees the bound PIP3. Once the Ca2+ concentration reaches its peak formation, intracellular Ca2+ begins to reload into the endoplasmic reticulum (ER) in the absence of further PLCγ activation, and the combination of Ca2+ reloading in the ER and binding to calmodulin causes the Ca2+ concentration to decrease. The Ca2+/calmodulin complex dissociates from RGS10 at the low Ca2+ concentration. Free PIP3 activates PLCγ and then binds RGS10 again without Ca2+/calmodulin complex competition. PLCγ activation triggers a release of Ca2+ from intracellular stores by generating IP3 to cause a second peak. This process continues to cycle, causing [Ca2+]i oscillations. In this way, RGS10 mediates PLCγ activation and [Ca2+]i oscillations through its [Ca2+]i-dependent dual interaction with Ca2+/calmodulin and PIP3. The RGS10-mediated intracellular [Ca2+]i oscillations activate calcineurin and NFATc1 expression for osteoclast terminal differentiation (Fig. 7G). Luo et al. (2001) proposed a model of [Ca2+]i oscillation regulation in pancreatic acinar cells based on the regulation of PLCβ activation by trimeric G protein Gqα signaling. However, trimeric G protein–PLCβ signaling has not been reported to be important in the osteoclast lineage. Our novel finding defined a new mechanism of [Ca2+]i oscillation regulation based on the regulation of PLCγ activation through RGS10 dual interaction with PIP3 and Ca2+/calmodulin complex. Our RGS10 working model (Fig. 7G) may not only represent the mechanism underlying RGS10-mediated modification of intracellular [Ca2+]i oscillations in osteoclast differentiation, but also the general mechanism of [Ca2+]i oscillations mediated by RGS proteins in differentiation of other cell types.
The osteopetrosis phenotype of the RGS10 knockout mice is very severe, and therefore our study is potentially of great importance and clinical relevance. General effects of some molecules that are essential to osteoclast differentiation and activity, such as NF-κB, c-Fos, and NFATc1 on bone mass, may not always be beneficial due to effects on the other cell types such as T cells and osteoblasts. Our result shows that RGS10-null mutation did not affect other RANKL-induced signaling pathways. Since RGS10 is predominantly expressed in osteoclasts and selectively involved in RANKL-induced [Ca2+]i oscillations, the essential role of RGS10 induction by RANKL in osteoclast differentiation described in this study may offer a very specific and powerful therapeutic target for treatment of bone diseases caused by excessive bone resorption.
Materials and methods
Cells and cell cultures
Human osteoclastoma tumors were obtained courtesy of Dr. Andrew Rosenberg (Department of Pathology, Massachusetts General Hospital, Boston, MA). The human osteoclast cells and stromal cells from the tumors were obtained as described previously (Li et al. 1995, 1996). Osteoblastic (HOS-TE85), myelomonocytic (U-937), and T lymphocyte (HSB-2) cell lines were purchased from American Type Culture Collection. The epithelial cell line Hep-2 was kindly provided by Dr. Margaret Duncan (Forsyth Institute, Boston, MA). Whole RNA from human tissues was purchased from Clontech.
GeneChip analysis
Total RNA was extracted from human osteoclastoma and stromal cells using Trizol reagent (Life Technologies, Inc.), as described by the manufacturer. GeneChip analysis was performed by the Microarray Core Facility at Harvard Medical School. The data were analyzed using an Affymetrix GeneChip scanner and accompanying gene expression software.
Northern blot analysis
Preparation of osteoclasts from human osteoclastoma, cell culture of different cell lines, and BMMs induced by RANKL at indicated times (Fig. 1B,D; Supplementary Fig. 1A,B) were performed as described in the Supplemental Material. Total RNA was isolated, as described previously (Li et al. 1995). Hybridization was performed as described previously (Yang et al. 2003). Human full-length RGS10 cDNAs (0.9 kb) were used as probes. Probes were radiolabeled with [α32P]dCTP using a random primer labeling kit (Stratagene).
RT–PCR and sequencing analysis
PCR primers were designed to hybridize with a sequence of the mouse RGS10 (mRGS10) gene (accession no. NM_026418) and c-Fos gene (accession no. NM_022197). The primer sequences for the full-length RGS10 gene are R10A-F1 (5′-ATGTTCAAC CGCGCCGTGA-3′) and R10A-R1 (5′-CATCCCATTGAAGGG TTTTG-3′). The primer sequences for the c-Fos gene are c-Fos-S (5′-CTGGTGCAGCCCACTCTGGTC-3′) and c-Fos-AS (5-GG AAGAAGACTCACCAGAAGC-3′). Total RNA from the normal mouse BMMs induced by RANKL or M-CSF for the indicated time (Fig. 5G) was isolated using Trizol reagent. One step RT–PCR was performed using the Access RT–PCR system (Promega). The identities of the amplified PCR products (659-base-pair [bp] fragment of RGS10 gene) were confirmed by direct sequencing.
Generation of RGS10−/− mice
Mouse RGS10 genomic DNA fragments were obtained from a 129/Sv genomic library (Stratagene) by screening with a radiolabeled DNA fragment corresponding to a 0.9-kb full-length RGS10 cDNA (GenBank accession no. NM_026418). A targeting vector that contained a 1.5-kb short arm and 5-kb long arm of homology flanking a PGK-neo cassette was constructed and electroporated into J1 ES cells (Li et al. 1992) and then selected in G418 (300 μg/mL). PCR and Southern blot analysis identified 19 clones with a single targeted allele in 151 G418/FIAU-resistant ES cell clones. Targeted cells were injected into fertilized blastocysts from C57BL/6J female mice. Chimeric male mice were crossed with C57BL/6J females for germline transmission. Following heterozygous matings, homozygotes were identified and distinguished from heterozygous and wild-type mice by Southern blot of SspI-digested genomic DNA hybridized to a flanking probe. To confirm the absence of RGS10 expression, we extracted total RNA from long bones of 3-d-old mice and used this RNA for Northern blot analysis. All mice were maintained in microisolator cages at the animal facility of the Forsyth Institute under specific pathogen-free conditions.
Histological and radiographic procedures
See the Supplemental Material.
Quantitative immunohistochemistry
Using both NIH ImageJ and Adobe Photoshop, random ×100 objective fields were analyzed by selecting a standardized color range for H&E, Von Kossa, TRAP, and immunohistochemical staining. After boundary delineation, the area under the pixilation histogram was calculated, comparing total stained areas to total tissue or cell areas.
Histomorphometric analysis
See the Supplemental Material.
In vitro osteoclastogenesis
Mouse BMMs, preosteoclasts, and osteoclasts were generated as described (Wang et al. 2003). Isolated BMMs from RGS10+/+ and RGS10−/− mice were cultured in α-MEM containing 10% FBS plus 10 ng/mL recombinant M-CSF. To generate preosteoclasts, BMMs were cultured in α-MEM containing 10% FBS in the presence of 10 ng/mL recombinant M-CSF and 10 ng/mL recombinant RANKL for 48–72 h To generate mature osteoclasts, 5 × 104 BMMs were plated in one well of a 24-well plate in α-MEM containing 10% FBS in the presence of 10 ng/mL recombinant M-CSF and 10 ng/mL recombinant RANKL. Mature osteoclasts began to form at 72 h of culture. For suppressive effects of a calcineurin inhibitior, 1 μg/mL FK506 was added to the medium for generating mature osteoclasts. To generate mature osteoclasts from the coculture system, BMMs and osteoblasts derived from normal calvaria cells were cultured in the presence of 10−8 M 1,25(OH)2 vitamin D3, and 10−6 M dexamethasone for 96–120 h, as described previously (Baumeister et al. 1998). The identity of osteoclasts was confirmed by TRAP staining. All data are expressed as mean ± SD (n = 6). TRAP+ MNCs were characterized by examining the bone-resorbing activity on dentine slices, as described previously (Horwood et al. 1999).
Calvarial osteogenic differentiation
Mouse calvariae were dissected aseptically from The Forsyth Institute mice postnatal day 1, and the connective tissue was removed. The calvariae were digested in Hanks’ balanced salt solution containing 0.02% type I collagenase (Sigma), 0.05% trypsin, and 0.53 mM EDTA (Invitrogen) for 10 min at 37°C with shaking. The digestion procedure was repeated five times, and the cells from the third to fifth digestions were pooled and resuspended in DMEM containing 10% (vol/vol) FBS and 1% penicillin/streptomycin. Twenty-four hours later, after the cells became confluent, differentiation medium α-MEM (Invitrogen) containing 10% FCS, 50 mg/mL ascorbic acid, and 5 mM β-glycerophosphate was used to maintain the cells for the duration of the experiment. Cells were harvested for analysis at different stages of differentiation.
Immunostaining
The cells were induced as indicated above. For the bone slides, the sections were deparaffinized. After fixing and blocking of nonspecific antibody-binding sites, cells and sections were washed and subsequently incubated in anti-RGS10 (goat, 1 μg/mL, Santa Cruz Biotechnology), anti-Apt6i-specific polyclonal antibody (rabbit), anti-cathepsin K polyclonal antibody (rabbit, from our laboratory), and anti-NFAT2 (mouse, 1 μg/mL, Santa Cruz Biotechnology) in PBS containing 1.5% normal rabbit, donkey, or goat serum for 60 min, followed by HRP-conjugated anti-goat, rabbit, or mouse IgG (1 μg/mL, Santa Cruz Biotechnology) with 1.5% normal rabbit, donkey, or goat serum for 60 min. HRP-coated antibodies were visualized using the VectaStain Elite ABC kit and DAB enzyme substrates (Vector Laboratories).
[Ca2+]i oscillation measurement
[Ca2+]i oscillation measurements were performed as described previously (Takayanagi et al. 2002). The cells were incubated with M-CSF or RANKL/M-CSF for 24, 48, or 72 h and then with 5 μM fluo-4 AM, 5 μM fura red AM, and 0.05% pluronic F127 for 30 min. The cells were post-incubated in DMEM medium with 10 ng/mL M-CSF for 20 min and were mounted on a confocal microscope (Leica). To estimate intracellular Ca2+ concentration in single cells, the ratio of the fluorescence intensity of fluo-4 to fura red was calculated. The increase in the ratio from the basal level was then divided by the maximum ratio increase obtained by adding 10 μM ionomycin and was expressed as the percent maximum ratio increase.
Western blot analysis
The cells were incubated with RANKL (10 ng/mL) and M-CSF (10 ng/mL) for the indicated time (Figs. 1F, 5E,F, 7E). Western blotting was performed as described (Yang et al. 2003) and visualized and quantified using a Fluor-S Multi-Imager and Multi-Analyst software (Bio-Rad).
Ca2+/calmodulin-binding assay
BMMs were stimulated with RANKL/M-CSF for 96 h and then washed with PBS and lysed in buffer containing 50 mM Tris-HCl (pH 8.0), 0.15 M NaCl, 1% Nonidet P-40, and phosphatase and protease inhibitor cocktail (Sigma) (Yang et al. 2003). Prior to incubation, 0.5 mM EGTA or 1 mM or 2 mM portions of CaCl2 was added to those lysates, incubated for 1 h at 4°C (Erickson-Viitanen and DeGrado 1987; Edlich et al. 2005), and then incubated for 2 h at 4°C with anti-RGS10 antibody (Santa Cruz Biotechnology, Inc.) and protein A/G-Sepharose beads (Amersham Biosciences). Calcium was added to the lysis buffer to mimic the cytoplasmic environment at high levels of Ca2+ and EGTA was added to the lysis buffer to mimic the cytoplasmic environment at levels of low or no Ca2+. The precipitates were separated by 4%–15% SDS-PAGE, followed by immunoblotting with anti-calmodulin antibody (Santa Cruz Biotechnology).
Phosphorylation of FcRγ/DAP12 and PLCγ
BMMs were stimulated by 10 ng/mL soluble RANKL/M-CSF or 10 ng/mL RANKLwith 20 uM LY294002 after 6 h of serum starvation. After various time periods, cell extracts were harvested from the cells using TNE buffer containing 10 mM Tris-HCl (pH 7.8), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 2 mM Na3VO4, 10 mM NaF, and 1% protease inhibitor cocktail. Cell extracts were incubated with 1 μg of anti-DAP12 or anti-FcRγ antibodies for 1 h at 4°C. Immune complexes were recovered with Protein A Sepharose, subjected to SDS-PAGE, and blotted with anti-phosphotyrosine antibody (4G10, Upstate Biotechnology) or the indicated antibodies. Activation of PLCγ was detected using anti-PLCγ1 and anti-phospho-PLCγ1 antibodies (Santa Cruz Biotechnology).
PIP3-binding assay
BMMs were treated with M-CSF for 24 h or RANKL/M-CSF for 96 h and suspended in 0.5 mL of binding assay buffer (100 mM KCl, 2 mM MgCl2, 0.5% Lubrol, and 20 mM Tris-Hcl at pH 7.5) in the presence of 1% protease inhibitor cocktail (Sigma). After brief sonication, cell lysates were centrifuged for 5 min at 1500g. The supernatant was collected and incubated with one of three reagents (2 mM EGTA, 1 mM CaCl2, or 10 uM calmodulin) and PIP3 beads (50 μL of slurry; Echelon Research Laboratories) for 12 h at 4°C. Then the mixture was centrifuged for 5 min at 1500g. The collected beads were washed with 1 mL of binding assay buffer three times, and the bound proteins were subjected to Western blot analysis (Lee et al. 2003; Tseng et al. 2004).
Apoptosis assay
Apoptosis was measured by Hoechst 33258 staining of condensed chromatin (Kameda et al. 1996) (see the Supplemental Material).
Lentiviral gene transfer
A 0.6-kb full-length mouse RGS10 gene was yielded by PCR and cloned into the pDONR221 vector (Invitrogen) according to the manufacturer’s instructions (Invitrogen). The LR recombination reaction was completed between the pDONR-RGS10 vector and plenti6/V5-Dest vector to generate plenti-RGS10, which is engineered to express RGS10. Plenti6/V5-LacZ (plenti-LacZ) was a control. Lentivirus packaging was performed according to the manufacturer’s instructions by cotransfection of these vectors and packaging mixtures (Invitrogen) into 293T cells. The viral supernatant was harvested after 48–72 h, and titers were determined. BMMs from RGS10−/− and RGS10+/+ mice were infected with the RGS10-expressing virus or control virus expressing LacZ. Protein expression of RGS10 was confirmed by immunostaining and Western blot in part of the plenti-RGS10-transfected BMMs. The rest of the cells were further cultured with M-CSF in the presence or absence of RANKL for the osteoclast formation assay. After 72–96 h, osteoclastogenesis was evaluated by TRAP staining and bone resorption assay.
Retroviral gene transfer
Retroviral vectors pBMN-GFP and pBMN-NFATc1 were constructed by inserting a full-length 2.1-kb NFATc1 cDNA (accession no. NM_016791) into the XhoI + NotI site of pBMN-I-GFP (Addgene), and packaging was performed as the protocol from Dr. Garry Nolan Laboratory, Stanford University, Stanford, CA. Briefly, Phoenix cells at 1.5–2 million cells per 6-cm plate were plated in producer cell growth medium. After 24 h, the Phoenix cells were transfected with retroviral vectors pBMN-GFP and pBMN-NFATc1 separately by the CaCl2 precipitation method. Retroviral supernatant was harvested and used for titer assay and to infect BMMs. The GFP and NFATc1 protein expression were confirmed by observation of GFP+ cells and performing immunostaining and Western blot. Two days after inoculation, BMMs were cultured with RANKL and M-CSF. After 4 d, osteoclastogenesis was evaluated by TRAP staining. The rescuing effect was normalized by measuring infection efficiency assessed by GFP expression as described (Takayanagi et al. 2002).
Statistical analysis
Where indicated, experimental data are reported as mean ± SD of triplicate independent samples. Data were analyzed by student’s t-test and one-way analysis of variance (ANOVA), followed by Tukey-Kramer multiple comparisons test to determine statistically significant differences between groups. P values <0.05 were considered significant.
Acknowledgments
We thank Drs. Wei Chen and Yoko Abe for ES cell culture, electroporation of ES cells, and ES cell cloning assistance. We thank Ms. Carrie Soltanoff for excellent manuscript assistance. We thank Dr. Douglas Hanson and Ms. Susan Orlando for critical reading of the manuscript. We thank Ms. Justine Dobeck for histological assistance. We thank Dr. Margaret A. Thompson and the Gene Manipulation Core of the Children’s Hospital, Boston Mental Retardation and Developmental Disabilities Research Center (P30 HD 18655), for technical assistance with the ES cell injections performed for this study. We thank Dr. Garry Nolan for use of the pBMN-I-GFP plasmid. This work was supported by NIH grants AR-44741 and AR-48133 (to Y.-P.L.), and DE016857 (to S.Y.).
Footnotes
Supplemental material is available at http://genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1544107
References
- Abramow-Newerly M., Roy A.A., Nunn C., Chidiac P., Roy A.A., Nunn C., Chidiac P., Nunn C., Chidiac P., Chidiac P. RGS proteins have a signalling complex: Interactions between RGS proteins and GPCRs, effectors, and auxiliary proteins. Cell. Signal. 2006;18:579–591. doi: 10.1016/j.cellsig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Appleton C.T., James C.G., Beier F., James C.G., Beier F., Beier F. Regulator of G-protein signaling (RGS) proteins differentially control chondrocyte differentiation. J. Cell. Physiol. 2006;207:735–745. doi: 10.1002/jcp.20615. [DOI] [PubMed] [Google Scholar]
- Baumeister W., Walz J., Zuhl F., Seemuller E., Walz J., Zuhl F., Seemuller E., Zuhl F., Seemuller E., Seemuller E. The proteasome: Paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Boyle W.J., Simonet W.S., Lacey D.L., Simonet W.S., Lacey D.L., Lacey D.L. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- Cunningham M.L., Waldo G.L., Hollinger S., Hepler J.R., Harden T.K., Waldo G.L., Hollinger S., Hepler J.R., Harden T.K., Hollinger S., Hepler J.R., Harden T.K., Hepler J.R., Harden T.K., Harden T.K. Protein kinase C phosphorylates RGS2 and modulates its capacity for negative regulation of Gα 11 signaling. J. Biol. Chem. 2001;276:5438–5444. doi: 10.1074/jbc.M007699200. [DOI] [PubMed] [Google Scholar]
- Edlich F., Weiwad M., Erdmann F., Fanghanel J., Jarczowski F., Rahfeld J.U., Fischer G., Weiwad M., Erdmann F., Fanghanel J., Jarczowski F., Rahfeld J.U., Fischer G., Erdmann F., Fanghanel J., Jarczowski F., Rahfeld J.U., Fischer G., Fanghanel J., Jarczowski F., Rahfeld J.U., Fischer G., Jarczowski F., Rahfeld J.U., Fischer G., Rahfeld J.U., Fischer G., Fischer G. Bcl-2 regulator FKBP38 is activated by Ca2+/calmodulin. EMBO J. 2005;24:2688–2699. doi: 10.1038/sj.emboj.7600739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson-Viitanen S., DeGrado W.F., DeGrado W.F. Recognition and characterization of calmodulin-binding sequences in peptides and proteins. Methods Enzymol. 1987;139:455–478. doi: 10.1016/0076-6879(87)39106-2. [DOI] [PubMed] [Google Scholar]
- Faccio R., Takeshita S., Zallone A., Ross F.P., Teitelbaum S.L., Takeshita S., Zallone A., Ross F.P., Teitelbaum S.L., Zallone A., Ross F.P., Teitelbaum S.L., Ross F.P., Teitelbaum S.L., Teitelbaum S.L. c-Fms and the αvβ3 integrin collaborate during osteoclast differentiation. J. Clin. Invest. 2003;111:749–758. doi: 10.1172/JCI16924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faccio R., Teitelbaum S.L., Fujikawa K., Chappel J., Zallone A., Tybulewicz V.L., Ross F.P., Swat W., Teitelbaum S.L., Fujikawa K., Chappel J., Zallone A., Tybulewicz V.L., Ross F.P., Swat W., Fujikawa K., Chappel J., Zallone A., Tybulewicz V.L., Ross F.P., Swat W., Chappel J., Zallone A., Tybulewicz V.L., Ross F.P., Swat W., Zallone A., Tybulewicz V.L., Ross F.P., Swat W., Tybulewicz V.L., Ross F.P., Swat W., Ross F.P., Swat W., Swat W. Vav3 regulates osteoclast function and bone mass. Nat. Med. 2005;11:284–290. doi: 10.1038/nm1194. [DOI] [PubMed] [Google Scholar]
- Gold S.J., Ni Y.G., Dohlman H.G., Nestler E.J., Ni Y.G., Dohlman H.G., Nestler E.J., Dohlman H.G., Nestler E.J., Nestler E.J. Regulators of G-protein signaling (RGS) proteins: Region-specific expression of nine subtypes in rat brain. J. Neurosci. 1997;17:8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler J.R., Berman D.M., Gilman A.G., Kozasa T., Berman D.M., Gilman A.G., Kozasa T., Gilman A.G., Kozasa T., Kozasa T. RGS4 and GAIP are GTPase-activating proteins for Gq α and block activation of phospholipase C β by γ-thio-GTP-Gq α. Proc. Natl. Acad. Sci. 1997;94:428–432. doi: 10.1073/pnas.94.2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heximer S.P., Watson N., Linder M.E., Blumer K.J., Hepler J.R., Watson N., Linder M.E., Blumer K.J., Hepler J.R., Linder M.E., Blumer K.J., Hepler J.R., Blumer K.J., Hepler J.R., Hepler J.R. RGS2/G0S8 is a selective inhibitor of Gqα function. Proc. Natl. Acad. Sci. 1997;94:14389–14393. doi: 10.1073/pnas.94.26.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan P.G., Chen L., Nardone J., Rao A., Chen L., Nardone J., Rao A., Nardone J., Rao A., Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes & Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- Horwood N.J., Kartsogiannis V., Quinn J.M., Romas E., Martin T.J., Gillespie M.T., Kartsogiannis V., Quinn J.M., Romas E., Martin T.J., Gillespie M.T., Quinn J.M., Romas E., Martin T.J., Gillespie M.T., Romas E., Martin T.J., Gillespie M.T., Martin T.J., Gillespie M.T., Gillespie M.T. Activated T lymphocytes support osteoclast formation in vitro. Biochem. Biophys. Res. Commun. 1999;265:144–150. doi: 10.1006/bbrc.1999.1623. [DOI] [PubMed] [Google Scholar]
- Iotsova V., Caamano J., Loy J., Yang Y., Lewin A., Bravo R., Caamano J., Loy J., Yang Y., Lewin A., Bravo R., Loy J., Yang Y., Lewin A., Bravo R., Yang Y., Lewin A., Bravo R., Lewin A., Bravo R., Bravo R. Osteopetrosis in mice lacking NF-κB1 and NF-κB2. Nat. Med. 1997;3:1285–1289. doi: 10.1038/nm1197-1285. [DOI] [PubMed] [Google Scholar]
- Ishii M., Inanobe A., Kurachi Y., Inanobe A., Kurachi Y., Kurachi Y. PIP3 inhibition of RGS protein and its reversal by Ca2+/calmodulin mediate voltage-dependent control of the G protein cycle in a cardiac K+ channel. Proc. Natl. Acad. Sci. 2002;99:4325–4330. doi: 10.1073/pnas.072073399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M., Fujita S., Yamada M., Hosaka Y., Kurachi Y., Fujita S., Yamada M., Hosaka Y., Kurachi Y., Yamada M., Hosaka Y., Kurachi Y., Hosaka Y., Kurachi Y., Kurachi Y. Phosphatidylinositol 3,4,5-trisphosphate and Ca2+/calmodulin competitively bind to the regulators of G-protein-signalling (RGS) domain of RGS4 and reciprocally regulate its action. Biochem. J. 2005;385:65–73. doi: 10.1042/BJ20040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameda T., Miyazawa K., Mori Y., Yuasa T., Shiokawa M., Nakamaru Y., Mano H., Hakeda Y., Kameda A., Kumegawa M., Miyazawa K., Mori Y., Yuasa T., Shiokawa M., Nakamaru Y., Mano H., Hakeda Y., Kameda A., Kumegawa M., Mori Y., Yuasa T., Shiokawa M., Nakamaru Y., Mano H., Hakeda Y., Kameda A., Kumegawa M., Yuasa T., Shiokawa M., Nakamaru Y., Mano H., Hakeda Y., Kameda A., Kumegawa M., Shiokawa M., Nakamaru Y., Mano H., Hakeda Y., Kameda A., Kumegawa M., Nakamaru Y., Mano H., Hakeda Y., Kameda A., Kumegawa M., Mano H., Hakeda Y., Kameda A., Kumegawa M., Hakeda Y., Kameda A., Kumegawa M., Kameda A., Kumegawa M., Kumegawa M. Vitamin K2 inhibits osteoclastic bone resorption by inducing osteoclast apoptosis. Biochem. Biophys. Res. Commun. 1996;220:515–519. doi: 10.1006/bbrc.1996.0436. [DOI] [PubMed] [Google Scholar]
- Kehrl J.H. Heterotrimeric G protein signaling: Roles in immune function and fine-tuning by RGS proteins. Immunity. 1998;8:1–10. doi: 10.1016/s1074-7613(00)80453-7. [DOI] [PubMed] [Google Scholar]
- Koga T., Inui M., Inoue K., Kim S., Suematsu A., Kobayashi E., Iwata T., Ohnishi H., Matozaki T., Kodama T., Inui M., Inoue K., Kim S., Suematsu A., Kobayashi E., Iwata T., Ohnishi H., Matozaki T., Kodama T., Inoue K., Kim S., Suematsu A., Kobayashi E., Iwata T., Ohnishi H., Matozaki T., Kodama T., Kim S., Suematsu A., Kobayashi E., Iwata T., Ohnishi H., Matozaki T., Kodama T., Suematsu A., Kobayashi E., Iwata T., Ohnishi H., Matozaki T., Kodama T., Kobayashi E., Iwata T., Ohnishi H., Matozaki T., Kodama T., Iwata T., Ohnishi H., Matozaki T., Kodama T., Ohnishi H., Matozaki T., Kodama T., Matozaki T., Kodama T., Kodama T., et al. Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature. 2004;428:758–763. doi: 10.1038/nature02444. [DOI] [PubMed] [Google Scholar]
- Lee S.J., Xu H., Kang L.W., Amzel L.M., Montell C., Xu H., Kang L.W., Amzel L.M., Montell C., Kang L.W., Amzel L.M., Montell C., Amzel L.M., Montell C., Montell C. Light adaptation through phosphoinositide-regulated translocation of Drosophila visual arrestin. Neuron. 2003;39:121–132. doi: 10.1016/s0896-6273(03)00390-8. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T.H., Jaenisch R., Bestor T.H., Jaenisch R., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Li Y.P., Alexander M., Wucherpfennig A.L., Yelick P., Chen W., Stashenko P., Alexander M., Wucherpfennig A.L., Yelick P., Chen W., Stashenko P., Wucherpfennig A.L., Yelick P., Chen W., Stashenko P., Yelick P., Chen W., Stashenko P., Chen W., Stashenko P., Stashenko P. Cloning and complete coding sequence of a novel human cathepsin expressed in giant cells of osteoclastomas. J. Bone Miner. Res. 1995;10:1197–1202. doi: 10.1002/jbmr.5650100809. [DOI] [PubMed] [Google Scholar]
- Li Y.P., Chen W., Stashenko P., Chen W., Stashenko P., Stashenko P. Molecular cloning and characterization of a putative novel human osteoclast-specific 116-kDa vacuolar proton pump subunit. Biochem. Biophys. Res. Commun. 1996;218:813–821. doi: 10.1006/bbrc.1996.0145. [DOI] [PubMed] [Google Scholar]
- Li Y.P., Chen W., Liang Y., Li E., Stashenko P., Chen W., Liang Y., Li E., Stashenko P., Liang Y., Li E., Stashenko P., Li E., Stashenko P., Stashenko P. Atp6i-deficient mice exhibit severe osteopetrosis due to loss of osteoclast-mediated extracellular acidification. Nat. Genet. 1999;23:447–451. doi: 10.1038/70563. [DOI] [PubMed] [Google Scholar]
- Liu J., Farmer J.D., Lane W.S., Friedman J., Weissman I., Schreiber S.L., Farmer J.D., Lane W.S., Friedman J., Weissman I., Schreiber S.L., Lane W.S., Friedman J., Weissman I., Schreiber S.L., Friedman J., Weissman I., Schreiber S.L., Weissman I., Schreiber S.L., Schreiber S.L. Calcineurin is a common target of cyclophilin–cyclosporin A and FKBP–FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- Luo X., Popov S., Bera A.K., Wilkie T.M., Muallem S., Popov S., Bera A.K., Wilkie T.M., Muallem S., Bera A.K., Wilkie T.M., Muallem S., Wilkie T.M., Muallem S., Muallem S. RGS proteins provide biochemical control of agonist-evoked [Ca2+]i oscillations. Mol. Cell. 2001;7:651–660. doi: 10.1016/s1097-2765(01)00211-8. [DOI] [PubMed] [Google Scholar]
- Maffucci T., Falasca M., Falasca M. Phosphoinositide 3-kinase-dependent regulation of phospholipase Cγ. Biochem. Soc. Trans. 2007;35:229–230. doi: 10.1042/BST0350229. [DOI] [PubMed] [Google Scholar]
- Mao D., Epple H., Uthgenannt B., Novack D.V., Faccio R., Epple H., Uthgenannt B., Novack D.V., Faccio R., Uthgenannt B., Novack D.V., Faccio R., Novack D.V., Faccio R., Faccio R. PLCγ2 regulates osteoclastogenesis via its interaction with ITAM proteins and GAB2. J. Clin. Invest. 2006;116:2869–2879. doi: 10.1172/JCI28775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov S.G., Krishna U.M., Falck J.R., Wilkie T.M., Krishna U.M., Falck J.R., Wilkie T.M., Falck J.R., Wilkie T.M., Wilkie T.M. Ca2+/Calmodulin reverses phosphatidylinositol 3,4, 5-trisphosphate-dependent inhibition of regulators of G protein-signaling GTPase-activating protein activity. J. Biol. Chem. 2000;275:18962–18968. doi: 10.1074/jbc.M001128200. [DOI] [PubMed] [Google Scholar]
- Saugstad J.A., Marino M.J., Folk J.A., Hepler J.R., Conn P.J., Marino M.J., Folk J.A., Hepler J.R., Conn P.J., Folk J.A., Hepler J.R., Conn P.J., Hepler J.R., Conn P.J., Conn P.J. RGS4 inhibits signaling by group I metabotropic glutamate receptors. J. Neurosci. 1998;18:905–913. doi: 10.1523/JNEUROSCI.18-03-00905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwable J., Choudhary C., Thiede C., Tickenbrock L., Sargin B., Steur C., Rehage M., Rudat A., Brandts C., Berdel W.E., Choudhary C., Thiede C., Tickenbrock L., Sargin B., Steur C., Rehage M., Rudat A., Brandts C., Berdel W.E., Thiede C., Tickenbrock L., Sargin B., Steur C., Rehage M., Rudat A., Brandts C., Berdel W.E., Tickenbrock L., Sargin B., Steur C., Rehage M., Rudat A., Brandts C., Berdel W.E., Sargin B., Steur C., Rehage M., Rudat A., Brandts C., Berdel W.E., Steur C., Rehage M., Rudat A., Brandts C., Berdel W.E., Rehage M., Rudat A., Brandts C., Berdel W.E., Rudat A., Brandts C., Berdel W.E., Brandts C., Berdel W.E., Berdel W.E., et al. RGS2 is an important target gene of Flt3-ITD mutations in AML and functions in myeloid differentiation and leukemic transformation. Blood. 2005;105:2107–2114. doi: 10.1182/blood-2004-03-0940. [DOI] [PubMed] [Google Scholar]
- Singleton P.A., Dudek S.M., Chiang E.T., Garcia J.G., Dudek S.M., Chiang E.T., Garcia J.G., Chiang E.T., Garcia J.G., Garcia J.G. Regulation of sphingosine 1-phosphate-induced endothelial cytoskeletal rearrangement and barrier enhancement by S1P1 receptor, PI3 kinase, Tiam1/Rac1, and α-actinin. FASEB J. 2005;19:1646–1656. doi: 10.1096/fj.05-3928com. [DOI] [PubMed] [Google Scholar]
- Sinnarajah S., Dessauer C.W., Srikumar D., Chen J., Yuen J., Yilma S., Dennis J.C., Morrison E.E., Vodyanoy V., Kehrl J.H., Dessauer C.W., Srikumar D., Chen J., Yuen J., Yilma S., Dennis J.C., Morrison E.E., Vodyanoy V., Kehrl J.H., Srikumar D., Chen J., Yuen J., Yilma S., Dennis J.C., Morrison E.E., Vodyanoy V., Kehrl J.H., Chen J., Yuen J., Yilma S., Dennis J.C., Morrison E.E., Vodyanoy V., Kehrl J.H., Yuen J., Yilma S., Dennis J.C., Morrison E.E., Vodyanoy V., Kehrl J.H., Yilma S., Dennis J.C., Morrison E.E., Vodyanoy V., Kehrl J.H., Dennis J.C., Morrison E.E., Vodyanoy V., Kehrl J.H., Morrison E.E., Vodyanoy V., Kehrl J.H., Vodyanoy V., Kehrl J.H., Kehrl J.H. RGS2 regulates signal transduction in olfactory neurons by attenuating activation of adenylyl cyclase III. Nature. 2001;409:1051–1055. doi: 10.1038/35059104. [DOI] [PubMed] [Google Scholar]
- Soriano P., Montgomery C., Geske R., Bradley A., Montgomery C., Geske R., Bradley A., Geske R., Bradley A., Bradley A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- Sun L., Peng Y., Zaidi N., Zhu L.L., Iqbal J., Yamoah K., Wang X., Liu P., Abe E., Moonga B.S., Peng Y., Zaidi N., Zhu L.L., Iqbal J., Yamoah K., Wang X., Liu P., Abe E., Moonga B.S., Zaidi N., Zhu L.L., Iqbal J., Yamoah K., Wang X., Liu P., Abe E., Moonga B.S., Zhu L.L., Iqbal J., Yamoah K., Wang X., Liu P., Abe E., Moonga B.S., Iqbal J., Yamoah K., Wang X., Liu P., Abe E., Moonga B.S., Yamoah K., Wang X., Liu P., Abe E., Moonga B.S., Wang X., Liu P., Abe E., Moonga B.S., Liu P., Abe E., Moonga B.S., Abe E., Moonga B.S., Moonga B.S., et al. Evidence that calcineurin is required for the genesis of bone resorbing osteoclasts. Am. J. Physiol. Renal Physiol. 2006;292:F285–F291. doi: 10.1152/ajprenal.00415.2005. [DOI] [PubMed] [Google Scholar]
- Takahashi N., Udagawa N., Tanaka S., Murakami H., Owan I., Tamura T., Suda T., Udagawa N., Tanaka S., Murakami H., Owan I., Tamura T., Suda T., Tanaka S., Murakami H., Owan I., Tamura T., Suda T., Murakami H., Owan I., Tamura T., Suda T., Owan I., Tamura T., Suda T., Tamura T., Suda T., Suda T. Postmitotic osteoclast precursors are mononuclear cells which express macrophage-associated phenotypes. Dev. Biol. 1994;163:212–221. doi: 10.1006/dbio.1994.1137. [DOI] [PubMed] [Google Scholar]
- Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Saiura A., Isobe M., Yokochi T., Inoue J., Isobe M., Yokochi T., Inoue J., Yokochi T., Inoue J., Inoue J., et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- Tondravi M.M., McKercher S.R., Anderson K., Erdmann J.M., Quiroz M., Maki R., Teitelbaum S.L., McKercher S.R., Anderson K., Erdmann J.M., Quiroz M., Maki R., Teitelbaum S.L., Anderson K., Erdmann J.M., Quiroz M., Maki R., Teitelbaum S.L., Erdmann J.M., Quiroz M., Maki R., Teitelbaum S.L., Quiroz M., Maki R., Teitelbaum S.L., Maki R., Teitelbaum S.L., Teitelbaum S.L. Osteopetrosis in mice lacking haematopoietic transcription factor PU.1. Nature. 1997;386:81–84. doi: 10.1038/386081a0. [DOI] [PubMed] [Google Scholar]
- Tseng P.H., Lin H.P., Hu H., Wang C., Zhu M.X., Chen C.S., Lin H.P., Hu H., Wang C., Zhu M.X., Chen C.S., Hu H., Wang C., Zhu M.X., Chen C.S., Wang C., Zhu M.X., Chen C.S., Zhu M.X., Chen C.S., Chen C.S. The canonical transient receptor potential 6 channel as a putative phosphatidylinositol 3,4,5-trisphosphate-sensitive calcium entry system. Biochemistry. 2004;43:11701– 11708. doi: 10.1021/bi049349f. [DOI] [PubMed] [Google Scholar]
- Wang Z.Q., Ovitt C., Grigoriadis A.E., Mohle-Steinlein U., Ruther U., Wagner E.F., Ovitt C., Grigoriadis A.E., Mohle-Steinlein U., Ruther U., Wagner E.F., Grigoriadis A.E., Mohle-Steinlein U., Ruther U., Wagner E.F., Mohle-Steinlein U., Ruther U., Wagner E.F., Ruther U., Wagner E.F., Wagner E.F. Bone and haematopoietic defects in mice lacking c-fos. Nature. 1992;360:741–745. doi: 10.1038/360741a0. [DOI] [PubMed] [Google Scholar]
- Wang Q., Xie Y., Du Q.S., Wu X.J., Feng X., Mei L., McDonald J.M., Xiong W.C., Xie Y., Du Q.S., Wu X.J., Feng X., Mei L., McDonald J.M., Xiong W.C., Du Q.S., Wu X.J., Feng X., Mei L., McDonald J.M., Xiong W.C., Wu X.J., Feng X., Mei L., McDonald J.M., Xiong W.C., Feng X., Mei L., McDonald J.M., Xiong W.C., Mei L., McDonald J.M., Xiong W.C., McDonald J.M., Xiong W.C., Xiong W.C. Regulation of the formation of osteoclastic actin rings by proline-rich tyrosine kinase 2 interacting with gelsolin. J. Cell Biol. 2003;160:565–575. doi: 10.1083/jcb.200207036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Wei D., Wang D., Phimphilai M., Krebsbach P.H., Franceschi R.T., Wei D., Wang D., Phimphilai M., Krebsbach P.H., Franceschi R.T., Wang D., Phimphilai M., Krebsbach P.H., Franceschi R.T., Phimphilai M., Krebsbach P.H., Franceschi R.T., Krebsbach P.H., Franceschi R.T., Franceschi R.T. In vitro and in vivo synergistic interactions between the Runx2/Cbfa1 transcription factor and bone morphogenetic protein-2 in stimulating osteoblast differentiation. J. Bone Miner. Res. 2003;18:705–715. doi: 10.1359/jbmr.2003.18.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Shao J., Chen W., Li Y.P., Shao J., Chen W., Li Y.P., Chen W., Li Y.P., Li Y.P. Osteoclast differentiation and gene regulation. Front. Biosci. 2007;12:2519– 2529. doi: 10.2741/2252. [DOI] [PubMed] [Google Scholar]